Abstract
Purpose
The cancerous inhibitor of protein phosphatase 2A (CIP2A) oncoprotein is overexpressed in colon cancer tissue compared to normal colon mucosa. We investigated the impact of CIP2A on colon cancer.
Methods
A tissue microarray consisting of 167 colon cancer specimens was investigated. The association between CIP2A and clinicopathological parameters was analyzed using the χ 2 test. Survival was analyzed using the Kaplan–Meier method. The impact of CIP2A on proliferation and drug resistance was evaluated using the 3-(4, 5-dimethylthiazolyl-2)-2, 5-diphenyltetrazolium bromide test. An anchorage-independent colony formation assay was also performed.
Results
CIP2A was an independent prognostic factor in colon cancer after controlling for other clinical confounding factors, such as stage and lymphovascular invasion, particularly in stages III and IV (hazard ratio = 2.974, P < 0.001). The knockdown of CIP2A reduced the proliferation and anchorage-independent colony formation of colon cancer cells. Knockdown of CIP2A decreased the resistance of the cells to 5-fluorouracil, oxaliplatin, and SN38 (an active metabolite of irinotecan). Treatment with 5-fluorouracil, oxaliplatin, and SN38 decreased CIP2A expression.
Conclusions
CIP2A is a prognostic factor in colon cancer. The knockdown of CIP2A reduced proliferation and anchorage-independent colony formation and increased 5-fluorouracil, oxaliplatin, and SN38 efficacy in colon cancer cell lines.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer is the third most common malignancy worldwide, with more than one million new cases and nearly 500,000 deaths each year.1 Despite advances in chemotherapy, including 5-fluorouracil (5-FU), irinotecan, oxaliplatin, bevacizumab, and cetuximab, the median overall survival in metastatic colorectal cancer is approximately 20–28 months.2–6 Thus, it is necessary to identify novel significant oncogenes or targets for the development of new cancer therapeutics.
Cancerous inhibitor of protein phosphatase 2A (PP2A) (CIP2A) is a recently indentified novel oncoprotein also referred to as KIAA1524 or p90 tumor-associated antigen. CIP2A is important for maintaining a malignant cellular phenotype, proliferation, and transformation.7,8 CIP2A has been demonstrated to inhibit PP2A activity toward the oncogenic transcription factor c-Myc, thereby preventing c-Myc proteolytic degradation, which is important for cell transformation and tumorigenesis in vivo and in vitro. Overexpression of CIP2A has been demonstrated in many common human malignancies, including leukemia, breast, gastric, prostate, lung, ovarian, and colon cancers, and head and neck carcinoma.8–16 With respect to colon cancer, CIP2A expression was analyzed using 43 human colon cancer specimens and five normal colon specimens using reverse transcriptase polymerase chain reaction (RT-PCR). CIP2A mRNA was found to be significantly overexpressed in human colon cancer specimens compared than in normal colon tissue.8 However, the associations among CIP2A, clinicopathological variables, survival, and chemotherapeutic drugs in patients with colon cancer are unclear.
In this study, we aimed to investigate whether CIP2A overexpression in colon cancer was associated with clinical cancer aggressiveness and to study the impact of CIP2A on cell proliferation, anchor-independent colony formation, and drug resistance including 5-FU, SN38 (an active metabolite of irinotecan), and oxaliplatin in colon cancer cell lines.
Materials and Methods
Patients
The protocol of this study was approved by the Institutional Review Board of the Taipei Veterans General Hospital. The study was a retrospective analysis of data compiled from the medical records of patients diagnosed with colon cancer at the Taipei Veteran General Hospital from January 2000 through January 2010. Patients were identified by searching the Department of Pathology and Laboratory Medicine database of pathology reports. The diagnosis of colon cancer was made according to the World Health Organization criteria. Patients were classified according to the American Joint Committee on Cancer (AJCC) staging system (version 6). The clinical data of all patients were obtained from the cancer registry and through chart review. Clinical–pathological staging and clinical course were determined by examining a computer database containing detailed information on all histology-proven colon cancer patients. Information regarding recurrence and death after hepatic resection was obtained from hospital records and the National Cancer Registry.
Left colon cancer was defined as a malignancy in the splenic flexure, descending colon, sigmoid, and/or rectosigmoid colon, and right colon cancer was defined as that occurring in the cecum, ascending colon, hepatic flexure, and/or transverse colon. The decision to perform hepatic resection was made by clinical physicians based on the extent of metastatic lesions and the condition of the individual patient.
Death from colon cancer was regarded as an event. Patients who were alive at the end of the follow-up period were not included in this study. Overall survival was defined as the time from primary resection to death.
The follow-up period in this study was considered to end in January 2010, or at the death of the patients. Patients were followed-up at least every 4 months from the time of primary resection for the first 2 years, followed by every 6 months for 5 years, and then annually until death.
Immunohistochemistry (IHC)
For the tissue microarray (TMA), hematoxylin and eosin-stained sections from each paraffin-embedded, formalin-fixed block were used to define diagnostic areas, and a representative 0.6 mm core was obtained from each case and inserted in a grid pattern into a recipient paraffin block. Sections (4 μm) were then deparaffinized in xylene and rehydrated in a descending ethanol series. To enhance immunoreactivity, sections were incubated in Tris–EDTA, pH 6.0, and boiled for 12 min. Endogenous peroxidase activity was eliminated by incubation in hydrogen peroxide. The antibody used in the study was a rabbit polyclonal antibody against human CIP2A (NB100-74663; Novus Biologicals, Littleton, CO, USA; dilution 1:900). Bound antibodies were visualized using the Envision Detection System (K500711; Dako Denmark A/S), and DAB (diaminobenzidine) was used as a chromogen. Omission of the primary antibody served as a negative control. Positive controls (normal liver) were stained in parallel with each set of TMA studied.
Immunopositivity was evaluated by two pathologists in our hospital who were blinded to the clinical information. Each cell was scored as 0, 1, 2, or 3, corresponding to negative, weak, moderate, and strong staining intensities, respectively. Percentages of stained cells were counted, and a final immunohistochemical score (H score) was calculated by summing the products of the staining intensities (0–3) and distributions (0–100%); H scores ranged from 0 to 300. An H score of more than or equal to 150 points was defined as strongly positive, and all others were scored as weakly positive.
Cell Lines
The HT29 and Caco2 human colon cell lines were from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 (Invitrogen, Carlsbad, CA, USA) containing 10% fetal calf serum (Invitrogen) and 100 IU/ml penicillin (Sigma, St. Louis, MO, USA). Cells were grown on sterilized culture dishes and were passaged every 3–4 days with 0.25% trypsin (Invitrogen).
Knockdown of CIP2A in Colon Cancer Cell Lines
The shRNA clones targeting CIP2A (pLKO.1-shCIP2A, TRCN0000135532, target sequence CCACAGTTTAAGTGGTGGAAA) and the entry clone expressing luciferase shRNA (pLKO.1-shLuc) as a non-targeting shRNA control were obtained from the National RNAi Core Facility, Institute of Molecular Biology, Academia Sinica, Taiwan (http://rnai.genmed.sinica.edu.tw/). Lentivirus production, infection, and cell line construction were performed as described previously.16
Western Blot Analysis
Total cellular protein was extracted using lysis buffer (Pierce, Rockville, MD, USA) and quantified using the Bradford method. Next, 35 μg of protein was separated using sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis (12%). The protein was transferred to a polyvinylidene fluoride membrane (Millipore, Billerica, MA, USA), and the membrane was incubated overnight at 4°C with antibodies against CIP2A (1:500; Novus Biologicals) and beta-actin (1:500, Santa Cruz Biotechnology, Santa Cruz, CA, USA). After incubation with peroxidase-coupled anti-mouse IgG (Santa Cruz Biotechnology) at 37°C for 2 h, bound proteins were visualized using the ECL reagent (Pierce) and detected using BioImaging Systems (UVP Inc., Upland, CA, USA). beta-Actin protein was used as a loading control.
Cell Proliferation Assay, Anchorage-Independent Colony Formation Assay, and Drug Resistance Studies
Cells were grown in 96-well plates (1,000 cells/well for proliferation and 4,000 cells/well for drug resistance studies). After incubation with or without reagents, the medium was removed and the cells were treated with 10 μl of 3-(4, 5-dimethylthiazolyl-2)-2, 5-diphenyltetrazolium bromide (MTT) for 2 h at 37°C. Subsequently, 200 μl of solubilization solution (10% SDS) was added and the mixture incubated overnight at 37°C. The solubilized formazan product was spectrophotometrically quantified using a microtiter plate reader from Power Wave XS (BioTek, Winooski, VT, USA) at 570 nm.
The following drugs were studied: 5-FU (1 g/20 ml, TTY Biopharm, Taipei, Taiwan), oxaliplatin (50 mg/10 ml, TTY Biopharm), and SN-38 (Irinotecan hydrochloride, Sigma, St. Louis, MO, USA). Drugs were dissolved and then diluted in the media for the drug resistance assay. The number of viable cells was determined by the MTT test 72 h after treatment with oxaliplatin, SN38, and 5-FU. The exposure times for oxaliplatin, SN38, and 5-FU were 24, 24, and 24 h, respectively.
The anchorage-independent colony formation assay was performed as follows: Each well of a six-well culture dish was coated with 1 ml bottom agar mixture [RPMI, 15% (v/v) fetal bovine serum (FBS), 0.5% (w/v) agar, 1% (v/v) penicillin–streptomycin]. After the bottom layer had solidified, 1 ml of top agar-medium mixture [RPMI or Dulbecco’s modified Eagle medium, 15% (v/v) FBS, 0.3% (w/v) agar, 1% (v/v) penicillin–streptomycin] containing 5,000 cells was added, and the dishes were incubated at 37°C for 2–4 weeks. Plates were stained with 0.5 ml of 0.005% crystal violet for 1 h before colony numbers were determined.17
RNA Extraction and Real-Time RT-PCR
The cellular RNA was extracted from cells using the RNeasy Plus Mini Kit from (Qiagen). Quantitative real-time polymerase chain reaction was arranged using SYBR Green PCR Master Mix (Applied Biosystems) on a 7900 Real-Time PCR System (Applied Biosystems): 50°C for 2 min, 95°C for 10 min, 35 cycles of 95°Cfor 15 s, and 60°C for 60 s. The primer pairs are: CIP2A forward, 5′-ATACTTCAGGACCCACGTTTGAT-3′, CIP2A reverse, 5′-TCTCCAAGTACTAAAGCAGGAAAATCT-3′; b-actin forward, 5′-ATAGCACAGCCTGGATAGCAACGTAC-3′, b-actin reverse, 5′-CACCTTCTACAATGAGCTGCGTGTG-3′. b-Actin was used as the reference gene. The relative levels of gene expression were represented as △C t–C t gene–C t reference, and the fold change of gene expression was calculated by the \( {{2}^{ - }}^{{\Delta \Delta {C_{\text{t}}}}} \) method. Experiments were repeated in triplicate.12
Statistical and Survival Analysis
Correlations between clinicopathological variables and immunopositivity in CIP2A were analyzed using the χ 2 test or Fisher’s exact test. Survival was estimated using the Kaplan–Meier method, and the log-rank test was used to compare survival curves as well as for univariate analysis. The t test was used to compare data from the densitometric analysis of foci numbers. The Cox proportional hazards model was applied for multivariate analysis. Variables with P values <0.05 in the log-rank test were entered in multivariate analysis. A two-sided P value <0.05 was regarded as statistically significant. SPSS software (version 16.00, SPSS, Chicago, IL, USA) was used for all statistical analyses.
Results
Patient Characteristics
A total of 167 patients with colon cancer were included (Table 1). The median age at diagnosis was 69.0 years (range, 30–89), with a male-to-female ratio of 1.93:1. The majority of the histopathological diagnoses were adenocarcinoma (98.8%). The percentage of patients with initial stage I, II, III, and IV disease were 10.2%, 34.1%, 29.9%, and 25.7%, respectively. Most cases were grade 1 or 2 (92.8%). Lymphovascular invasion accounted for 19.8% (n = 33) of patients. Fifty-three specimens (31.7%) were defined as exhibiting strong expression of CIP2A, and 114 specimens (68.3%) were defined as exhibiting weak expression. We also compared 15 paired specimens (tumor n = 15 vs. paired normal colon n = 15). Fourteen of 15 paired specimens exhibited strong expression of CIP2A in the tumors than paired normal colon specimens. Selected IHC images are shown in Fig. 1a–f. The bar plot of all H scores from 15 paired specimens is showed in Fig. 1g.
CIP2A protein expression in colon cancer specimens as detected by immunohistochemistry. a Positive control; normal liver tissue. b Negative control; without anti-CIP2A antibody. c Strong expression of CIP2A. d Weak expression of CIP2A. e Weak expression of CIP2A in the paired normal colon. f Strong expression of CIP2A in the paired tumor. g The bar plot of all immunohistochemical scores (H scores) from 15 paired specimens
CIP2A Is an Independent Prognostic Factor in Patients with Colorectal Cancer
Five-year survival rates of the patients were 100% for those in stage I, 93.0% in stage II, 66.0% in stage III, and 21.0% in stage IV (P < 0.001) (Fig. 2a). This result is similar to those described in previous AJCC reports. In our study, overexpression of CIP2A was associated with significant poor prognosis. The 5-year survival rates of patients with strong and weak CIP2A expression were 43.4% and 78.1%, respectively (P < 0.001; Fig. 2b).
The variables affecting survival were examined using univariate analysis of stage, histology grade, lymphovascular invasion, and CIP2A (Table 2). Among these factors, stage (hazard ratio = 5.162, P < 0.001), lymphovascular invasion (hazard ratio = 3.344, P < 0.001), and CIP2A (hazard ratio = 3.344, P < 0.001) were shown to be significantly predictive of poor prognosis. CIP2A was an independent prognostic factor after controlling for stage and lymphovascular invasion (hazard ratio = 3.378, P < 0.001; Table 2).
CIP2A Is Associated with More Advanced Colon Cancer
CIP2A was shown to be an independent prognostic factor after controlling for other clinical variables. Table 3 shows the association of CIP2A and other potential confounding prognostic factors. The presence of advanced stage status, T4 invasion, high lymph node (LN) involvement (n > 3), and lymphovascular invasion were associated with strong CIP2A expression. However, there were no differences in histological grade and location between the two subgroups.
The effect of CIP2A expression on survival in each stage was further analyzed, and the survival curve is shown in Fig. 3. CIP2A overexpression was a significant predictor of poor prognosis in stage III and IV colon cancer.
Overall survival (OS) of patients with colon cancer in each stage according to CIP2A expression status. CIP2A was a significant predictor of poor prognosis in patients with stage III and IV colon cancer. a Stage I, not shown due to the absence of death events, b stage II, c stage III, and d stage IV
Knockdown of CIP2A Decreases Cell Proliferation and Anchorage-Independent Colony Formation in Colon Cancer Cells
We examined whether knockdown of CIP2A in the colon cancer cell line HT29 decreased the proliferation and anchorage-independent colony formation of colon cancer cells (Fig. 4), as was reported previously.8 A significant reduction was observed in the proliferation rate (Fig. 4b) and anchorage-independent colony formation (Fig. 4c) of HT29 cells transfected with CIP2A siRNA (shCIP2A) than in those transfected with negative control (shLuc).
Knockdown of CIP2A in HT29 cells decreased their proliferation and anchorage-independent colony formation. a Immunoblot analysis of CIP2A protein expression in control (shLuc) and CIP2A knockdown cells (shCIP2A). b Knockdown of CIP2A decreased the proliferation rate. c Knockdown of CIP2A significantly decreased anchorage-independent colony formation
Effect of CIP2A Expression on Drug Resistance in Colon Cancer Cells
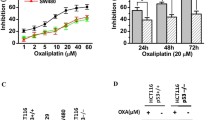
We examined the impact of CIP2A expression on the sensitivity of cancer cells to 5-FU, oxaliplatin, and SN38 (Fig. 5). Caco2 (Fig. 5a) and HT-29 (Fig. 5b) cells showing elevated CIP2A expression were used for knockdown of CIP2A by shRNA, resulting in decreased CIP2A expression. The reduction in CIP2A expression promoted the inhibitory effect of 5-FU, oxaliplatin, and SN38 on the proliferation of colon cancer cell lines (Fig. 5c–h). We also arranged the study about the effect of chemotherapy on CIP2A mRNA expression. The 24- and 48-h CIP2A mRNA expression decreased after exposure to oxaliplatin, 5-FU, and SN38 in both HT29 and Caco2 cell lines. This phenomenon was similar to the previous report that treatment with doxorubicin decreased the CIP2A expression in the HCT116 colon cancer cell lines (Fig. 6).18
Impact of CIP2A on drug resistance in Caco2 and HT-29 cells. Immunoblot analysis of CIP2A protein expression in control (shLuc) and CIP2A knockdown (shCIP2A) in a Caco2 and b HT29 cells. Caco2 and HT29 cells with CIP2A knockdown had significantly weaker resistance to 5-fluorouracil (5-FU), SN38, and oxaliplatin (c–h) (*P < 0.05)
Effect of chemotherapy on CIP2A expression. a HT-29 cells were treated with 5-fluorouracil (5-FU), SN38, and oxaliplatin; the 24- and 48-h CIP2A mRNA expression decreased after treatment with 5-FU, SN38, and oxaliplatin. b Caco2 cells were treated with 5-FU, SN38, and oxaliplatin, the 24- and 48-h CIP2A mRNA expression decreased after treatment with 5-FU, SN38, and oxaliplatin (*P < 0.05)
Discussion
The clinical role of CIP2A in colon cancer has not been reported thus far. After adjusting for stage, histological grade, and lymphovascular involvement, CIP2A was found to be an independent prognostic factor, particularly in stage III and IV colon cancer. Furthermore, CIP2A overexpression was correlated with aggressiveness, such as T4 tumor, LN involvement greater than 3, lymphovascular involvement, and advanced stage of colon cancer. Our results indicate that the CIP2A is an oncoprotein involved in colon cancer. The role of CIP2A in colon cancer appears to be the same as in other cancers such as leukemia, breast, gastric, prostate, lung, ovarian, and head and neck carcinomas.8–12,14–16
To further support our clinical observations, we carried out studies in colon cancer cell lines. Knockdown of CIP2A reduced the proliferation and anchorage-independent colony formation of these cells, similar to previous reports.8 The possible explanations for the above results are as follows: Colon cancer is a human malignancy associated with the microtubule-associated protein kinase/extracellular signal-regulated kinases (MAPK/ERK) pathway.19 Key components of the MAPK/ERK pathway include epidermal growth factor receptor, v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (RAS), v-raf murine sarcoma viral oncogene homolog (RAF), ERK, and MAPK; this pathway may regulate nearly all aspects of tumorigenesis, particularly in colon cancer.20,21 Interestingly, CIP2A can promote RAS-elicited tumor formation in mouse embryo fibroblasts and transform human cells, particularly those with high expression of KRAS.7,8 Moreover, the activated MAPK pathway further elevates CIP2A expression.22 CIP2A is one of most important endogenous inhibitors associated with PP2A, which is the major serine–threonine phosphatase in mammalian cells. Inactivation of PP2A by viral oncoproteins, mutations in specific subunits, or overexpression of endogenous inhibitors such as CIP2A contributes to cell transformation.23 For example, CIP2A inhibits c-Myc-associated PP2A activity and protects c-Myc S62 from dephosphorylation, which leads to cancer cell transformation and proliferation.7,15 Taken together, CIP2A is a very important oncoprotein in colon cancer.
In our cell line studies, we also demonstrated that knockdown of CIP2A reduces resistance to 5-FU, oxaliplatin, and SN38, all of which are important chemotherapeutic agents in the treatment of colon cancer. A similar phenomenon was also observed in a previous study, which found that CIP2A overexpression was associated with doxorubicin resistance.18 One of the possible mechanisms is that BRAF is inhibited by PP2A, which is inhibited by CIP2A.24 BRAF is a significant predictor of poor prognosis in colon cancer, and the BRAF mutation leads to resistance to chemotherapy.7,8,13,20,21 Thus, further elucidation of the molecular function of CIP2A in drug resistance and in the MAPK/ERK pathway is important. The above observations indicate that CIP2A plays an important role in drug resistance with respect to colon cancer chemotherapy.
There are some limitations to our study. Our specimens did not include rectal cancer because the one of the standard treatments for this group is neoadjuvant concurrent chemoradiation therapy (CCRT). Thus, our results could not be applied to rectal cancer after CCRT to investigate the association between CIP2A and radiation efficacy. However, our current findings provide direct evidence that CIP2A is a prognostic factor in colon cancer and indirectly suggest that it plays a role in drug resistance. Additionally, because few death events were observed in stages I and II, more cases should be analyzed to determine the impact of CIP2A on stage I and II colon cancer.
Conclusion
CIP2A was an independent prognostic factor in colon cancer patients. The knockdown of CIP2A reduced proliferation and anchorage-independent colony formation and increased 5-FU, oxaliplatin, and SN38 efficacies in colon cancer cell lines.
References
Naishadham D, Lansdorp-Vogelaar I, Siegel R, Cokkinides V, Jemal A. State disparities in colorectal cancer mortality patterns in the United States. Cancer Epidemiol Biomarkers Prev 2011; 20: 1296-1302
Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, Bets D, Mueser M, Harstrick A, Verslype C, Chau I, Van Cutsem E. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med 2004; 351: 337–345
Saltz LB, Clarke S, Diaz-Rubio E, Scheithauer W, Figer A, Wong R, Koski S, Lichinitser M, Yang TS, Rivera F, Couture F, Sirzen F, Cassidy J. Bevacizumab in combination with oxaliplatin-based chemotherapy as first-line therapy in metastatic colorectal cancer: a randomized phase III study. J Clin Oncol 2008; 26: 2013–2019
Novak R, Zeng Y, Shuga J, Venugopalan G, Fletcher DA, Smith MT, Mathies RA. Single-cell multiplex gene detection and sequencing with microfluidically generated agarose emulsions. Angew Chem Int Ed Engl 2011; 50: 390–395
Hoving JL, de Vet HC, Twisk JW, Deville WL, van der Windt D, Koes BW, Bouter LM. Prognostic factors for neck pain in general practice. Pain 2004; 110: 639–645
Montagnani F, Chiriatti A, Turrisi G, Francini G, Fiorentini G. A systematic review of FOLFOXIRI chemotherapy for the first-line treatment of metastatic colorectal cancer: improved efficacy at the cost of increased toxicity. Colorectal Dis 2011; 13: 846–852
Junttila MR, Westermarck J. Mechanisms of MYC stabilization in human malignancies. Cell Cycle 2008; 7: 592–596
Junttila MR, Puustinen P, Niemela M, Ahola R, Arnold H, Bottzauw T, Ala-aho R, Nielsen C, Ivaska J, Taya Y, Lu SL, Lin S, Chan EK, Wang XJ, Grenman R, Kast J, Kallunki T, Sears R, Kahari VM, Westermarck J. CIP2A inhibits PP2A in human malignancies. Cell 2007; 130: 51–62
Vaarala MH, Vaisanen MR, Ristimaki A. CIP2A expression is increased in prostate cancer. J Exp Clin Cancer Res 2010; 29: 136
Come C, Laine A, Chanrion M, Edgren H, Mattila E, Liu X, Jonkers J, Ivaska J, Isola J, Darbon JM, Kallioniemi O, Thezenas S, Westermarck J. CIP2A is associated with human breast cancer aggressivity. Clin Cancer Res 2009; 15: 5092–5100
Li W, Ge Z, Liu C, Liu Z, Bjorkholm M, Jia J, Xu D. CIP2A is overexpressed in gastric cancer and its depletion leads to impaired clonogenicity, senescence, or differentiation of tumor cells. Clin Cancer Res 2008; 14: 3722–3728
Dong QZ, Wang Y, Dong XJ, Li ZX, Tang ZP, Cui QZ, Wang EH. CIP2A is overexpressed in non-small cell lung cancer and correlates with poor prognosis. Ann Surg Oncol 2010; 18: 857–865.
Chen KF, Liu CY, Lin YC, Yu HC, Liu TH, Hou DR, Chen PJ, Cheng AL. CIP2A mediates effects of bortezomib on phospho-Akt and apoptosis in hepatocellular carcinoma cells. Oncogene 2010; 29: 6257–6266.
Coenen EA, Zwaan CM, Meyer C, Marschalek R, Pieters R, van der Veken LT, Beverloo HB, van den Heuvel-Eibrink MM. KIAA1524: a novel MLL translocation partner in acute myeloid leukemia. Leuk Res 2010; 35: 133–135.
Khanna A, Bockelman C, Hemmes A, Junttila MR, Wiksten JP, Lundin M, Junnila S, Murphy DJ, Evan GI, Haglund C, Westermarck J, Ristimaki A. MYC-dependent regulation and prognostic role of CIP2A in gastric cancer. J Natl Cancer Inst 2009; 101: 793–805
Bockelman C, Lassus H, Hemmes A, Leminen A, Westermarck J, Haglund C, Butzow R, Ristimaki A. Prognostic role of CIP2A expression in serous ovarian cancer. Br J Cancer 2011; 105: 989–995
Yang MH, Chiang WC, Chou TY, Chang SY, Chen PM, Teng SC, Wu KJ. Increased NBS1 expression is a marker of aggressive head and neck cancer and overexpression of NBS1 contributes to transformation. Clin Cancer Res 2006; 12: 507–515
Choi YA, Park JS, Park MY, Oh KS, Lee MS, Lim JS, Kim KI, Kim KY, Kwon J, Yoon do Y, Moon EY, Yang Y. Increase in CIP2A expression is associated with doxorubicin resistance. FEBS Lett 2011; 585: 755–760
Fang JY, Richardson BC. The MAPK signalling pathways and colorectal cancer. Lancet Oncol 2005; 6: 322–327
McCubrey JA, Steelman LS, Abrams SL, Lee JT, Chang F, Bertrand FE, Navolanic PM, Terrian DM, Franklin RA, D'Assoro AB, Salisbury JL, Mazzarino MC, Stivala F, Libra M. Roles of the RAF/MEK/ERK and PI3K/PTEN/AKT pathways in malignant transformation and drug resistance. Adv Enzyme Regul 2006; 46: 249–279
Siena S, Sartore-Bianchi A, Di Nicolantonio F, Balfour J, Bardelli A. Biomarkers predicting clinical outcome of epidermal growth factor receptor-targeted therapy in metastatic colorectal cancer. J Natl Cancer Inst 2009; 101: 1308–1324
Zhao D, Liu Z, Ding J, Li W, Sun Y, Yu H, Zhou Y, Zeng J, Chen C, Jia J. Helicobacter pylori CagA upregulation of CIP2A is dependent on the Src and MEK/ERK pathways. J Med Microbiol 2010; 59: 259–265
Eichhorn PJ, Creyghton MP, Bernards R. Protein phosphatase 2A regulatory subunits and cancer. Biochim Biophys Acta 2009; 1795: 1–15
Dhillon AS, Meikle S, Yazici Z, Eulitz M, Kolch W. Regulation of Raf-1 activation and signalling by dephosphorylation. EMBO J 2002; 21: 64–71
Acknowledgments
The authors wish to thank Dr. Po-Min Chen for his help in discussions regarding the data. This work was supported in part by the Division of Experimental Surgery of the Department of Surgery, Taipei Veterans General Hospital and by the Taipei Veterans General Hospital, Taiwan Clinical Oncology Research Foundation, Department of Health, Taiwan (Center of Excellence for Cancer Research at Taipei Veterans General Hospital, DOH99-TD-C-111-007 and DOH99-TD-C-111-007; Taipei Veterans General Hospital, 100DHA0100416, 99DHA0100446, 101DHA0100661, and 101DHA0100369; CSH-2012-C-030).
Disclosure Statement
None declared.
Author information
Authors and Affiliations
Corresponding author
Additional information
Hsei-Wei Wang and Kuen-Feng Chen contributed equally to the manuscript.
Rights and permissions
About this article
Cite this article
Teng, HW., Yang, SH., Lin, JK. et al. CIP2A Is a Predictor of Poor Prognosis in Colon Cancer. J Gastrointest Surg 16, 1037–1047 (2012). https://doi.org/10.1007/s11605-012-1828-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11605-012-1828-3