Abstract
In an effort to develop a sustainable protocol for the micropropagation of a shy suckering elite chrysanthemum cv. Arka Swarna (yellow pompon type), in vitro cultures were established using surface-sterilized nodal microcuttings (1–1.5 cm) from polyhouse-grown plants on MS medium containing 3% sucrose, 0.25% phytagel, and 5 μM benzyl adenine (BA) or kinetin. Microbial contamination in the range of 6–24% was encountered during the first in vitro passage. Apparently clean cultures after one passage on MS basal medium were transferred to medium with BA or kinetin (0, 1, 5, 10, or 20 μM) in culture bottles, and were monitored for eight in vitro passages (1 mo. each) for growth and microbial contamination. Plant growth regulator (PGR)-free medium was the best for sustainable micropropagation over successive in vitro passages yielding a single shoot from cultured microcuttings. Higher cytokinin levels inhibited rooting and induced one or more shorter shoots with close nodes resulting in low propagation rates. All apparently clean stocks revealed covert endophytic bacteria during tissue-indexing using bacteriological media. Three distinct bacterial morphotypes were isolated from such stocks, identified based on 16S rRNA gene sequence analysis as different morphotypes of Curtobacterium citreum. The endophytes tended to show obvious growth on chrysanthemum culture medium with increase in cytokinin levels (5–20 μM), but such growth was not noticed in inoculations on MS medium without plants. Sustainable micropropagation of cv. Arka Swarna for more than 2 yr with the resident endophytic bacteria in covert form was realized on PGR-free MS medium giving a net propagation rate of three to four times over a subculture cycle of 2–3 wk.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chrysanthemum (Dendranthema × grandiflora (Ramat.) Kitamura is highly valued as a cut flower worldwide with its diverse floral types and colors (Teixeira da Silva 2004). It is generally propagated using suckers and terminal cuttings (Rout and Das 1997). This approach, however, is inadequate to attain fast multiplication particularly for shy suckering varieties. Micropropagation would be a promising alternative for such varieties. Micropropagation of chrysanthemum has been reported by various researchers using shoot apex and axillary bud explants, callus-derived stem, and leaf explants and floral parts (Ben-Jaacov and Langhans 1972; Earle and Langhans 1974; Bhattacharya et al. 1990; Kaul et al. 1991; Kumari and Verghese 2003). This generally involves separation of shootlets and their rooting with or without an intermediate shoot elongation phase, making it multistep process (Rout and Das 1997; Teixeira da Silva 2004). For faster multiplication, use of bioreactors with liquid medium is proposed (Hahn and Paek 2005; Sivakumar et al. 2005).
The prime requirements for a viable micropropagation protocol include simple media formulation, minimum steps or stages in production, consistent propagation rates, long-term maintainability of stocks without the need for initiating fresh cultures from time to time and a good control over microbial contamination (Cassells 2000; Thomas and Prakash 2004; Thomas et al. 2006). Micropropagation studies often do not mention about the performance and stability of cultures upon continuous in vitro culturing. Besides, microbial contamination is a serious threat to tissue cultures, admitted by all workers but is seldom reported in research publications while discussing otherwise “efficient” protocols. There are several routes of microbial entry in plant tissue cultures (Leifert and Woodward 1998; Leifert and Cassells 2001; Thomas 2006a, b), one of which is endophytes (Leifert and Cassells 2001; Thomas et al. 2007a, b). Endophytes colonize plants internally without doing any apparent damage to the host, but under the modified conditions in vitro, they may become pathogenic, overriding the cultures (Herman 2004; Thomas et al. 2007a). The reason for high incidence of contamination at culture initiation is widely believed as resulting from inefficient surface sterilization or endophytic organisms that are not eliminated by the disinfectants. It is often assumed that the organisms that escaped the initial decontamination procedure would express on the culture medium allowing quick identification and culling of such cultures. Our recent observations, on the other hand, have indicated widespread prevalence of bacteria in covert form in the medium or as specific tissue-colonizers in the cultures of different crop plants, which would go unnoticed in the absence of specific tests aimed at detecting them (Thomas 2004a, b, c). Based on the above observations, a three-step screening procedure involving visual examination, indexing of medium, and testing of tissue on two bacteriological media at different temperatures was evolved (Thomas 2004a), and it has been instrumental for the reliable identification of covertly contaminated cultures and in the cleansing of such stocks (Thomas and Prakash 2004; Thomas et al. 2006).
We have taken up the present study against this background with a view to evolve a sustainable micropropagation strategy for the shy suckering elite “pompon” chrysanthemum cv. Arka Swarna developed at the Indian Institute of Horticultural Research, Bangalore (Janakiram and Rao 2001) and to assess if the cultures harbored any bacteria in covert form.
Materials and Methods
Culture initiation.
Nodal cuttings from actively growing tender shoots of polyhouse-grown healthy plants of chrysanthemum (Dendranthema grandiflora) cv. Arka Swarna were used for culture establishment based on the outcome of a preliminary trial comparing them with shoot tips. Tender shoot cuttings (1.5–2 cm) with one to three nodes were surface-sterilized using 1,000 mg l−1 cetrimide (Hi-Media, Mumbai, India) for 15 min followed by NaOCl (4% available chlorine; Sd-fine Chemicals, Mumbai, India) for 10 min. After six rinses in sterile distilled water, the cut ends were removed and 0.8–1.0 cm microcuttings with 1–2 nodes were cultured singly in glass culture tubes (150 × 25 mm) containing 15 ml of MS-based (Murashige and Skoog 1962) culture medium (pH 5.8, adjusted using 0.1 M NaOH) with 3% sucrose, 2.5 g l−1 Phytagel® (Sigma Chemical Co., St. Louis, MO), and 5 μM of either benzyl adenine (BA) or kinetin (Kn) (50 cuttings each per treatment). Each tube was closed with a cotton bung and the bung was wrapped with waxed paper to protect from dust and microarthropod vectors. The last two wash solutions (100 μl) after NaOCl treatment were plated on two bacteriological indexing media (BIM), namely, BIM1 and BIM2 (Thomas 2004a), and the plates were observed for 2 wk for any microbial growth at 37 and 25°C, respectively, to ascertain proper surface sterilization.
Monitoring culture growth over subcultures.
Visibly clean nodal cultures that showed shoot growth on establishment medium were transferred to MS basal medium (3% sucrose) for one cycle to defuse the plant growth regulator (PGR) effect, and the stocks from BA and Kn treatments were transferred, respectively, onto medium with five different BA/Kn levels (0, 1, 5, 10, or 20 μM). A completely randomized design (CRD) experiment was laid out with five replicate bottles (110 × 65 mm; 50 ml medium) per treatment each with four in vitro-derived shoot-tip or nodal microcuttings. Cultures were monitored for percent shoot and root growth response and other growth variables (Table 1) and any obvious microbial contamination. Propagation rate (PR) was worked out based on the number of microcuttings that could be prepared from a stock shoot and net PR was worked out as the product of PR and percent growth response (Thomas 2004b). Cultures were transferred to fresh medium of the same composition at regular intervals of 1 mo. for eight passages. Wherever paucity of stocks owing to microbial contamination was encountered (usually at higher cytokinin levels; see “Results”), stocks from 0 or 1 μM BA or Kn treatment(s) were used to maintain the specified number of replications in each passage.
Monitoring cultures for covert endophytic bacteria.
At the end of each in vitro passage in the above experiment, visibly clean cultures were subjected to medium- and tissue-indexing for any bacteria using two BIMs as per Thomas (2004a). For this, tissue segments (4–5 mm) from different plant parts were placed on BIM1 and BIM2 and the plates were incubated at 37 and 25°C, respectively. A culture was identified as index-positive if bacterial growth was detected in any of the tested spots during the 2 wk following indexing. Covert bacteria-harboring cultures were carried forward whereas all visibly contaminated cultures were discontinued. Culture indexing was repeated with two new batches of visibly clean stocks (25 each) that were maintained singly in glass culture tubes during their second in vitro passage.
Isolation and identification of associated bacteria.
Bacterial growth that emerged from visibly clean stocks on BIMs during the eighth in vitro passage was dilution-plated and three distinct colony types (Ch.AS.a, Ch.AS.b, and Ch.AS.c) were selected based on colony color and morphology. Bacterial identification was undertaken through PCR amplification of 16S rRNA gene and sequence analysis as described elsewhere (Thomas 2004c). Briefly, DNA was extracted from a single colony in 0.1 × TE and 16S rDNA was PCR-amplified using primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′). The cleaned PCR product was single-end-sequenced at Macrogen Inc., Seoul, Korea (http://www.macrogen.com) using 27F, which yielded a sequence read of ∼700 bp. Based on the observation that all these isolates yielded identical nucleotide data, sequencing was repeated at MWG Biotech, Bangalore, India (http://www.mwgdna.com) using the PCR product from freshly purified single colonies. Further, these isolates were taken up for primer-walk sequencing using Ch.AS.CurtFP1 primer (5′-TTGGAATTCCTGGTGTAGC-3′). Similarity of derived partial 16S rDNA nucleotide sequences (approx. 1.4 kb) with known sequences in the NCBI GenBank database (http://www.ncbi.nlm.nih.gov/) was determined using BLASTn version 2.2.14 as of November 2006, and the identification results were validated with the Ribosomal Database Project II of Michigan State University (http://rdp.cme.msu.edu/). Percent similarity between two sequences was assessed as described elsewhere (Thomas 2006b). The partial 16S rRNA gene sequence data of these isolates have been deposited with the NCBI GenBank (accession nos. EF197914–EF197916). Standard microbiological tests were carried out as described earlier (Thomas 2006a).
Effect of cytokinin levels on growth of bacterial endophytes.
This experiment was taken up following the frequent observation of visible bacterial growth in cultures grown at higher BA/Kn levels with a view to ascertain if the same was contributed by higher PGR levels or the plant growth pattern at these levels. In the first trial, a dilute bacterial suspension of the above three isolates was prepared in sterile distilled water using 2-d-old colony growth on BIM1 after adjusting the optical density (OD) at 550 nm to 0.1. The suspension was spotted (1 μl × 10 spots per isolate) on phytagel-gelled MS medium (3% sucrose) in single-use petri dishes containing different levels of BA or Kn (0, 1, 5, 10, or 20 μM). There were five replicate plates per treatment. The plates after sealing in sterile polypropylene bags were incubated under similar conditions as for chrysanthemum stocks (26 ± 1°C) for 1 mo. and observed for colony growth. In the second trial, MS liquid medium (1 ml each in 2 ml sterile microfuge tubes) with different cytokinin levels as above was inoculated with bacterial suspension (100 μl of OD550 = 1.0) prepared as above, and the tubes were incubated at 26 ± 1°C with agitation (160 rpm) in a rotary shaker. There were five replications per treatment per isolate. The bacterial growth was assessed on the seventh day by checking the OD550 using a Biomate-3 spectrophotometer (Thermo Spectronic, Shelton, CT) employing 1-ml disposable cuvettes (Greiner, Kremsmunster, Austria).
Acclimatization of micropropagated plants.
The rooted plantlets derived from MS basal medium or lower BA/Kn treatments were acclimatized employing sachet technique (Ravindra and Thomas 1995), as described previously for grapes (Thomas 1998). Briefly, plantlets of 3–5 cm with 1–5 or more roots were washed under tap water and were planted singly in a potting mixture (2:1:1 of autoclaved sand, soil, and Soilrite TC®) in polythene bags (9″height × 5″ width), filled to one third capacity. The planting mixture was drenched to field capacity and the bags were closed after planting one rooted plantlet per bag. Incubation conditions were as described earlier (Thomas 1998). Planting of one, two, or five rooted plantlets per polythene bag was tried. The ex vitro establishment was recorded 1 mo. after planting and the sachets were shifted to a glasshouse or the plants were transplanted to a nursery bed under shade (400–500 μmol m−2 s−1).
Extended culture monitoring.
Bacteria-harboring apparently clean cultures were monitored for growth and PR on PGR-free MS medium for over 2 yr with regular subculturing at 2–3 wk.
Sterile practices.
The cultures at each transfer were handled on a single-vessel basis using a fresh sterile petri dish and sterile forceps for each culture. Stringent sterile practices as described elsewhere (Thomas 2004a, 2006b; Thomas and Prakash 2004), including the sealing of each culture bottle in polypropylene bags during culture incubation were followed.
Statistical analysis.
The data on growth of cultures and other variable at different levels of the two cytokinins were analyzed in factorial CRD (Gomez and Gomez 1984) for each in vitro cycle for eight passages. The percent values were subjected to arcsine transformation before ANOVA. Other experiments were laid out in CRD.
Results
Culture establishment.
Preliminary studies indicated that nodal microcuttings were superior to shoot tips based on explant survival after disinfectant treatment (80 and 6% survival, respectively, on medium with 1 μM BA), and that 5 μM BA/Kn was superior to lower or higher levels based on initial shoot growth response.
In the current study, 20 and 5% microbial contamination was encountered in BA and Kn (5 μM) supplemented sets, respectively, during the first passage. Both shoot tip and nodal microcuttings from these in vitro stocks served as the propagules for the subsequent evaluation after one passage in PGR-free MS medium.
Monitoring culture growth over subcultures.
In PGR-free medium as well as in low BA/Kn treatments (1 μM), the cultured microcuttings gave rise to a single shoot together with rooting. With increase in PGR levels, the cultures showed a reduction in rooting (1 μM) or no rooting (5–20 μM) and they tended to produce one or more shorter shoots with close-set nodes. The PR and net PR appeared to be the best in PGR-free medium followed by 1 μM BA or Kn, whereas explants on higher PGR levels appeared less responsive during the first passage (Table 1).
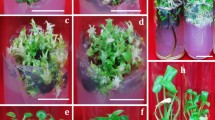
At the end of the first cycle, some cultures at higher BA/Kn levels (>5 μM) exhibited visible bacterial contamination. Such obvious contamination was not observed at low or zero BA/Kn level. Indexing the apparently clean cultures revealed bacteria in all explants at 5–20 μM BA/Kn levels and in 60–80% of the 0–1 μM BA/Kn treatments. Observations over the eight subculture passages indicated frequent activation of associated bacteria to visibly detectable form on the medium at higher PGR levels (Fig. 1 a, b). These obviously contaminated cultures were not useful for subculturing whereas the covert bacteria-harboring cultures appeared normal.
Monitoring the growth of cultures over consecutive passages revealed the best shoot growth response (%) coupled with rooting, proper shoot elongation, and best PR in PGR-free medium in both BA (Fig. 2 a–d) and Kn (Fig. 3 a–d) treatments with consistent results over eight cycles. There was no significant difference in shoot growth response between different cycles at a particular growth regulator level (P > 0.05 in all cases) but in each passage there existed significant differences between different levels (P < 0.05 in all cases). In the trial involving different BA levels, net PR was significantly higher in MS basal medium (2.6–3.9×) followed by 1 μM (1.4–2.4×), 5 μM (0.5–1.3×), 10 μM (0.2–0.5×), and 20 μM (0.1–0.2×) in that order and the results were holding good over the in vitro regimes. In the experiment relating to different Kn levels also, a similar trend was observed with the best net PR (2.6–3.8×) in PGR-free medium followed by 1 μM (1.2–2.4×), 5 μM (0.5–1.3×), 10 μM (0.2–0.5×), and 20 μM (0.1×). Overall, shoot and root growth were the best in PGR-free MS medium followed by 1, 5, 10, and 20 μM BA/Kn as indicated by the culture performance after 10 (2 + 8) passages (Fig. 4).
Screening additional cultures for covert bacteria.
Besides the above cultures, 50 additional stocks in culture tubes were subjected to visual screening followed by indexing of medium and tissue during the first passage. None of these showed indications of bacteria such as cloudiness of medium, hazy patches, or halo around the base of plantlets (Fig. 5 a). Ninety-five percent of such clean cultures turned out to be index-positive during medium indexing whereas all of them proved to be tissue-index-positive in one or both BIM (Fig. 5 b). Cultures that initially appeared index-positive on only one BIM turned out to be index-positive on both BIM after one or more cycles.
Isolation and identification of endophytic bacteria.
Three distinct single colony morphotypes (yellow, light orange, and cream) were picked up after dilution plating and restreaking of bacterial growth formed on BIM1 and BIM2 from index-positive cultures. PCR on bacterial DNA employing 16S rRNA gene universal primers yielded a 1.5-kb (approx.) band. Single end sequencing of the PCR product using 27F yielded 906, 830, and 784 bp for Ch.AS.a, Ch.AS.b, and Ch.AS.c isolates, respectively. Clustal analysis showed the three sequences to be quite identical, and BLAST search of NCBI database indicated the three organisms to be gram-negative Curtobacterium sp. Repeat sequencing undertaken using fresh single-colony-derived PCR product confirmed the earlier observations, indicating that the three isolates were morphological variants or different strains of the same organism. Primer walk sequencing yielded 1,350, 1,389, and 1,373 base sequence data for the above three isolates, which indicated that isolates Ch.AS.b and Ch.AS.c are exactly identical in their sequences, but differing from Ch.AS.a isolate in just two nucleotide substitutions. BLAST search of NCBI GenBank showed the highest similarity of these sequences to Curtobacterium citreum strain Z10zhy isolated from deep sea (AM411064) (99.5%) followed by C. citreum-type strain DSM 20528 (AM410690), Curtobacterium flaccumfaciens pv. basellae isolated as a pathogen of Malabar spinach in China (AY273210), an uncultured Micrococcineae bacterium (AB114608), and Curtobacterium luteum DSM20542 (X77437) (99%). Searching the Ribosomal database Project II of Michigan State University confirmed the identity of these isolates as C. citreum of class Actinobacteria, order Actinomycetales, and family Microbacteriaceae with 99.2% sequence similarity to strain Z10zhy (AM411064).
Testing the growth of bacterial isolates at different PGR levels.
No obvious bacterial growth was observed in spotting tests employing plain phytagel-gelled MS medium containing different BA or Kn levels for up to 1 mo. In the trial where MS liquid medium with different BA/Kn levels was inoculated with these isolates, some amount of bacterial growth was observed based on OD550 readings but there was no significant difference between the different treatments (P > 0.05 in all cases) (data not presented).
Acclimatization of micropropagated plants.
Employing sachet method, 90–100% establishment of rooted plantlets was attained in different batches. The sachets could be opened 2 wk after planting followed by shifting the plants to polyhouse or transplanting to a secondary nursery giving 80–90% net establishment. Planting as many as five plantlets per sachet was feasible followed by their separation at transplanting to the secondary nursery.
Extended culture monitoring.
Bacteria-harboring apparently clean cultures showed consistent growth and PR in the range of 3–4× on PGR-free medium for over 2 yr. Delaying the subculturing beyond 6–8 wk induced necrosis of lower leaves and suboptimal PR. In such instances, growth could be restored with a surface disinfection treatment before subculturing.
Discussion
The preset study demonstrates a simple micropropagation strategy for the shy suckering elite pompon chrysanthemum “Arka Swarna” employing microcuttings on PGR-free MS medium (except for one initial passage during culture establishment) offering a satisfactory multiplication rate and meeting other prime requirements for a viable micropropagation protocol (Cassells 2000; Thomas and Prakash 2004; Thomas et al. 2006). Earlier studies have demonstrated the potential for micropropagation of this crop but the approach often involved induction of multiple shoots on a PGR-supplied medium, separation of shoots, and their rooting.
Microbial contamination is a universal problem affecting plant tissue culture work (Leifert and Woodward 1998; Leifert and Cassells 2001; Herman 2004). A good control over contamination is essential for a sustainable micropropagation protocol. It is often assumed that organisms that are not eliminated by the disinfectants would express on the culture medium, allowing the quick identification and elimination of such cultures. This study indicates the widespread presence of covertly surviving endophytic bacteria in apparently clean cultures, which was brought out through culture indexing, employing bacteriological media. It appeared initially that at least three different organisms were associated with Arka Swarna stocks but the identification of different colony types employing the powerful tool of 16S rDNA sequence technique (Brenner et al. 2000) revealed that they were morphological variants or different strains of the same organism, identified as C. citreum, an actinobacterium. It is pertinent to mention that this organism did not obviously interfere with culture performance as long as the cultures were growing actively with rooting, but tended to display active colony growth on culture medium overriding the stocks at higher PGR levels when there was no vigorous shoot or root growths. However, such active microbial growth was observed in the presence of plants only. The results from the tests undertaken with plain MS medium indicated that the cause for the active bacterial growth at higher BA/ Kn levels was not merely the PGR level but the modified growth pattern or stress experienced by the cultures. Earlier studies have indicated such bacterial activation resulting from change in pH of culture medium or because of tissue breakdown products (Thomas 2004b). Supplying the tissue culture medium with host tissue extract enhanced the growth of various papaya endophytes (Thomas et al. 2007a).
Actinobacteria are known to produce phytohormones (Conn and Franco 2004) and this may be a possible explanation for the normal culture growth observed in PGR-free medium. It warrants stocks devoid of any bacteria to ascertain this probability. The endophytic survival of the bacteria in NaOCl- or HgCl2-treated shoots indicated the need for antibiotic challenge and extensive tissue indexing for cleansing the cultures as experienced with grape and watermelon stocks (Thomas and Prakash 2004; Thomas et al. 2006). This aspect is beyond the scope of this paper and will be addressed later. Curtobacterium citreum to our knowledge has not been documented as an endophyte. It is also a matter of concern that the organism in this study has shown high sequence similarity to C. flaccumfaciens pv. basellae, which has been isolated as a pathogen of Malabar spinach. Recently, there is an emerging interest in endophytes as agents of plant growth promotion or stress alleviation. There is also a school of thought that they may be latent or emerging future pathogens (Hallmann 2001). It is essential to have a proper documentation of common endophytes of different crop plants to ascertain this possibility and to be prepared for such an eventuality.
Our observations with the cultures of watermelon (Thomas 2004b; Thomas et al. 2006), grape, banana, capsicum, brinjal (Thomas 2004a, c, 2006a), and papaya (Thomas et al. 2007a) have all indicated widespread prevalence of endophytic bacteria in covert form in the medium or as tissue colonizers in apparently clean cultures that would go unnoticed in the absence of extensive tests aimed at detecting them (Thomas 2004a). The results were found true with another chrysanthemum variety, Arka Ravi, from which three different endophytes, namely, Microbacterium, Enterobacter, and Methylobacterium spp. have been isolated (Panicker 2005). In chrysanthemum, being a clonally propagated crop, it is possible that the endophytes survive in a persistent form. It is possible that visibly clean cultures of this crop elsewhere too might be harboring endophytic bacteria in covert form and the micropropagation going on unhindered with the resident endophytic microflora. The cultures upon continuous in vitro culturing in this study occasionally showed fungal contamination in spite of the extreme care taken, the source of which is being investigated now. More recent observations have indicated the presence of fastidious or normally nonculturable bacteria prevalent in index-negative cultures of papaya (Thomas et al. 2007b).
In conclusion, the present study demonstrates a sustainable micropropagation protocol for shy suckering pompon chrysanthemum using microcuttings MS on medium devoid of any PGRs, facilitating long-term maintenance of cultures with satisfactory multiplication rates avoiding the need for frequent initiation of cultures. The study has also indicated the frequent association of endophytic bacteria with in vitro cultures of Arka Swarna, which remained in covert form in actively growing stocks without any obvious interference with culture performance.
References
Ben-Jaacov, J.; Langhans, R. W. Rapid multiplication of chrysanthemum plants by stem-tip proliferation. HortScience 7: 289–290; 1972.
Bhattacharya, P; Dey, S.; Das, N.; Bhattacharya, B. C.; Bhattacharya, P. Rapid mass propagation of Chrysanthemum morifolium by callus derived stem and leaf explants. Plant Cell Rep. 9: 439–442; 1990.
Brenner, D.; Staley, J. T.; Krieg. N. Classification of prokaryotic organisms and the concept of bacterial speciation. In Boone, D. R.; Castenholz, R. W.; Garrity, G. M., ed., Bergey’s manual of systematic bacteriology, 2nd ed., vol. 1. New York: Springer Verlag; 2000: 27–38.
Cassells, A. C. Contamination detection and elimination in plant cell culture. In: Spier, R. E., ed. Encyclopedia of cell technology. New York: John Wiley & Sons, Inc.; 2000: 577–586.
Conn, V.M.; Franco, C.M.M. Effect of microbial inoculants on the indigenous actinobacterial endophyte population in the roots of wheat as determined by terminal restriction fragment length polymorphism. Appl. Environ. Microbiol. 70: 6407–6413; 2004.
Earle, E. D.; Langhans, R. W. Propagation of Chrysanthemum in vitro. I. Multiple plantlets from shoot tips and the establishment of tissue cultures. J. Am. Soc. Hort. Sci. 99: 128–131; 1974.
Gomez, A. K.; Gomez, A.A. Statistical Procedures for Agricultural Research 2nd Edition, New York: John Wiley & Sons Publication; 1984.
Hallmann, J. Plant interactions with endophytic bacteria. In Jeger, M. J.; Spence, N. J., ed. Biotic interactions in plant–pathogen associations. Oxon, Wallingford: CABI Publishing; 2001: 87–119.
Hahn, E-J; Paek, K-Y. Multiplication of Chrysanthemum shoots in bioreactors as affected by culture method and inoculation density of single node stems. Plant Cell Tissue Org. Cult. 81: 301–306; 2005.
Herman, E. B. Recent advances in plant tissue culture VIII. Microbial contaminants in plant tissue cultures: solutions and opportunities 1996–2003. Shrub Oak, USA; Agritech Consultants, Inc.; 2004.
Janakiram, T.; Rao, T. M. Chrysanthemum. Technical bulletin. Bangalore, India: Indian Institute of Horticultural Research; 2001. 18p.
Kaul, V.; Miller, R. M.; Hutchinson, J. F.; Richards, D. Shoot regeneration from stem and leaf explants of Dendranthema grandiflora Tzvelev. (syn. Chrysanthemum morifolium Ramat.).Plant Cell Tissue Org. Cult. 21: 21–30.
Kumari, M; Verghese, T. M. Efficient in vitro regeneration of plantlets from capitulum explant in chrysanthemum cultivars Miss Universe and Snowball. J. Ornam. Hortic. 6: 316–2321; 2003.
Leifert, C.; Cassells, A. C. Microbial hazards in plant tissue and cell cultures. In Vitro Cell. Dev. Biol. Plant 37: 133–138; 2001.
Leifert, C.; Woodward, S. Laboratory contamination management: the requirement for microbial quality assurance. Plant Cell Tissue Org. Cult. 52: 83–88; 1998.
Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15: 473–497; 1962.
Panicker, B. Studies on in vitro propagation in chrysanthemum (Dendranthema grandiflora Tzvelev). PhD Thesis, University of Agricultural Sciences, Bangalore, India; 2005.
Sivakumar, G.; Kim, S. J.; Hahn, E. J.; Paek, K. Y. Optimizing environmental factors for large-scale multiplication of chrysanthemum (Chrysanthemum grandiflorum) in balloon-type bioreactor culture. In Vitro Cell. Dev. Biol. Plant. 41: 822–825; 2005.
Ravindra, M. B.; Thomas, P. Sachet technique—an efficient method for the acclimatization of micropropagated grapes (Vitis vinifera L.). Curr. Sci. 68; 546–548; 1995.
Rout, G. R.; Das, P. Recent trends in the biotechnology of Chrysanthemum: a critical review. Sci. Hortic. 69: 239–257; 1997.
Teixeira da Silva, J. A. Ornamental chrysanthemums: improvement by biotechnology. Plant Cell Tissue Org. Cult. 79: 1–18; 2004.
Thomas, P. Humid incubation period and plantlet age influence acclimatization and establishment of micropropagated grapes. In Vitro Cell. Dev. Biol. Plant 34: 52–56; 1998.
Thomas, P. A three-step screening procedure for detection of covert and endophytic bacteria in plant tissue cultures. Curr. Sci. 87: 67–72; 2004a.
Thomas, P. In vitro decline in plant cultures: detection of a legion of covert bacteria as the cause for degeneration of long-term micropropagated triploid watermelon cultures. Plant Cell Tissue Org. Cult. 77: 173–179; 2004b.
Thomas, P. Isolation of Bacillus pumilus from in vitro grapes as a long-term alcohol-surviving and rhizogenesis inducing covert endophyte. J. Appl. Microbiol. 97: 114–123; 2004c.
Thomas, P. Isolation of an ethanol-tolerant endospore-forming gram-negative Brevibacillus sp as a covert contaminant in grape tissue cultures. J. Appl. Microbiol.101: 764–774; 2006a.
Thomas, P. Reemergence of covert bacteria Bacillus pumilus and Brevibacillus sp. in microbe-freed grape and watermelon stocks attributable to occasional autoclaving-defying residual spores from previous cycles. Plant Cell Tissue Org. Cult. 87: 155–165; 2006b.
Thomas, P.; Prakash, G. S. Sanitizing long-term micropropagated grapes from covert and endophytic bacteria and preliminary field testing of plants after eight years in vitro. In Vitro Cell. Dev. Biol. Plant 40: 603–607; 2004.
Thomas, P.; Prabhakara, B. S.; Pitchaimuthu, M. Cleansing the long-term micropropagated triploid watermelon cultures from covert bacteria and field testing the plants for clonal fidelity and fertility during the 7–10 year period in vitro. Plant Cell Tissue Org. Cult. 85:317–329; 2006.
Thomas, P.; Kumari, S.; Swarna, G. K.; Gowda, T. K. S. Papaya shoot tip associated endophytic bacteria isolated from in vitro cultures and host–endophyte interaction in vitro and in vivo. Can. J. Microbiol.; 53 (3); 2007a (doi: 10.1139/W06-141).
Thomas, P.; Kumari, S.; Swarna, G. K; Prakash, D. P.; Dinesh, M. R. Ubiquitous presence of fastidious endophytic bacteria in field shoots and index-negative apparently clean shoot-tip cultures of papaya. Plant Cell Rep. 2007b (doi: 10.1007/s00299-007-0363-2).
Acknowledgement
This study was financed under the project “Identification of covert endophytic microbes in plant tissue cultures and their management and control” from the Department of Biotechnology, Government of India, and it formed a part of the Ph.D. thesis of Ms. Bindu Panicker, submitted to the University of Agricultural Sciences, Bangalore.
Author information
Authors and Affiliations
Corresponding author
Additional information
Editor: M. E. Kane
Rights and permissions
About this article
Cite this article
Panicker, B., Thomas, P., Janakiram, T. et al. Influence of cytokinin levels on in vitro propagation of shy suckering chrysanthemum “Arka Swarna” and activation of endophytic bacteria. In Vitro Cell.Dev.Biol.-Plant 43, 614–622 (2007). https://doi.org/10.1007/s11627-007-9061-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11627-007-9061-6