Abstract
Objective
The aim of this study is to evaluate the clinical significance of S100A4 mRNA expression in pancreatic cancer.
Materials and Methods
We obtained invasive ductal carcinoma (IDC) cells from ten lesions, intraductal papillary mucinous neoplasm (IPMN) cells from 20 lesions, and normal ductal cells from 20 normal pancreatic tissues by laser microdissection of frozen tissues. S100A4 expression was examined in the microdissected cells and in formalin-fixed paraffin-embedded (FFPE) samples of 87 pancreatic cancers by quantitative reverse transcription-polymerase chain reaction.
Results
IDC cells expressed higher levels of S100A4 than IPMN cells (P = 0.002) and normal ductal cells (P < 0.001), although the difference between IPMN cells and normal ductal cells was not statistically significant (P = 0.070). Analysis of FFPE samples revealed that high S100A4 expression was significantly associated with a shorter overall survival (P = 0.023). In immunohistochemical analysis, the extent of S100A4 mRNA expression was significantly correlated with the expression of S100A4 protein (P = 0.028).
Conclusion
S100A4 could be a marker for malignancy in pancreatic tumors and for poor prognosis in patients with pancreatic cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pancreatic cancer is one of the most lethal tumors and is the fourth leading cause of tumor-related deaths in the industrialized world.1,2 Only 10–20% of patients with pancreatic cancer have a chance of curative resection because most patients are at advanced stages of the disease at the time of diagnosis.3,4 Therefore, early diagnosis of pancreatic cancer is critical to improve survival. On the other hand, many asymptomatic pre-invasive pancreatic neoplasms with cystic lesions have been found as a result of recent advances of diagnostic tools and screening strategy. This poses a dilemma for clinicians because it is often difficult to distinguish between pancreatic cancers and nonhazardous tumors. Intraductal papillary mucinous neoplasm (IPMN), which is recognized as a precursor of pancreatic ductal adenocarcinoma, is representative of such neoplasms. Prognosis is favorable for patients with IPMN without invasion but poor for those with invasion, which accounts for a rate of death of about 30% of patients with IPMNs.5 To determine the nature of pancreatic lesions preoperatively, novel modalities are needed. A promising approach is to measure molecular markers that could classify patients into different risk categories and aid clinicians in choosing suitable treatments for individual patients. To date, p53, transforming growth factor-β, basic fibroblast growth factor,6 Bcl-2,7–9 matrix-metalloproteinases,10 β-catenin/E-cadherin,11 vascular endothelial derived growth factor,12,13 platelet-derived endothelial growth factor,14,15 and human equilibrative nucleoside 116 have been suggested as biomarkers to predict the prognosis of pancreatic cancer patients. However, there are conflicting findings with regard to their validity as prognostic markers,6 and none of the markers described above are used in clinical practice.
S100A4 is a member of the S100 family of calcium-binding proteins, which is characterized by two distinct EF-hand structural motifs.17,18 S100A4 is known to be overexpressed in many solid tumors, including breast carcinoma,19 gastric carcinoma,20 and colorectal adenocarcinoma,21 while S100A4 has historically been referred to as fibroblast-specific protein 1 (FSP1), as a marker of fibroblasts.22 There are also alternative names for S100A4 including mts1, pEL-98, 18A2, p9Ka, CAPL, and calvasculin. S100A4 promotes cell motility and invasion in cancer21,23–25 and induces remodeling of the extracellular matrix,26–29 suggesting that S100A4 is a mediator of tumor metastasis.30 S100A4 has also been reported to be a prognostic marker in a number of human cancers, including esophageal-squamous cancers,31 non-small-cell lung cancers,32 gastric cancers,20 and bladder cancers.33 In pancreatic cancers, it was reported that S100A4 overexpression is associated with poor differentiation34 and poor prognosis.35,36 Recently, Mahon et al.37 showed that S100A4 contributed to chemoresistance and the inhibition of apoptosis in pancreatic cancer.
The aim of this study was to evaluate the clinical significance of S100A4 mRNA expression in pancreatic cancers as a diagnostic and prognostic marker. Using quantitative reverse transcription-polymerase chain reaction (qRT-PCR), we evaluated S100A4 mRNA expression in invasive ductal carcinoma (IDC) cells, nonmalignant IPMN cells, and normal ductal cells of pancreatic tissues obtained by laser microdissection. Moreover, we investigated the association between S100A4 expression and the prognosis of patients with pancreatic cancers using formalin-fixed paraffin-embedded (FFPE) samples.
Materials and Methods
Patients and Pancreatic Tissues
Tissue samples were obtained from primary pancreatic tumors at the time of surgery at Kyushu University Hospital (Fukuoka, Japan) between 1992 and 2007. Normal pancreatic tissues were taken from peripheral tissues away from the tumor or from nonneoplastic pancreas resected due to bile duct disease. The tissue samples were removed as quickly as possible after resection, and a part of each sample was embedded in ornithine carbamyl transferase compound (Sakura, Tokyo, Japan), snap-frozen for analysis by microdissection, and stored at −80°C. The remainder was fixed in formalin and embedded in paraffin for pathological diagnosis. Tissues adjacent to the specimens were evaluated histologically according to the criteria of the World Health Organization.38 Two pathologists were in agreement with regard to the pathological features of all cases, and the diagnoses were confirmed. In IPMNs, main-duct IPMNs or branch-duct IPMNs which were larger than 3 cm in diameter were removed on suspicion of being high-risk lesions. We only used IPMNs diagnosed with nonmalignant cystic tumors, which were confirmed to be intraductal papillary mucinous adenocarcinoma or intraductal papillary mucinous borderline tumor, not intraductal papillary mucinous carcinoma (IPMC), by pathological examination. Overall survival analysis was conducted for 87 patients who underwent pancreatic resection for pancreas cancer (85 ductal adenocarcinomas and two adenosquamous cell carcinomas). The patients comprised 53 men and 34 women with a median age of 65 years (range, 36–86 years). Survival was measured from the time of pancreatic resection, with death as the endpoint. Prognosis was examined in October 2008. The median observation time for overall survival was 16.3 months, ranging from 1 to 108 months. Sixty-four patients died during the follow-up, and the other patients were alive and censored. This study was approved by the Ethics Committee of Kyushu University and conducted according to the Ethical Guidelines for Human Genome/Gene Research enacted by the Japanese Government and the Helsinki Declaration.
RNA Isolation from Microdissected Samples and FFPE Samples
Frozen tissue samples were cut into 8-μm-thick sections. One section was stained with hematoxylin and eosin (H&E) for histological examination, and the diagnosis of target cells was confirmed by the expert pathologist. Target cells (IDC cells from ten lesions; IPMN cells from 20 lesions, excluding IPMCs; and normal ductal epithelial cells from 20 tissues with the histological appearance of normal pancreas) were isolated selectively with a laser-microdissection and pressure catapulting system (P.A.L.M. Microlaser Technologies, Bernried, Germany) in accordance with the manufacturer’s protocol.39 We microdissected 500–1,000 target cells to perform reliable and reproducible measurements of mRNA levels. We obtained 20 μl of RNA per lesion with the concentration of 10–50 ng/μl. The 28S/18S rRNA ratios ranged from 0.5 to 2.5. Total RNA was extracted from microdissected cells by a microdissection technique using a High Pure RNA Isolation Kit (Roche Diagnostics, Mannheim, Germany) and treated with DNase I (Roche Diagnostics) according to the manufacturer’s instructions. The total RNA derived from FFPE samples was isolated using the RNeasy FFPE kit (Qiagen, Tokyo, Japan), as previously described.40 We used FFPE samples from 87 IDC patients with available prognostic data. After a review of representative H&E-stained slides, four to seven sections of 5-μm thickness were obtained from FFPE blocks of pancreatic cancers for macrodissection. Adjacent normal tissues, including normal acinar tissues and adipose tissues, were removed macroscopically using a scalpel. Only the cancerous parts of the sections were used for the isolation of mRNA. The extracted RNA was quantified by reading the absorbance of 260 nm and 280nm (A260/280) with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Rockland, DE, USA). RNA integrity was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Palo Alto, CA, USA).
Quantitative Reverse Transcription-Polymerase Chain Reaction
Quantitative RT-PCR was performed with a Chromo4 Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA) for 40 cycles of 15 s at 94°C and 30 s at 55°C with a QuantiTect SYBR Green Reverse Transcription-PCR kit (Qiagen) according to the manufacturer’s instructions.
We designed specific primers for S100A4 (forward, 5′-atcgccatgatgtgtaacga-3′; reverse, 5′-cccaaccacatcagaggagt-3′) and β-actin (forward, 5′-aaatctggcaccacaccttc-3′; reverse, 5′-ggggtgttgaaggtctcaaa-3′) using primer 3 and performed BLAST searches to confirm primer specificity. The PCR product sizes of these primers are small (S100A4, 85 base pairs (bp); β-actin, 139 bp, respectively), which allowed accurate and sensitive qRT-PCR despite the fragmented RNA extracted from FFPE tissue specimens.41,42 The S100A4 and β-actin expression levels were calculated for all cases using a standard curve constructed with total RNA from SUIT-2, a pancreatic cancer cell line. One microliter of RNA was used in qRT-PCR despite the concentration of RNA. S100A4 mRNA expression levels were normalized using β-actin as an internal control and expressed as the ratio of expression of S100A4 mRNA to that of β-actin mRNA. All samples were run in triplicate. The accuracy and integrity of the PCR products were confirmed with an Agilent 2100 Bioanalyzer (Agilent Technologies Inc.).
Immunohistochemical Procedures and Evaluation
Sections (4 μm thick) were cut from paraffin-embedded tissues, deparaffinized in xylene, and rehydrated through a graded ethanol series. Endogenous peroxidase activity was blocked by incubation with 3% hydrogen peroxide in methanol for 30 min. Antigen retrieval was achieved by autoclaving the sections in citrate buffer at pH 6.0. A Histofine SAB-PO(R) kit (Nichirei, Tokyo, Japan) was used for immunohistochemical labeling. Each section was exposed to 10% non-immunized goat serum for 10 min to block nonspecific antibody binding, followed by incubation with a rabbit polyclonal anti-S100A4 antibody (NeoMarkers, Fremont, CA, USA; 1:100 dilution) at 4°C overnight. The sections were then sequentially incubated with a biotinylated anti-rabbit immunoglobulin solution for 20 min followed by peroxidase-labeled streptavidin for 20 min. The reaction products were visualized using 3,3′-diaminobenzidine as a chromogen, followed by nuclear counterstaining with hematoxylin. Cells were considered positively immunostained when nuclei and cytoplasm were stained. The distribution of stained S100A4 was evaluated as the percentage of stained cells, which was scored as 0, <5%; 1, 5–25%; 2, 26–50%; and 3, >51%, and as staining intensity, which was scored as 0, no staining; 1, weak; 2, moderate; and 3, strong. When the multiplication product of the two scores was greater than 2, S100A4 was considered positively stained. In the immunohistochemical staining, we performed additional staining without primary antibodies in parallel to confirm that no staining was seen. All slides were evaluated independently by two investigators (NI and KN) without any knowledge of the background of each case.
Statistical Analysis
Data were analyzed using the Kruskal–Wallis test if comparisons involved three groups and the Mann–Whitney U test if comparisons involved two groups. S100A4 expression was split into high- and low-level groups using recursive descent partition analysis, as described by Hoffmann et al.43 Survival curves were constructed with the Kaplan–Meier product-limit method and compared by log-rank test. The statistical significance was defined as a P value <0.05. All statistical analyses were performed with JMP 7.01 software (SAS Institute, Cary, NC, USA).
Results
Quantitative Analysis of S100A4 mRNA Expression in IDC, Nonmalignant IPMN, and Normal Ductal Epithelial Cells
We measured the S100A4 mRNA expression levels in IDC cells, nonmalignant IPMN cells, and normal ductal epithelial cells by qRT-PCR after laser-microdissection from frozen sections to determine whether S100A4 is differentially expressed between pancreatic cancer cells and cells from nonmalignant tumors or normal ductal cells. S100A4 mRNA expression was significantly higher in IDC cells than in IPMN (P = 0.002) and normal ductal cells (P < 0.001), as shown in Fig. 1. IPMNs tended to express higher levels of S100A4 compared with normal ductal cells, although the difference did not reach statistical significance (P = 0.070).
qRT-PCR analysis of S100A4 mRNA expression in IDC, nonmalignant IPMNs, and normal ductal epithelial cells. IDC cells expressed higher levels of S100A4 compared with IPMNs (P = 0.002) and normal ductal cells (P < 0.001). IPMNs tended to express higher levels of S100A4 compared with normal ductal cells, although the difference did not reach statistical significance (P = 0.070). The expression of S100A4 was normalized to that of β-actin. The scale is logarithmic.
S100A4 mRNA Expression Was Correlated with Prognosis of Patients with Pancreatic Cancers
To investigate the correlation between S100A4 expression and prognosis in patients with pancreatic ductal carcinomas, we isolated total RNA from FFPE samples from 87 patients with pancreatic cancers and measured the levels of S100A4 expression. After normalizing S100A4 mRNA expression to β-actin expression, we obtained two groups with high versus low S100A4 expression (cutoff value, 20.5). The high- and low-expression S100A4 groups comprised 21 and 66 cases, respectively. High S100A4 expression was significantly associated with a shorter overall survival (P = 0.023, Fig. 2). The median survival time of the patients with high and low S100A4 expression was 12 and 23 months, respectively.
S100A4 mRNA Expression Was Correlated with the Expression of S100A4 Protein
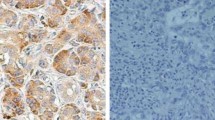
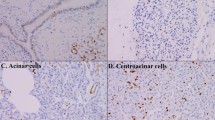
S100A4 was immunoreactive in cytoplasm and nuclei of cancer cells (Fig. 3a, b). Cancer cells were highly stained with S100A4 compared with that in fibroblast in the stroma, even though S100A4 has been called “fibroblast-specific protein 1”. The level of S100A4 mRNA expression was significantly correlated with the expression of S100A4 protein, as shown in Fig. 3 (P = 0.028).
Discussion
We measured the S100A4 mRNA expression levels in IDC cells, nonmalignant IPMN cells, and normal ductal cells with qRT-PCR and found that IDC cells expressed the highest levels of S100A4 among the cell types analyzed in the present study. To our knowledge, this is the first study to evaluate the correlation of S100A4 expression in pancreatic cancers and IPMN. We have previously reported that IDC cells expressed higher levels of S100A2, another S100 family member, than premalignant cells and that IPMN cells with high-grade atypia expressed higher levels of S100A2 than IPMN with low-grade atypia and normal ductal cells.40 In the present study, a trend for a stepwise increase in S100A4 mRNA expression from normal ductal cells to IDC cells was shown, suggesting that S100A4 may also be involved in pancreatic carcinogenesis, similar to S100A2.
We quantitatively measured S100A4 mRNA expression by qRT-PCR using FFPE samples of surgically resected pancreatic cancers. We found that high S100A4 expression was significantly associated with a shorter overall survival, suggesting that S100A4 mRNA could be a prognostic marker in pancreatic cancers. This finding supports a report of an immunohistochemical analysis of 62 surgical cases with pancreatic cancers, in which overexpression of S100A4 was significantly correlated with tumor size, tumor–node–metastases stage, and poor prognosis.35 These consistent results also indicate that quantitative analysis of S100A4 mRNA by qRT-PCR could be a reliable modality to contribute to the prediction of the prognosis of patients with pancreatic cancer. In fact, S100A4 mRNA expression was correlated with the expression of S100A4 protein. The measurement of S100A4 mRNA expression by qRT-PCR offers a high level of objectivity and quantitative performance compared with immunohistochemical examination. Additionally, the evaluation of S100A4 mRNA expression of the tumor could be also performed from tiny tissue samples, resulting in a clinically informative technique.
Cytological specimens obtained by endoscopic ultrasound-guided fine-needle aspiration (EUS-FNA) and by endoscopic retrograde cholangiopancreatography (ERCP) have played an important role in the diagnosis of pancreatic cancer. However, cytological interpretation of clinical specimens obtained by these techniques is often difficult because samples are scant and bloody.44–47 Therefore, molecular markers are needed to aid the diagnosis in indeterminate cytological samples.48 The present study revealed that S100A4 mRNA expression level was significantly higher in cancer cells than in nonmalignant IPMN cells or normal ductal cells. The merit of the analysis used in the present study is that we can sensitively and accurately measure the mRNA expression levels using gene-specific primers that generate short PCR products, even for tiny tissue samples or fragmented RNA obtained by EUS-FNA or ERCP. The measurement of S100A4 mRNA for clinical samples could give clinicians important information, including tumor nature and the patient’s prognosis, because S100A4 expression was correlated with prognosis, although further studies are required to confirm this clinical application.
In summary, S100A4 mRNA was expressed at higher levels in pancreatic cancer cells than in cells derived from nonmalignant tumors or nonneoplastic epithelium. The level of S100A4 expression was significantly correlated with the prognosis of patients with pancreatic cancer. Thus, S100A4 could be a marker of malignancy in pancreatic tumors and for poor prognosis in patients with pancreatic cancer.
References
Eskelinen MJ, Haglund UH. Prognosis of human pancreatic adenocarcinoma: review of clinical and histopathological variables and possible uses of new molecular methods. Eur J Surg 1999;165:292–306.
Yeo TP, Hruban RH, Leach SD, Wilentz RE, Sohn TA, Kern SE, Iacobuzio-Donahue CA, Maitra A, Goggins M, Canto MI, Abrams RA, Laheru D et al. Pancreatic cancer. Curr Probl Cancer 2002;26:176–275.
Sener SF, Fremgen A, Menck HR, Winchester DP. Pancreatic cancer: a report of treatment and survival trends for 100,313 patients diagnosed from 1985–1995, using the National Cancer Database. J Am Coll Surg 1999;189:1–7.
American gastroenterological association medical position statement: epidemiology, diagnosis, and treatment of pancreatic ductal adenocarcinoma. Gastroenterology 1999;117:1463–1484.
Adsay NV, Conlon KC, Zee SY, Brennan MF, Klimstra DS. Intraductal papillary-mucinous neoplasms of the pancreas: an analysis of in situ and invasive carcinomas in 28 patients. Cancer 2002;94:62–77.
Garcea G, Neal CP, Pattenden CJ, Steward WP, Berry DP. Molecular prognostic markers in pancreatic cancer: a systematic review. Eur J Cancer 2005;41:2213–2236.
Sinicrope FA, Evans DB, Leach SD, Cleary KR, Fenoglio CJ, Lee JJ, Abbruzzese JL. bcl-2 and p53 expression in resectable pancreatic adenocarcinomas: association with clinical outcome. Clin Cancer Res 1996;2:2015–2022.
Friess H, Lu Z, Andren-Sandberg A, Berberat P, Zimmermann A, Adler G, Schmid R, Buchler MW. Moderate activation of the apoptosis inhibitor bcl-xL worsens the prognosis in pancreatic cancer. Ann Surg 1998;228:780–787.
Nio Y, Dong M, Iguchi C, Yamasawa K, Toga T, Itakura M, Tamura K. Expression of Bcl-2 and p53 protein in resectable invasive ductal carcinoma of the pancreas: effects on clinical outcome and efficacy of adjuvant chemotherapy. J Surg Oncol 2001;76:188–196.
Yamamoto H, Itoh F, Iku S, Adachi Y, Fukushima H, Sasaki S, Mukaiya M, Hirata K, Imai K. Expression of matrix metalloproteinases and tissue inhibitors of metalloproteinases in human pancreatic adenocarcinomas: clinicopathologic and prognostic significance of matrilysin expression. J Clin Oncol 2001;19:1118–1127.
Joo YE, Rew JS, Park CS, Kim SJ. Expression of E-cadherin, alpha- and beta-catenins in patients with pancreatic adenocarcinoma. Pancreatology 2002;2:129–137.
Seo Y, Baba H, Fukuda T, Takashima M, Sugimachi K. High expression of vascular endothelial growth factor is associated with liver metastasis and a poor prognosis for patients with ductal pancreatic adenocarcinoma. Cancer 2000;88:2239–2245.
Niedergethmann M, Hildenbrand R, Wostbrock B, Hartel M, Sturm JW, Richter A, Post S. High expression of vascular endothelial growth factor predicts early recurrence and poor prognosis after curative resection for ductal adenocarcinoma of the pancreas. Pancreas 2002;25:122–129.
Ikeda N, Adachi M, Taki T, Huang C, Hashida H, Takabayashi A, Sho M, Nakajima Y, Kanehiro H, Hisanaga M, Nakano H, Miyake M. Prognostic significance of angiogenesis in human pancreatic cancer. Br J Cancer 1999;79:1553–1563.
Kuwahara K, Sasaki T, Kuwada Y, Murakami M, Yamasaki S, Chayama K. Expressions of angiogenic factors in pancreatic ductal carcinoma: a correlative study with clinicopathologic parameters and patient survival. Pancreas 2003;26:344–349.
Giovannetti E, Del Tacca M, Mey V, Funel N, Nannizzi S, Ricci S, Orlandini C, Boggi U, Campani D, Del Chiaro M, Iannopollo M, Bevilacqua G et al. Transcription analysis of human equilibrative nucleoside transporter-1 predicts survival in pancreas cancer patients treated with gemcitabine. Cancer Res 2006;66:3928–3935.
Donato R. S100: a multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int J Biochem Cell Biol 2001;33:637–668.
Tarabykina S, Kriajevska M, Scott DJ, Hill TJ, Lafitte D, Derrick PJ, Dodson GG, Lukanidin E, Bronstein I. Heterocomplex formation between metastasis-related protein S100A4 (Mts1) and S100A1 as revealed by the yeast two-hybrid system. FEBS Lett 2000;475:187–191.
Rudland PS, Platt-Higgins A, Renshaw C, West CR, Winstanley JH, Robertson L, Barraclough R. Prognostic significance of the metastasis-inducing protein S100A4 (p9Ka) in human breast cancer. Cancer Res 2000;60:1595–1603.
Yonemura Y, Endou Y, Kimura K, Fushida S, Bandou E, Taniguchi K, Kinoshita K, Ninomiya I, Sugiyama K, Heizmann CW, Schafer BW, Sasaki T. Inverse expression of S100A4 and E-cadherin is associated with metastatic potential in gastric cancer. Clin Cancer Res 2000;6:4234–4242.
Takenaga K, Nakamura Y, Sakiyama S. Expression of antisense RNA to S100A4 gene encoding an S100-related calcium-binding protein suppresses metastatic potential of high-metastatic Lewis lung carcinoma cells. Oncogene 1997;14:331–337.
Strutz F, Okada H, Lo CW, Danoff T, Carone RL, Tomaszewski JE, Neilson EG. Identification and characterization of a fibroblast marker: FSP1. J Cell Biol 1995;130:393–405.
Grigorian M, Ambartsumian N, Lykkesfeldt AE, Bastholm L, Elling F, Georgiev G, Lukanidin E. Effect of mts1 (S100A4) expression on the progression of human breast cancer cells. Int J Cancer 1996;67:831–841.
Lloyd BH, Platt-Higgins A, Rudland PS, Barraclough R. Human S100A4 (p9Ka) induces the metastatic phenotype upon benign tumour cells. Oncogene 1998;17:465–473.
Maelandsmo GM, Hovig E, Skrede M, Engebraaten O, Florenes VA, Myklebost O, Grigorian M, Lukanidin E, Scanlon KJ, Fodstad O. Reversal of the in vivo metastatic phenotype of human tumor cells by an anti-CAPL (mts1) ribozyme. Cancer Res 1996;56:5490–5498.
Bjornland K, Winberg JO, Odegaard OT, Hovig E, Loennechen T, Aasen AO, Fodstad O, Maelandsmo GM. S100A4 involvement in metastasis: deregulation of matrix metalloproteinases and tissue inhibitors of matrix metalloproteinases in osteosarcoma cells transfected with an anti-S100A4 ribozyme. Cancer Res 1999;59:4702–4708.
Miyamori H, Hasegawa K, Kim KR, Sato H. Expression of metastasis-associated mts1 gene is co-induced with membrane type-1 matrix metalloproteinase (MT1-MMP) during oncogenic transformation and tubular formation of Madin Darby canine kidney (MDCK) epithelial cells. Clin Exp Metastasis 2000;18:51–56.
Schmidt-Hansen B, Klingelhofer J, Grum-Schwensen B, Christensen A, Andresen S, Kruse C, Hansen T, Ambartsumian N, Lukanidin E, Grigorian M. Functional significance of metastasis-inducing S100A4(Mts1) in tumor-stroma interplay. J Biol Chem 2004;279:24498–24504.
Semov A, Moreno MJ, Onichtchenko A, Abulrob A, Ball M, Ekiel I, Pietrzynski G, Stanimirovic D, Alakhov V. Metastasis-associated protein S100A4 induces angiogenesis through interaction with Annexin II and accelerated plasmin formation. J Biol Chem 2005;280:20833–20841.
arrett SC, Varney KM, Weber DJ, Bresnick AR. S100A4, a mediator of metastasis. J Biol Chem 2006;281:677–680.
Ninomiya I, Ohta T, Fushida S, Endo Y, Hashimoto T, Yagi M, Fujimura T, Nishimura G, Tani T, Shimizu K, Yonemura Y, Heizmann CW et al. Increased expression of S100A4 and its prognostic significance in esophageal squamous cell carcinoma. Int J Oncol 2001;18:715–720.
Kimura K, Endo Y, Yonemura Y, Heizmann CW, Schafer BW, Watanabe Y, Sasaki T. Clinical significance of S100A4 and E-cadherin-related adhesion molecules in non-small cell lung cancer. Int J Oncol 2000;16:1125–1131.
Davies BR, O’Donnell M, Durkan GC, Rudland PS, Barraclough R, Neal DE, Mellon JK. Expression of S100A4 protein is associated with metastasis and reduced survival in human bladder cancer. J Pathol 2002;196:292–299.
Rosty C, Ueki T, Argani P, Jansen M, Yeo CJ, Cameron JL, Hruban RH, Goggins M. Overexpression of S100A4 in pancreatic ductal adenocarcinomas is associated with poor differentiation and DNA hypomethylation. Am J Pathol 2002;160:45–50.
Ai KX, Lu LY, Huang XY, Chen W, Zhang HZ. Prognostic significance of S100A4 and vascular endothelial growth factor expression in pancreatic cancer. World J Gastroenterol 2008;14:1931–1935.
Oida Y, Yamazaki H, Tobita K, Mukai M, Ohtani Y, Miyazaki N, Abe Y, Imaizumi T, Makuuchi H, Ueyama Y, Nakamura M. Increased S100A4 expression combined with decreased E-cadherin expression predicts a poor outcome of patients with pancreatic cancer. Oncol Rep 2006;16:457–463.
Mahon PC, Baril P, Bhakta V, Chelala C, Caulee K, Harada T, Lemoine NR. S100A4 contributes to the suppression of BNIP3 expression, chemoresistance, and inhibition of apoptosis in pancreatic cancer. Cancer Res 2007;67:6786–6795.
Hamilton SR and Aaltonen LA. (eds): World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of the Digestive System. Lyon: IARC Press, 2000.
Tachikawa T, Irie T. A new molecular biology approach in morphology: basic method and application of laser microdissection. Med Electron Microsc 2004;37:82–88.
Ohuchida K, Mizumoto K, Miyasaka Y, Yu J, Cui L, Yamaguchi H, Toma H, Takahata S, Sato N, Nagai E, Yamaguchi K, Tsuneyoshi M et al. Over-expression of S100A2 in pancreatic cancer correlates with progression and poor prognosis. J Pathol 2007;213:275–282.
Godfrey TE, Kim SH, Chavira M, Ruff DW, Warren RS, Gray JW, Jensen RH. Quantitative mRNA expression analysis from formalin-fixed, paraffin-embedded tissues using 5′ nuclease quantitative reverse transcription-polymerase chain reaction. J Mol Diagnostics 2000;2:84–91.
Abrahamsen HN, Steiniche T, Nexo E, Hamilton-Dutoit SJ, Sorensen BS. Towards quantitative mRNA analysis in paraffin-embedded tissues using real-time reverse transcriptase-polymerase chain reaction: a methodological study on lymph nodes from melanoma patients. J Mol Diagnostics 2003;5:34–41.
Hoffmann AC, Mori R, Vallbohmer D, Brabender J, Klein E, Drebber U, Baldus SE, Cooc J, Azuma M, Metzger R, Hoelscher AH, Danenberg KD et al. High expression of HIF1a is a predictor of clinical outcome in patients with pancreatic ductal adenocarcinomas and correlated to PDGFA, VEGF, and bFGF. Neoplasia 2008;10:674–679.
McCarthy DM, Maitra A, Argani P, Rader AE, Faigel DO, Van Heek NT, Hruban RH, Wilentz RE. Novel markers of pancreatic adenocarcinoma in fine-needle aspiration: mesothelin and prostate stem cell antigen labeling increases accuracy in cytologically borderline cases. Appl Immunohistochem Mol Morphol 2003;11:238–243.
Khalid A, Pal R, Sasatomi E, Swalsky P, Slivka A, Whitcomb D, Finkelstein S. Use of microsatellite marker loss of heterozygosity in accurate diagnosis of pancreaticobiliary malignancy from brush cytology samples. Gut 2004;53:1860–1865.
Govil H, Reddy V, Kluskens L, Treaba D, Massarani-Wafai R, Selvaggi S, Gattuso P. Brush cytology of the biliary tract: retrospective study of 278 cases with histopathologic correlation. Diagn Cytopathol 2002;26:273–277.
Selvaggi SM. Biliary brushing cytology. Cytopathology 2004;15:74–79.
Buchholz M, Kestler H, Gress TM. Differential diagnosis of pancreatic tumors by molecular analysis of clinical specimens. Pancreatology 2008;8:551–557.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supported in part by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan and grants from the Takeda Science Foundation, Pancreas Research Foundation of Japan, and Nakajima Foundation.
Rights and permissions
About this article
Cite this article
Ikenaga, N., Ohuchida, K., Mizumoto, K. et al. S100A4 mRNA is a Diagnostic and Prognostic Marker in Pancreatic Carcinoma. J Gastrointest Surg 13, 1852–1858 (2009). https://doi.org/10.1007/s11605-009-0978-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11605-009-0978-4