Abstract
Tension force-induced bone formation is a complex biological process altered by various factors, for example miRNAs and gene regulatory network. However, we know little about critical gene regulators and their functional consequences on this complex process. The aim of this study was to determine the integrated relation between microRNA and mRNA expression in tension force-induced bone formation in periodontal ligament cells by a system biological approach. We identified 818 mRNAs and 32 miRNAs differentially expressed between cyclic tension force-stimulated human periodontal ligament cells and control cells by microarrays. By using miRNA/mRNA network analysis, protein-protein interactions network analysis, and hub analysis, we found that miR-195-5p, miR-424-5p, miR-1297, miR-3607-5p, miR-145-5p, miR-4328, and miR-224-5p were core microRNAs of tension force-induced bone formation. WDR33, HSPH1, ERBB3, RIF1, IKBKB, CREB1, FGF2, and PAG1 were identified as hubs of the PPI network, suggesting the biological significance in this process. The miRNA expression was further examined in human PDLC and animal samples by using quantitative real-time PCR. Thus, we proposed a model of tension force-induced bone formation which is co-regulated through integration of the miRNA and mRNA. This study illustrated the benefits of system biological approaches in the analysis of tension force-induced bone formation as a complex biological process. We used public information and our experimental data to do comprehensive analysis and revealed the coordination transcriptional control of miRNAs of tension force-induced bone formation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Orthodontic tooth movement (OTM) is the result of mechanical loading applied to a tooth and usually provided by an orthodontic appliance. Orthodontic force leads to the deposition or resorption of the supporting alveolar bone, depending on whether the tissues are exposed to a tension or compression strain, respectively. Bone formation at tension sites (TS) is of great importance to tooth stability during tooth movement (Krishnan and Davidovitch 2006; Wise and King 2008). Located between the tooth and alveolar bone, the periodontal ligament (PDL) plays a pivotal role in tension force-induced bone formation (Krishnan and Davidovitch 2006). It has been shown that periodontal ligament cells (PDLC) participate in tension force-induced bone formation by conducting the force to TS. Moreover, PDLC has been found to exhibit osteogenic differentiation and to produce osteogenesis-related factors, such as BMP2, BMP6, alkaline phosphatase (ALP), osteocalcin (OCN), SOX9, MSX1, and VEGFA, when cultured under tension force condition (Yang et al. 2005; Wescott et al. 2007).
Previous research has suggested that bone formation at TS is under the control of several signaling molecules, such as runt-related transcription factor 2, osterix, OCN, ALP, and fibroblast growth factors (FGFs) (Huang et al. 2009; Ku et al. 2009; Zainal Ariffin et al. 2011). However, their interactions and regulation process at the post-transcriptional level are poorly characterized.
MicroRNAs (miRNAs) represent a large group of small (approximately 22 nt or less), conserved, non-coding RNA. The miRNAs, acting as post-transcriptional suppression factors, bind to the specific mRNA recognition sequences (seed sequences) located in the 3’ untranslated regions (3’ UTRs) of the target mRNA, leading to mRNA degradation or protein translation interference (Friedman et al. 2009). Emerging evidence has indicated that miRNA biogenesis is indispensable to osteogenic differentiation of PDLC. miR-146a is reported to induce the differentiation in PDLC by targeting the NF-kappa B signaling (Hung et al. 2010). miR-21 and miR-101 are reported to directly target periodontal ligament-associated protein-1 expression during the osteogenic differentiation process of PDLC (Li et al. 2012). Downregulation of miR-17 expression in inflammatory periodontal ligament tissue-derived mesenchymal stem cells (PDLSCs) results in Smad ubiquitin regulatory factor one (Smurf1) activation, thus inhibiting the osteogenic differentiation of PDLSCs (Liu et al. 2011). miR-218 targets RUNX2 and decreases RUNX2 expression in undifferentiated human DSCs, such as PDLSC, DPSC, GSC, and BMSC (Gay et al. 2014). However, few miRNAs were reported to be associated with tension force-induced bone formation. Recent studies suggested that miR-21 increased stretch-induced osteogenic differentiation of PDLSCs by regulating ACVR2B protein expression (Wei et al. 2014) and miR-132 probably regulates PDLC osteogenic differentiation under the fluid shear stress through the mTOR signaling pathway (Chen et al. 2014). Computational prediction revealed that a miRNA can regulate hundreds of genes, and a target gene may be regulated by several miRNAs (Selbach et al. 2008). Although miRNA in tension force-induced bone formation has been partially addressed, the understanding of miRNAs and their regulatory networks in tension force-induced bone formation are still unclear.
In this study, we performed microarray profiling assays on miRNA and mRNA expression of PDLC osteogenic differentiation with tension force stimulation. We carried out the study to analyse microRNAs and mRNAs co-regulatory network in tension force-induced bone formation. We uncovered core miRNAs and achieved miRNA-centered biological regulation networks of tension force-induced bone formation in PDLC.
Materials and Methods
This study was approved by the Ethics Committee of the School of Stomatology, Wuhan University (2013. No. 17) and in accordance with local laws and regulations.
Cell culture.
Human PDLC were isolated and cultured according to widely used protocols. Periodontal ligament tissues were isolated from premolars that were extracted for orthodontic purposes at the Wuhan University Stomatology Hospital. All patients gave their informed consent before providing the samples. Tissues were scraped from the middle of the tooth roots and placed into 25-cm2 flasks with 1 mm × 1 mm × 1 mm size and 10 mm spacing (Corning Glass Works, Corning, NY). PDL tissues were then cultured in αMEM (HyClone, Auckland, New Zealand) supplemented with 10% (v/v) fetal bovine serum (FBS; HyClone, Logan, UT), 100 U ml−1 penicillin, and 100 mg ml−1 streptomycin (HyClone) at 37°C in a humidified atmosphere of 5% CO2. After 5 to 7 d in culture, PDLC from the explants were observed with a microscope, and the medium was refreshed every 2 d from then on. When the cells reached 80% confluence, they were separated and spread.
In vitro tension force-applying system.
The technique used to apply tension force to PDLC was based on previously reported protocols (Wescott et al. 2007). Equal numbers of P3 PDLC (approximately 1.0 × 106 cells) were seeded onto 6-well BioFlex plates coated with type I collagen (BioFlex, Flexcell International Corp, Hillsborough, NC). When cells were grown to 80% confluence, the medium was refreshed, and the plates were fixed on a metal tank connected to the strain unit. Cells were flexed with a cyclic tension force of 6 cycles/min (5 s on and 5 s off) and a 12% deformation rate at six different points in time (6, 12, 24, 48, or 72 h).
Cell morphology and cell orientation analysis.
Cell morphology was observed and photographed in randomly selected vision fields using an inverted microscope (Olympus, Tokyo, Japan) with a cell image system. The OriginPro 9 software (Northampton, MA) was used to analyze cell orientation based on cell photographs. Briefly, cell orientation was classified as follows: cells with a long axis parallel to the tension force direction were classified as the 0 group, and different angles of the cell long axis compared to the force direction were classified into different groups. After cell counting, cell orientation was calculated as percentages in each photograph. Different lines represented the directions, especially red line was parallel to tension force direction, and the length of each line represented the percentages of cells in each direction.
Microarray.
Both the miRNA and mRNA expression profiles of PDLC which had been stimulated with tension force for 72 h were compared with the profiles of untreated PDLC using a microarray. The total RNA of each sample was isolated using the Qiazol miRNA Isolation Kit (Qiagen, Hamburg, Germany). The mRNA microarray was performed by KangChen Bio-tech Inc. (KangChen, Shanghai, China) using Agilent Whole Genome Oligo Microarrays (Agilent, Santa Clara, CA) while the miRNA microarray was performed using an Exiqon miRNA Array (Exiqon, Vedbaek, Denmark).
The mRNA microarray: Total RNA from each sample was quantified and assessed for integrity. After being labeled, total RNA were hybridized onto the Whole Human Genome Oligo Microarray (4 × 44 K, Agilent Technologies). After the slides were washed, arrays were scanned with Agilent Scanner G2505C. The resulting images were analyzed using the Agilent Feature Extraction software (version 11.0.1.1). Quantile normalization was performed using GeneSpring GX v11.5.1, and low-intensity genes were filtered out. Genes that showed detected flags in at least three of the six samples were chosen for further analysis.
The miRNA microarray: Total RNA was harvested using the Qiagen miRNeasy mini kit according to manufacturer’s instructions. After passing the RNA quantity measurement, samples were labeled using the miRCURY™ Hy3™/Hy5™ Power kit and hybridized with the miRCURY™ LNA Array (v.18.0). After being washed, the slides were scanned using the Axon GenePix 4000B (Sunnyvale, CA) microarray scanner. Scanned images were then imported into the GenePix Pro 6.0 software (Axon) for grid alignment and data extraction. Replicated miRNAs were averaged, and those with intensities ≥30 in all samples were chosen to calculate a normalization factor. Expressed data were normalized using the median normalization. After normalization, significant differentially expressed miRNAs were identified through volcano plot filtering. Finally, hierarchical clustering was performed to show distinguishable miRNA expression profiling among samples.
Integrated miRNA/mRNA network analysis.
Initially, the miRNA target prediction was presented using online software MIRDB (http://mirdb.org/miRDB/). The predicted targets of 15 downregulated miRNAs were compared with those of the upregulated mRNAs from the mRNA microarray to detect overlaps. The miRNAs were ranked based on the number of overlap targets that they could regulate, and the top 20% were selected. The interactive network of selected miRNAs and actually deregulated target mRNAs was visualized using the Cytoscape software (San Diego, CA).
Network analysis of protein-protein interactions.
To further understand the function of these miRNAs, a network analysis of protein-protein interactions (PPI) depending on the PPI database was performed. Firstly, we searched the protein uniprot of these mRNAs in Uniprot (http://www.uniprot.org/). Secondly, by using MAS (http://bioinfo.capitalbio.com/mas3/), a highly reliable protein functional interaction network annotation system, we searched for the protein-protein interactions based on four commonly used databases: MINT (http://mint.bio.uniroma2.it/mint/Welcome.do), DIP (http://dip.doe-mbi.ucla.edu/dip/), IntAct (http://www.ebi.ac.uk/intact/), and HPRD (http://www.hprd.org/). Thirdly, the protein-protein interactions network was constructed based on related proteins of these and proteins in the PPI database. PPI network was visualized using the Cytoscape software.
Hubs in protein-protein interactions.
Highly connected nodes are crucial to the proper function of a PPI network. CytoHubba, a Cytoscape plug in, was used to identify the hubs of the PPIs network.
Animals.
The animal model for studying bone remodeling stimulated with tension force was established according to Silvana Taddei’s method (Taddei et al. 2012). Briefly, 8-wk-old female C57BL/6 mice were anesthetized with an intraperitoneal injection of 0.4 mg/kg chloral hydrate (Sigma, St. Louis, MO) and were placed in supine position on a surgical table. A stereomicroscope with a light system and a mouth opener were used to fully visualize the intraoral structures. After being carefully cleaned, the right first maxillary molar and incisors had their surfaces etched with a self-etching primer (Unitek/3M, Minneapolis, MN). The distal end of a nickel–titanium open-coil spring (Tomy, Tokyo, Japan) was bonded to the occlusal surface of the right first maxillary molar by using a light-cured resin (Transbond, Unitek/3M). The anterior portion of the coil was bonded to both upper incisors. A tension gauge was used to calibrate the force magnitude in the mesial direction. No reactivation was performed during the experimental period. The left side of the maxilla, without an orthodontic appliance, was used as a control (Supplementary Fig. 1). Mice were sacrificed at the following times: 3 d (for real-time PCR analysis) or 3, 12, and 18 d (for histological analysis). For every set of experiments, five animals were used at each time point.
Tooth movement measured by X-ray.
Mice were sacrificed after 3, 12, or 18 d. The entire right and left sides of the maxillary alveolar process (including three molars and alveolar bone) were isolated and taken X-rays detection (Bruker, Madison, WI). The distance between the cemento-enamel junctions (CEJs) of the mechanical-loaded first molars and the second molars was measured with an image tool, and the OTM ratio of tension force-stimulated sites (TS) was compared with that of contralateral sites (CS).
HE staining and mineral deposition rate assay.
Both the right and left sides of the maxillary alveolar process were fixed with 4% paraformaldehyde (pH 7.4) overnight. After demineralisation in 10% EDTA (pH 7.4) for 6 wk and dehydration in increasing concentrations of methanol, the samples were embedded in paraffin. Sagittal sections (5 μm) were cut and stained with hematoxylin and eosin (HE).
A mineral deposition rate assay was carried out using Calcein/Alizarin red fluorochrome labeling to examine the bone formation at tension sites. Briefly, 20 mg/kg Alizarin red (Sigma) was injected into the abdominal cavities of mice at day 7, while 5 mg/kg calcein (Sigma) was injected at day 14. The mice were sacrificed at day 18, and their right maxillary alveolar bones were dehydrated and embedded in methyl methacrylate. Then, 10-μm sections were cut and viewed under a fluorescent illumination microscope.
Immunohistochemistry and immunofluorescence staining.
Immunohistochemistry and immunofluorescence staining was performed as previously described (Song et al. 2013). Anti-CREB antibody (Cell Signaling Technology, Danvers, MA) was used for immunohistochemistry staining. Sections were counterstained with hematoxylin and used for the histological examinations. For immunofluorescence staining, FGF2 antibody (sc-79) (Santa Cruz Biotechnology, Santa Cruz, CA) was used as primary antibody. Nuclear counterstaining was performed using DAPI.
Total RNA isolation and quantitative real-time PCR.
The PDL and surrounding alveolar bone were extracted using a stereomicroscope. The gingival tissue, oral mucosa, and teeth were discarded. The tissues that were extracted from the distal area of the distal buccal and mesial roots of the upper first molar stimulated with tension force were considered as tension tissues. Tissues extracted from the other side of the maxilla were designated as controls. The tension tissues and control tissues were submitted to RNA extraction using a Qiagen miRNeasy micro kit (217084; Qiagen). The PDLC stimulated with tension force at specified times (6, 12, 24, 48, or 72 h) were washed three times with PBS, and the extracted total RNA was measured using Qiagen miRNeasy mini kit (217004; Qiagen). For miRNA expression assays, complementary DNA (cDNA) was synthesized using total RNA through a reverse transcription reaction (218061; miScript RT-II, Qiagen). Then, quantitative real-time PCR was performed using the ABI PRISM 7900 Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA) with All-in-One™ miRNA qPCR Primer (Genecopoeia, Rockville, MD) and the miScript SYBR Green PCR kit (218073; Qiagen). The U6 snRNA level was used as an internal normalization control. For mRNA expression, cDNA was synthesized using the Transcriptor First Strand cDNA Synthesis Kit (Roche, Basel, Switzerland), and quantitative real-time PCR was carried out with a SYBR Green PCR kit (Roche). The GAPDH mRNA level was used as an internal normalization control. Primer sequences were listed in Table 1. The mean Ct values from duplicate measurements were used to calculate the expression levels of the target gene, with normalization to an internal control.
Statistical analysis.
All values were expressed as the mean ± standard deviation (SD). Statistical significance was determined using the unpaired Student’s t test. P values <0.05 were considered statistically significant.
Results
Orthodontic tension force-induced PDLC osteogenic differentiation.
In order to investigate the microRNA and mRNA co-regulatory networks in tension force-induced bone formation, we used a strain unit to apply tension force to PDLC and evaluated the osteogenic differentiation level. Each well of cell culture plate had a flexible silicon membrane with a total growth area of 9.62 cm2 and a thickness of 0.05 mm of cell culture matrix. Flexible membrane deformation was produced by the strain unit, which used a computer-controlled vacuum to strain the flexible membranes. The deformation of the flexible membrane can cause attached cells to deform due to applied tension force (Fig. 1a ). PDLC were stimulated with tension force for 6, 12, 24, 48, and 72 h. The analysis of cell morphology and orientation showed a slender cell and a uniform alignment along the tension force direction (Fig. 1b ). The mRNA levels of OCN and ALP were significantly upregulated at 24 and 48 h, reaching a peak at 72 h. The optimal time of tension force-induced PDLC osteogenic differentiation was determined to be 72 h (Fig. 1c ).
Orthodontic tension force-induced PDLC osteogenic differentiation. a Schematic diagram of strain unit applying tension force on periodontal ligament cells. b Periodontal ligament cells had tension force stimulated for 6, 12, 24, 48, and 72 h. Cell morphology and orientation were recorded by phase-contrast microscopy (microscope, ×100). Cell orientation tends to be consistent with tension direction. c The expression of OCN and ALP in tension force-stimulated PDLC was determined by real-time PCR. GAPDH was used as a normalization control. Scale bar 40 μm.
miRNA expression profile during tension force-induced PDLC osteogenic differentiation.
To determine the miRNA expression differences, miRNA microarray was conducted using total RNA extracted from PDLC that underwent 72 h of tension force stimulation compared with PDLC that did not undergo any mechanical loading. The results revealed that among the 3100 screened miRNAs, 17 miRNAs were upregulated and 15 miRNAs were downregulated after 72 h of mechanical loading. The threshold used to screen up- or downregulated miRNAs was fold change > =2.0 and P value < =0.05 (Fig. 2a, b ).
miRNA expression profiling in tension force-stimulated PDLC. a miRNA volcano plot. Red point in the plot represents the differentially expressed miRNAs with statistical significance. b Heat map and cluster analysis of the 32 differentially expressed miRNAs in tension force-stimulated PDLC. Red represents high relative expression levels; green represents low relative expression levels. T tension force-stimulated PDLC, C untreated PDLC.
The mRNA microarray was performed with the same samples used in the miRNA microarray using Agilent gene expression profiling. Fold change > =3.5 and P value < =0.05 were set as the threshold. Three hundred and forty-four mRNAs were downregulated, whereas 818 mRNAs were upregulated in PDLC undergoing mechanical loading.
Network analysis of miRNA/mRNA protein-protein interactions.
To find correlation between different expression of miRNAs and their predicted target mRNAs in tension force-induced bone formation, a series of bioinformatic analyses were performed. miRNAs were usually negatively correlated with target mRNAs. It has been found that the expression of 818 mRNAs was upregulated while the expression of 344 mRNAs was significantly downregulated. Therefore, we chose upregulated expression mRNA and downregulated expression of miRNAs as analysis objects. To identify potential miRNA-regulated target genes, the datasets of all miRNAs and transcripts differentially expressed were integrated. We set two selection criteria for the potential targets. The target mRNAs should be: (1) reciprocal expressed to miRNA and (2) predicted as gene target by reliable miRNA target prediction program.
Putative targets of the 15 downregulated miRNAs were predicted by MIRDB and matched with the 818 upregulated mRNAs. miRNAs that could regulate these overlap mRNAs were ranked based on the number of deregulated target mRNAs and the top 20% were selected. The top seven miRNAs that had the most overlap with mRNAs were hsa-miR-195-5p, hsa-miR-424-5p, hsa-miR-1297, hsa-miR-3607-5p, hsa-miR-145-5p, hsa-miR-4328, and hsa-miR-224-5p, which collectively targeted a total of 150 mRNAs. The interactive network of these 7 miRNAs and 150 upregulated mRNAs was visualized using Cytoscape (Fig. 3a ). Round nodes represented mRNAs, and pentagons represented miRNAs. Larger nodes represented mRNAs regulated by more than three miRNAs in this network.
The miRNA/mRNA interactive network. a miRNA/mRNA network was constructed between the selected seven miRNAs and 150 mRNAs by using Cytoscape software. b The PPI network of genes related to tension force-induced PDLC osteogenic differentiation was visualized by using Cytoscape. Green nodes represented genes that were predicted been targeted and actually upregulated in mRNA profiling. Gray nodes represented interactive genes, based on protein databases (HPRD, MINT, DIP, and IntAct). Interactions between genes are shown as solid lines. c Hubs of PPI network were visualized by CytoHubba.
Network analysis of protein-protein interactions and hubs.
In the miRNA/mRNA network, there were hundreds of mRNAs. In order to further understand the function of miRNAs, we use protein-protein interactions analysis and hub analysis to comb protein interaction networks and further find the hubs. Protein-protein interactions (PPI) is the relation between the protein molecules in cells or organs. MAS is a highly reliable protein functional interaction network annotation system, storing the network relationship between protein and protein. mRNAs in miRNA/mRNA network were entered into MAS; interactions were found out based on four commonly used databases: MINT, DIP, IntAct, and HPRD. A sixty-four node PPI network was constructed based on twenty-six proteins from the miRNA/mRNA network and others closely related to them from the databases. Proteins were represented as nodes in network; interaction relations were represented as edges (Fig. 3b ).
Nodes in the network often have one or more interactions to the others; hubs are a small number of highly connected protein nodes in the protein-protein interaction (PPI) network, suggesting the biological significance of the network (He and Zhang 2006). We ranked the nodes in our PPI network according to their node degrees (the number of edges). Hubs of the PPI network were WDR33, HSPH1, ERBB3, RIF1, IKBKB, CREB1, FGF2, and PAG (Fig. 3c ).
Validation of the miRNA and mRNA expression in vitro.
To further validate the results of profilings and bioinformatic analysis, miRNA and mRNA were extracted from PDLC after 72 h with mechanical loading or without loading. Quantitative real-time PCR showed that these seven miRNAs were significantly downregulated in tension force-stimulated PDLC (Fig. 4a–g ), similar to the microarray results.
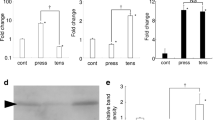
Validation of differentially expressed miRNAs and mRNA in vitro. a–g The seven miRNAs expression in tension force-stimulated PDLC was determined by real-time PCR and compared with miRNA microarray. U6 was used as a normalization control. All data were based on three independent experiments. Asterisks indicate statistical significance between experimental and control groups at P < 0.05. h Representative miRNA/mRNA binding sites of CREB1 or FGF2 for each of the three miRNAs. i–j CREB1 and FGF2 expression in tension force-stimulated PDLC was determined by real-time PCR and compared with the mRNA microarray (*p < 0.05).
Among the eight hubs, FGF2 and CREB1 have been widely accepted as key factors in osteogenic differentiation. FGF2 was a putative target of miR-195-5p, miR-424-5p, and miR-3607-5p. CREB1 was a putative target of miR-4328, miR-1297, and miR-224-5p (Fig. 4h ). Representative quantitative real-time PCR for FGF2 and CREB1 showed that they were upregulated in tension force-stimulated PDLC (Fig. 4i, j ).
Orthodontic tension force-induced tooth movement and bone formation at TS in mice.
In order to provide more evidence to support our findings, we established a mouse model of orthodontic tooth movement for studying miRNA different expression at TS in vivo. The right first maxillary molars of female C57BL/6 mice received 35 g forward tension force for 3, 12, and 18 d. We did morphological analysis to identify the tension force-induced bone formation in this model. First, X-ray photographs showed an increasing OTM of the first molars at days 3, 12, and 18 (Fig. 5a, b ). Second, HE staining showed that after 18 d of orthodontic mechanical loading, there was more bone tissue volume at TS than at control sites (Fig. 5c ). Third, obvious golden and red lines in the hard tissue sections of calcein and alizarin red double staining showed active bone formation at TS compared with the contralateral sites (Fig. 5d ).
Tooth movement and TS bone formation in mice. a Thirty-five-gram tension force was loaded on the mouse’s first maxillary molar for 0, 3, 12, or 18 d, and the maxilla was pictured by high-resolution X-ray. b The OTM ratio of tension force-stimulated sites (TS) was compared with that of contralateral sites (CS), measuring the distance between the CEJs of the first molars and the second molars. c Alveolar bone mass in the distal area to the first molar in the 18-d and control groups were shown with HE staining. d Hard tissue sections of calcein and alizarin red double staining showed a different mineral deposition rate at TS and CS at day 18. ( R root, AB alveolar bone). e–i miRNAs expression at tension sites of orthodontic tooth movement mice molar at day 3 was determined by real-time PCR. All data were based on three independent experiments; asterisks indicate statistical significance between experimental and control groups. *P < 0.05. Scale bar: a 500 μm; c, d 100 μm.
Validation of the miRNA expression in mouse.
These results proved that obvious bone formation existed at tension sites of the mice moving tooth. Tissues were isolated at day 3 from TS and contralateral sites. RNA extracted from them was prepared to reverse transcription and quantitative real-time PCR. The expression of miR-195-5p, miR-424-5p, miR-1297, miR-145-5p, and miR-224-5p at tension sites of orthodontic tooth movement mice molar was downregulated at day 3 (Fig. 5e–i ). These results confirmed our bioinformatic analysis.
Discussion
In the present study, we employed the miRNA microarray and the mRNA microarray to profile the miRNA and mRNA expression pattern during tension force-induced PDLC osteogenic differentiation. The miRNA expression profiling and mRNA expression profiling showed that 32 miRNAs and thousands of mRNAs were differently expressed during the process. Furthermore, we critically exploited the miRNAs’ and mRNAs’ relationships in tension force-induced bone formation by using a systems biology approach. In this way, we found seven core miRNAs and eight core proteins such as FGF2 and CREB1. The results of our analysis were further confirmed in vitro and in vivo.
PDLC cannot only play a crucial role in conducting tension force but also exhibit osteogenic differentiation under tension force stimulation (Yang et al. 2006; Wescott et al. 2007). It had been validated that the cyclical tension force-stimulated periodontal ligament cells could lead to certain characteristics of osteoblasts in vitro, such as the high upregulation of ALP, the interleukin family, and some growth factors (Yamaguchi et al. 1996; Pinkerton et al. 2008). In this study, PDLC became more slender and exhibited a uniform alignment along the tension force direction and a high upregulation of OCN and ALP, indicating obvious osteogenic differentiation under tension force stimulation, which is similar to previous studies.
Tension force-induced bone formation is a complex process with varying degrees of genes expression. In our study, 32 miRNAs and thousands of mRNAs were found to be likely related to tension force-induced bone formation by microarrays. A few studies had reported the miRNAs expression profiling under force stimulation, for example, fluid shear stress stimulation (Chen et al. 2014). However, these studies were lacking a comprehensive function analysis of the differential expression of miRNAs.
By integrating different data sources on the network level, miRNAs and mRNAs network analysis is able to provide a common and appropriate way to investigate the physiological mechanism and interactions between the miRNAs and mRNAs. Through a series of analyses, we found that miR-195-5p, miR-424-5p, miR-1297, miR-3607-5p, miR-145-5p, miR-4328, and miR-224-5p were core miRNAs and may regulate abundant genes. There are also connections between these genes, thus forming a complicated network in this process.
MiR-195-5p, activated in multiple diseases such as cancers, heart failure, and schizophrenia, regulates a number of target proteins, which are involved in the cell cycle, apoptosis, and proliferation (He et al. 2011). Meanwhile, miR-195-5p was found to be downregulated in osteosarcoma cell lines (Namlos et al. 2012). In this study, our data and analysis showed that it was related to tension force-induced bone formation. What is more, this function may depend on the suppressive effect on related genes.
Both miR-195-5p and miR-424-5p are members of the miR-15/107 group of microRNA, which are defined based on the included AGCAGC sequence near the mature miRNA 5’ end. This miRNA group is increasingly noted to serve key functions in humans, to regulate gene expression involved in cell division, metabolism, stress response, and angiogenesis (Finnerty et al. 2010). In our study, miR-424-5p was found to be downregulated in tension force-induced osteogenesis, similar to a previous study in osteogenic differentiation of human mesenchymal stromal cells (Gao et al. 2011).
The role of miR-145-5p in tumor progression, cardiovascular system disease, and nervous system disease has been stated. The miR-224-5p was also reported to be related to tumor development. Several studies have revealed a decreased level of miR-224-5p in osteogenic differentiation in different cells (Palmieri et al. 2008; Schaap-Oziemlak et al. 2010; Mosakhani et al. 2013). Similarly, miR-145-5p decreased and served as a repressor, inhibiting the osteogenic differentiation of C2C12 and MC3T3-E1 cells and odontoblast differentiation during tooth development (Jia et al. 2013; Liu et al. 2013). Our study revealed that the downregulated of miR-145-5p and miR-224-5p expression was related to tension force-induced bone formation.
The research on miR-1297, miR-3607-5p, and miR-4328 is much less revealing. It has been shown that these miRNAs could affect cell proliferation, progression, or metastasis in tumor cells or tumor-like skin scars (Li et al. 2013; Saini et al. 2014; Yang et al. 2014). Our study revealed their expression in tension force-induced bone formation. This finding indicated that these miRNAs may affect this process.
The relationships among target genes were visualized, called PPI network. WDR33, HSPH1, ERBB3, RIF1, IKBKB, CREB1, FGF2, and PAG1 were found to be hubs of the PPI network. Among the hubs, ERBB3, IKBKB, CREB1, FGF2, and PAG1 have been found to be involved in bone metabolism (Lin et al. 2008; Matsubara et al. 2010; Otero et al. 2010).
The bioinformatic analysis revealed that these miRNAs and mRNAs might be key biomarkers to tension force-induced bone formation. To confirm these results, these seven miRNAs’ expression was validated in tension force-induced PDLC osteogenic differentiation by using quantitative real-time PCR. Except for miR-3607-5p and miR-4328 which have not been reported to exist in mice, the expression of miR-195-5p, miR-424-5p, miR-1297, miR-145-5p, and miR-224-5p were proved to be downregulated at mice TS at day 3 by using quantitative real-time PCR, compared with contralateral sites without tension force. Our further study revealed that the mRNAs expression of WDR33, RIF1, IKBKB, CREB1, FGF2, and PAG1 at TS of orthodontic tooth movement mice molar were significantly upregulated at day 3. But, HSPH1 and ERBB3 expression did not show obvious change at mRNA level (Supplementary Fig. 2).
FGF2 and CREB1 were chosen for representative validation for four reasons. First, FGF2 and CREB1 mRNA expression was upregulated in tension force-induced bone formation. Second, both FGF2 and CREB1 were key upregulated genes in hub network analysis. Third, FGF2 was the putative target gene of miR-195-5p, miR-424-5p, and miR-3607-5p, whereas miR-224-5p, miR-4328, and miR-1297 were predicted to suppress CREB1. Fourth, previous studies have revealed that FGF2 induced osteogenic differentiation through Runx2 activation or by modulating the WNT pathway, whereas CREB1 was identified as a transcription factor that can bind to the BMP2 promoter, promote BMP2 expression in osteoblasts (Zhang et al. 2011; Byun et al. 2014). FGF2 and CREB1 were found to be overexpressed at mice TS in immunohistochemistry or immunofluorescence experiments. FGF2 and CREB1 were strongly localized at tension sites of first molar but were weakly localized at the compression sites or contralateral sites at day 3. (Supplementary Fig. 3). These results further confirmed the robustness of our analysis.
In summary, the present study revealed that miR-195-5p, miR-424-5p, miR-1297, miR-3607-5p, miR-145-5p, miR-4328, and miR-224-5p are core miRNAs related to tension force-induced bone formation in PDLC by downregulating the expression of target genes. Proteins such as WDR33, HSPH1, ERBB3, RIF1, IKBKB, CREB1, FGF2, and PAG1 may be key proteins in this process. The systemic, biology-based miRNA and mRNA expression analysis result of our research is a useful reference to further study of the regulation network in tension force-induced bone formation.
References
Byun MR, Kim AR, Hwang JH, Kim KM, Hwang ES, Hong JH (2014) FGF2 stimulates osteogenic differentiation through ERK induced TAZ expression. Bone 58:72–80
Chen L, Holmstrom K, Qiu W, Ditzel N, Shi K, Hokland L, Kassem M (2014) MicroRNA-34a inhibits osteoblast differentiation and in vivo bone formation of human stromal stem cells. Stem Cells 32(4):902–912
Finnerty JR, Wang WX, Hebert SS, Wilfred BR, Mao G, Nelson PT (2010) The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol 402(3):491–509
Friedman RC, Farh KKH, Burge CB, Bartel DP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19(1):92–105
Gao J, Yang T, Han J, Yan K, Qiu X, Zhou Y, Fan Q, Ma B (2011) MicroRNA expression during osteogenic differentiation of human multipotent mesenchymal stromal cells from bone marrow. J Cell Biochem 112(7):1844–1856
Gay I, Cavender A, Peto D, Sun Z, Speer A, Cao H, Amendt BA (2014) Differentiation of human dental stem cells reveals a role for microRNA-218. J Periodontal Res 49(1):110–120
He JF, Luo YM, Wan XH, Jiang D (2011) Biogenesis of MiRNA-195 and its role in biogenesis, the cell cycle, and apoptosis. J Biochem Mol Toxicol 25(6):404–408
He X, Zhang J (2006) Why do hubs tend to be essential in protein networks? PLoS Genet 2(6):e88
Huang XF, Zhao YB, Zhang FM, Han PY (2009) Comparative study of gene expression during tooth eruption and orthodontic tooth movement in mice. Oral Dis 15(8):573–579
Hung PS, Chen FC, Kuang SH, Kao SY, Lin SC, Chang KW (2010) miR-146a induces differentiation of periodontal ligament cells. J Dent Res 89(3):252–257
Jia J, Tian Q, Ling S, Liu Y, Yang S, Shao Z (2013) miR-145 suppresses osteogenic differentiation by targeting Sp7. FEBS Lett 587(18):3027–3031
Krishnan V, Davidovitch Z (2006) Cellular, molecular, and tissue-level reactions to orthodontic force. Am J Orthod Dentofac Orthop 129(4):469, e461–432
Ku SJ, Chang YI, Chae CH, Kim SG, Park YW, Jung YK, Choi JY (2009) Static tensional forces increase osteogenic gene expression in three-dimensional periodontal ligament cell culture. BMB Rep 42(7):427–432
Li C, Bai Y, Liu H, Zuo X, Yao H, Xu Y, Cao M (2013) Comparative study of microRNA profiling in keloid fibroblast and annotation of differential expressed microRNAs. Acta Biochim Biophys Sin 45(8):692–699
Li C, Li C, Yue J, Huang X, Chen M, Gao J, Wu B (2012) miR-21 and miR-101 regulate PLAP-1 expression in periodontal ligament cells. Mol Med Rep 5(5):1340–1346
Liu H, Lin H, Zhang L, Sun Q, Yuan G, Zhang L, Chen S, Chen Z (2013) miR-145 and miR-143 regulate odontoblast differentiation through targeting Klf4 and Osx genes in a feedback loop. J Biol Chem 288(13):9261–9271
Lin SH, Cheng CJ, Lee YC, Ye X, Tsai WW, Kim J, Pasqualini R, Arap W, Navone NM, Tu SM, Hu M, Yu-Lee LY, Logothetis CJ (2008) A 45-kDa ErbB3 secreted by prostate cancer cells promotes bone formation. Oncogene 27(39):5195–5203
Liu Y, Liu W, Hu C, Xue Z, Wang G, Ding B, Luo H, Tang L, Kong X, Chen X, Liu N, Ding Y, Jin Y (2011) MiR-17 modulates osteogenic differentiation through a coherent feed-forward loop in mesenchymal stem cells isolated from periodontal ligaments of patients with periodontitis. Stem Cells 29(11):1804–1816
Matsubara T, Ikeda F, Hata K, Nakanishi M, Okada M, Yasuda H, Nishimura R, Yoneda T (2010) Cbp recruitment of Csk into lipid rafts is critical to c-Src kinase activity and bone resorption in osteoclasts. J Bone Miner Res 25(5):1068–1076
Mosakhani N, Pazzaglia L, Benassi MS, Borze I, Quattrini I, Picci P, Knuutila S (2013) MicroRNA expression profiles in metastatic and non-metastatic giant cell tumor of bone. Histol Histopathol 28(5):671–678
Namlos HM, Meza-Zepeda LA, Baroy T, Ostensen IH, Kresse SH, Kuijjer ML, Serra M, Burger H, Cleton-Jansen AM, Myklebost O (2012) Modulation of the osteosarcoma expression phenotype by microRNAs. PLoS ONE 7(10):e48086
Otero JE, Dai S, Alhawagri MA, Darwech I, Abu-Amer Y (2010) IKKbeta activation is sufficient for RANK-independent osteoclast differentiation and osteolysis. J Bone Miner Res 25(6):1282–1294
Palmieri A, Pezzetti F, Brunelli G, Martinelli M, Lo Muzio L, Scarano A, Degidi M, Piattelli A, Carinci F (2008) Peptide-15 changes miRNA expression in osteoblast-like cells. Implant Dent 17(1):100–108
Pinkerton MN, Wescott DC, Gaffey BJ, Beggs KT, Milne TJ, Meikle MC (2008) Cultured human periodontal ligament cells constitutively express multiple osteotropic cytokines and growth factors, several of which are responsive to mechanical deformation. J Periodontal Res 43(3):343–351
Saini S, Majid S, Shahryari V, Tabatabai ZL, Arora S, Yamamura S, Tanaka Y, Dahiya R, Deng G (2014) Regulation of SRC kinases by microRNA-3607 located in a frequently deleted locus in prostate cancer. Mol Cancer Ther 13(7):1952–1963
Schaap-Oziemlak AM, Raymakers RA, Bergevoet SM, Gilissen C, Jansen BJ, Adema GJ, Kogler G, le Sage C, Agami R, van der Reijden BA, Jansen JH (2010) MicroRNA hsa-miR-135b regulates mineralization in osteogenic differentiation of human unrestricted somatic stem cells. Stem Cells Dev 19(6):877–885
Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455(7209):58–63
Song X, Liu W, Xie S, Wang M, Cao G, Mao C, Lv C (2013) All-transretinoic acid ameliorates bleomycin-induced lung fibrosis by downregulating the TGF-beta1/Smad3 signaling pathway in rats. Lab Investig 93(11):1219–1231
Taddei SR, Moura AP, Andrade I Jr, Garlet GP, Garlet TP, Teixeira MM, da Silva TA (2012) Experimental model of tooth movement in mice: a standardized protocol for studying bone remodeling under compression and tensile strains. J Biomech 45(16):2729–2735
Wei FD, Liu C, Feng F, Zhang S, Yang Y, Hu G Ding and Wang S (2014) MicroRNA-21 mediates stretch-induced osteogenic differentiation in human periodontal ligament stem cells. Stem Cells Dev
Wescott D, Pinkerton M, Gaffey B, Beggs K, Milne T, Meikle M (2007) Osteogenic gene expression by human periodontal ligament cells under cyclic tension. J Dent Res 86(12):1212–1216
Wise GE, King GJ (2008) Mechanisms of tooth eruption and orthodontic tooth movement. J Dent Res 87(5):414–434
Yamaguchi M, Shimizu N, Shibata Y, Abiko Y (1996) Effects of different magnitudes of tension-force on alkaline phosphatase activity in periodontal ligament cells. J Dent Res 75(3):889–894
Yang NQ, Zhang J, Tang QY, Guo JM, Wang GM (2014) miRNA-1297 induces cell proliferation by targeting phosphatase and Tensin homolog in testicular germ cell tumor cells. Asian Pac J Cancer Prev 15(15):6243–6246
Yang Y, Li X, Rabie A, Fu M, Zhang D (2005) Human periodontal ligament cells express osteoblastic phenotypes under intermittent force loading in vitro. Front Biosci J Virtual Libr 11:776–781
Yang YQ, Li XT, Rabie AB, Fu MK, Zhang D (2006) Human periodontal ligament cells express osteoblastic phenotypes under intermittent force loading in vitro. Front Biosci 11:776–781
Zainal Ariffin SH, Yamamoto Z, Zainol Abidin IZ, Megat Abdul Wahab R, Zainal Ariffin Z (2011) Cellular and molecular changes in orthodontic tooth movement. ScientificWorldJournal 11:1788–1803
Zhang RJ, Edwards R, Ko SY, Dong S, Liu H, Oyajobi BO, Papasian C, Deng HW, Zhao M (2011) Transcriptional regulation of BMP2 expression by the PTH-CREB signaling pathway in osteoblasts. PLoS ONE 6(6):e20780
Acknowledgment
This study was supported by the National Natural Science Foundation of China (NSFC); NSFC number: 81371169.
Author information
Authors and Affiliations
Corresponding author
Additional information
Editor: T. Okamoto
Guangli Han holds a PhD degree, Wuhan University.
Maolin Chang and Heng Lin contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(XLS 3517 kb)
ESM 2
(XLS 56 kb)
Supplementary Fig. 1
Description of in vivo orthodontic force applying system. (a) Mice positioned in the supine position on a surgical table. (b) Surgical table placed under a stereomicroscope with light system. (c) Use of a mouth-opener in a mouse. (d) Occlusal view of the maxillary molar region. The distal end of a nickel–titanium open-coil spring (Tomy, Japan) was bonded to the occlusal surface of the right first maxillary molar using a light-cured resin. (e) A tension gauge was attached to the surgical table to show the force magnitude. (f) The coil spring bonded to both upper incisors. Stereomicroscope (S), mouth-opener (O), coil spring (C), upper first molar (M), tension gauge (T). (JPEG 1223 kb)
Supplementary Fig. 2
Validation of differentially expressed mRNAs in vivo. (a-h) The mRNAs expression of WDR33, RIF1, IKBKB, CREB1, FGF2, PAG1 and HSPH1 at tension sites of orthodontic tooth movement mice molar at day 3 was determined by real-time PCR. β-actin was used as a normalization control. All data were based on three independent experiments. Asterisks indicate statistical significance between experimental and control groups at P < 0.05. (JPEG 354 kb)
Supplementary Fig. 3
Strongly expression of FGF2 and CREB1 at tension sites of orthodontic tooth movement mice molar. (a) FGF2 expression was detected by immunofluorescence staining. FGF2 was strongly localized at tension site of first molar but were weakly localized at the contralateral sites at day 3. TS: tension site; CS: contralateral site. (Original magnification, 200×) (b) CREB1 expression was detected by immunohistochemistry staining. CREB1 was strongly localized at tension sites of first molar but were weakly localized at the compression sites at day 3. Scale bar for A, 200 μm; scale bar for B and C, 100 μm. TS: tension site; CS: compression site. (JPEG 914 kb)
Rights and permissions
About this article
Cite this article
Chang, M., Lin, H., Luo, M. et al. Integrated miRNA and mRNA expression profiling of tension force-induced bone formation in periodontal ligament cells. In Vitro Cell.Dev.Biol.-Animal 51, 797–807 (2015). https://doi.org/10.1007/s11626-015-9892-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11626-015-9892-0