Abstract
Tuber brumale (winter truffle) is one of the most controversial true truffles, not only in regard to its ecological and economical role but also its taxonomy. Multilocus phylogenetic analyses have revealed that specimens identified earlier as T. brumale belong to two species. These species were deemed cryptic right away, because preliminary morphological measurements did not show any phenotypical differences. In this study, we measured the morphology of 119 T. brumale agg. specimens, identified by DNA-based phylogenetic tools. We found several continuous morphological characters which show strong statistical differences between the two species, albeit not without overlap. Using a combination of these characters, we show that efficient separation of the two species is possible. We describe T. cryptobrumale sp. nov. and present the environmental demands and the potential area reconstruction of both species. We argue that non-representative sampling is a major culprit in most failures to detect both the existence of morphologically similar species and their morphological differences. Our findings illustrate the benefits of integrative taxonomy: the use of a combination of molecular, morphological and ecological tools.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A fundamental endeavour of biology is the classification of living things, delimiting species in the process. Recently, molecular phylogenetics has overwritten the traditional morphology-based classification—quite remarkably in many cases. For example, phylogenetic species recognition (PSR), focusing on nucleotide divergence, caused a ca. 300% increase in the number of fungal species (Agapow et al. 2004; see also GCPSR in Taylor et al. 2000). Most of these changes were attributable to the previous practice of using morphological characters for species delimitation, and the arbitrariness in their selection. Accordingly, these characters were often unable to explain evolution. Morphology-based taxonomy’s intrinsic difficulty with reconstructing evolution has other causes as well, namely convergent evolution, pleomorphy (morphologically different forms in a life cycle) and the cryptic nature of some of the species (Hibbett et al. 2011).
Species are cryptic if morphological studies are unable to distinguish their evolutionary lineages identified by molecular methods (Lundholm et al. 2012). This can be quite common in recently split species, since genetic changes can be detected earlier by molecular studies than through the study of phenotypic differences (Cai et al. 2011). When molecular studies lead to the partitioning of a previously described species, the resulting two or more species will at first be considered cryptic. If morphological differences are found between similar species, but only in the wake of molecular studies, those species are instead regarded as pseudocryptic (Sáez et al. 2003). So when the discovery of cryptic species spurs rigorous morphological re-investigation, resulting in the recognition of morphological differences between the species, cryptic species will turn pseudocryptic. The separating characters found a posteriori are often subtle and hardly appreciable, or they require specialists (e.g. the use of electron microscopy, Sáez et al. 2003; Orive et al. 2013, or shape-recognition software, Arribas et al. 2013).
In taxa with simple architecture, a high rate of species can be cryptic/pseudocryptic (Medina et al. 2012). Simulations show that, in algal groups, species from character-poor lineages are more difficult to distinguish from one another than species from character-rich lineages (Verbruggen et al. 2014). Most species of Kingdom Fungi are relatively character-poor and have a simple structure (Balasundaram et al. 2015), which is even more true for hypogeous fungi (Reynolds 2011). This has led to their increasingly neglected morphological investigation (which is time-consuming and labour-intensive) and, consequently, to the absence of, or their erroneous, species descriptions.
The economical importance of edible ectomycorrhizal mushrooms, especially the gastronomically cherished true truffles (Tuber Micheli ex Wiggers) is undoubted (Hall and Zambonelli 2012). Due to their ability to form ectomycorrhizae with a variety of tree species, truffles are also important ecologically. The predicted species richness of the genus has been estimated to be around 180–230 (Bonito et al. 2010a), though some species are yet to be found and others await taxonomic revision. Since 2010, with the growth of truffle research, more than 47 new Tuber species have been described worldwide, particularly from un- or understudied regions such as China (e.g. Deng et al. 2012; Fan et al. 2012, 2013, 2015). Also, in well-studied regions, the existence of several phylogenetic lineages have been detected by molecular phylogenetic methods. Examples are found in the groups of Excavatum (Bonito et al. 2010a; Urban et al. 2013), Puberulum (Bonuso et al. 2009), Rufum (Bonito et al. 2010a; Merényi 2014), Gibborum (Bonito et al. 2010b), and Melanosporum (Chen et al. 2011; Merényi et al. 2014). The reason that these species and potential species stayed hidden from taxonomists’ eyes for so long might be that they have related taxa without (conspicuous) phenotypic differences—with which they form (pseudo-)cryptic species complexes (Bonuso et al. 2009; Chen et al. 2011; Merényi et al. 2014).
The present study deals with the recently reexamined Tuber brumale agg. (Merényi et al. 2014), which contains a sibling species pair. Tuber brumale Vittad. was first described by Vittadini in 1831 prope Milano, Italy (Vittadini 1831). However, a survey of T. brumale and T. melanosporum collections attributed to Vittadini himself in six major European herbaria showed variations in morphology or even misidentifications, and none of these could be confirmed as Vittadini types (Trappe, unpublished data). Later in the nineteenth century, a new species was described with the name of Tuber moschatum Bull., which was similar to T. brumale but varied in its odour and colour (Ceruti et al. 2003). Taxonomic confusion surrounding these species persisted for centuries (Ceruti et al. 2003; Merényi et al. 2016). In our previous multiloci phylogenetic work (Merényi et al. 2014), we genotyped 141 specimens using two different methods: a phylogenetic analysis based on the nuclear internal transcribed spacer (ITS), the large subunit (LSU) regions of the ribosomal DNA repeat, and the variable segment of the protein kinase C (PKC) nucleotide sequences, and the ITS restriction fragment length polymorphism (RFLP). Based on broad geographical sampling, it showed that T. brumale should indeed be treated as a species aggregate, containing two phylogenetically distinct species (clades A and B), but these do not correspond to the earlier division of T. brumale and T. moschatum (or T. brumale f. moschatum). And although we found a clear molecular division between the members of the aggregate, our preliminary morphological measurements did not permit a similarly suitable partitioning of the aggregate and the description of the new species. Thus, we suggested treating them temporarily as a cryptic species complex. We argued that the species originally described by Vittadini had to be clade A—thus we refer to it as T. brumale sensu stricto (s.str.). Also, we have shown the existence of two significantly segregated ITS haplogroups (AI and AII) within T. brumale s.str. (Merényi et al. 2014).
In this article, we present a novel, more detailed morphological examination and reconsider the aggregate’s cryptic status. We describe clade B as a new species, Tuber cryptobrumale sp. nov. We find that the aggregate’s quantitative characters, though they overlap among the two species to various degrees, are different enough to enable a morphology-based identification. We perform Linear Discriminant Analyses (LDA) to find how best to separate the two species, first based on morphological characters, then soil characters, and, finally, coenological data. We characterize aspects of the two species’ ecology, including their potential area reconstruction, and the relationship of each species and its habitat (soil and plant coenology). We discuss the pitfalls of non-representative sampling, both in terms of the species’ geographic distribution and the characters’ variability. We recommend the use of multiple characters for species delimitation, especially in the case of morphologically similar species.
Materials and methods
Specimens and survey data
In the last 25 years, members of the First Hungarian Truffling Association (FHTA) collected numerous hypogeous fungus specimens, which were subsequently organized into a mycotheca (Bratek et al. 2013). Related botanical and pedological data were arranged into a database (Merényi et al. 2010). The former consists of information on the species identity and abundance of all plants in a 100-m2 (∼10 × 10 m) area around the truffle beds, and inferred from this their community type (coenosis; classified according to Borhidi 2003). The latter is based on soil samples collected in the immediate vicinity of the ascocarps, from the surface (layer A), analysed by the laboratories of the Hungarian Central Agricultural Office. Also, through collaborative efforts, the FHTA mycotheca and database has been supplemented with specimens from around Europe.
Molecular identification
We also have information on the genetic identity of the above specimens (see “introduction”). As part of this study, we characterized an additional 28 specimens by ITS-RFLP using MboI and HinfI (Fermentas) restriction enzymes or specific primers. On this basis, we can classify all examined specimens into 3 groups: clade A haplogroups AI and AII, and clade B. Also, we sequenced the ITS-LSU region of two clade B specimens (for a detailed method, see Merényi et al. 2014). In addition, we developed a multiplex PCR assay for the separation of the two clades (A and B). Two forward primers (TbrA and TbrB) were designed for the T. brumale agg. specific insertion of ITS1 region (about insertion, see Merényi et al. 2016). The target of TbrA (5′- CGTTAGACTGTATCGGTGTC-3′) is in the middle of the ∼300-bp insertion (from the 258th bp of ITS1) and planned for Clade A (both Haplogroups I and II), while the target of TbrB (5′- GCTACCTTGTACTGCCTGCC-3′) is in the 3′ end of the insertion (from the 134th bp of ITS1) and planned for clade B. The choice of primers were checked using the online OligoAnalyser 3.1 (IDT, Coralville, IA, USA, www.IDTDNA.com) tool, to exclude primer-primer and self interactions. The multiplex PCR method was tested with gradient PCR reactions, involving at least five specimens from each clade, which were all sequenced earlier.
Morphological description and measurements
We had intended to base our description of T. brumale on a holotype or lectotype, but we have not succeeded in finding a collection so designated. Vittadini sent portions of many of his collections to colleagues at many European or British universities, and at his death in 1865 his collections remaining at the University of Milan were dispersed to other herbaria. With generous contributions from many of these herbaria, Mattirolo (1907) later reassembled much of Vittadini’s original herbarium at what is now the Herbarium of the Department of Plant Biology (TO) University of Turin, Italy. One of us (JMT) spent a year’s sabbatical there in 1967–1969 and wrote detailed descriptions of all collections attributed to Vittadini. The Vittadini collection packets of T. brumale were variously labelled “collezione Vittadini” or “autoptico Vittadini”, respectively “collection of Vittadini” or “seen by Vittadini”. No other information was recorded. The updated description of T. brumale by Ceruti et al. (2003) is said to be based on a “sintipo” as defined by Kirk et al. (2003): "...syntype... one of several elements cited by an author when originally proposing a name but where no holotype was selected. " Vittadini did not select a holotype, the concept of types being formulated decades later. It is not clear whether any of the collections attributed to Vittadini were listed in his original description or were collected later, because his collections have no data other the attribution to him. Hence, if a type is to be designated, it would need to be a neotype. Trappe examined all of the T. brumale packets labelled ‘Vittadini “. Some contained missidentified species or mixes of T. brumale and other specimens. For our purposes, he picked one that had T. brumale that matched Vittadini’s description precisely, and that he described macro- and microscopically in detail to serve as our standard to identify our recent collections and provide DNA sequences. Its Vittadini packet was labelled “Collezione Vittadini del Museo de Pavia”, i.e., a collection from his own herbarium that had been donated to Pavia but that Pavia had later sent to Mattirolo for deposit at TO.
We measured the morphology of 119 specimens. Most of them originated from the Carpathian Basin (87) where the three genetic groups coexist. A total of 18 specimens originated from northern and western Europe (Spain, France, Italy, Poland), and 14 specimens came from southern and eastern Europe (Romania, Greece, Turkey) (Online Resource 1). We chose a total of 16 quantitative morphological characters, among them both measured and derived ones. We measured the length and width of the ascospores (SPL and SPW, respectively), as these are generally used characters for truffles. We also made more labour-intensive measurements, resulting in characters such as the diameter of peridial warts (Warts), the average size of peridial cells (ASPC), and the height of spines on the spores (Spine). From the size and the number of the spores, we derived characters such as the estimated volume of the spore (Vol), and the frequency of asci with 1, 2, …, 8 number of spores (R1–R8). Detailed descriptions of the morphological measurements are included in Online Resource 2. We succeeded in exploring the value of all 16 characters for 83 specimens (n = 51 for clade A and n = 32 for clade B; see Online Resource 3).
Multicharacter species delimitation
We wanted to determine whether the two clades differed significantly in their morphological characters. Since the distribution of some of the characters was non-normal (determined by the Shapiro–Wilks test), we checked the equality of the two clades’ values with the nonparametric Brunner–Munzel test (Online Resource 3).
We have seen that the characters show significant overlap between the two species, therefore we tested if character combinations can separate them. Finding the best discriminating combination requires the elimination of the redundant and poorly explaining characters. For this task, we used variable selection: stepwise Linear Discriminant Analysis (sLDA). This method starts with the single best discriminating character, and adds new characters to it one by one, as long as they significantly increase the explained variance. We assigned weights to the resulting subset of characters using LDA. LDA finds the linear combination of given variables which displays the largest variance between the groups relative to the intragroup variance. The resulting combination can be used as a linear classifier, enabling in our case classification based on mophological and soil characters.
We calculated several linear classifiers, differing in the features they consider as input (morphology only, soil only, and both) and the groups they were chosen to predict in their output (clade A and B vs. haplogroups AI, AII, and clade B). For the morphology-based classification, we used 16 characters of 83 specimens [haplogroups AI (n = 29) and AII (n = 22), and clade B (n = 32)]. For classifications considering soil data, we used 39 specimens, as only they had all the required measurements.
The best linear classifiers are not always flawless: a morphology- and/or soil-based prediction of specimens’ genetic clade identity may not be possible without errors. Internal validation measures the extent of misclassification within the training dataset, the very data used for constructing the classifier. This “internal error” informs us of the relative utility of the different linear classifiers, and thus helps us assess the importance of measuring the different characters.
External validation is based on novel data, and it quantifies the expected predictive power of a classifier. To illustrate the potential (absolute) utility of classifying T. brumale agg. species in the absence of genetic data, we externally validated a linear classifier having a small “internal error”. For this purpose, we collected a dozen more T. brumale agg. specimens, and measured three of their characters (to be used by the selected linear classifier) and their genetic clade.
For LDA and sLDA calculations, we used the statistical software R (v.3.0.2; R core team 2016) packages MASS (Venables and Ripley 2002) and klaR (Weihs et al. 2005), respectively.
Ecological analyses
We compared the surrounding vegetation, the properties of the soil, and the fruiting periods of the two species. The vegetation survey had a sample size of 59 (36 of clade A, 23 of clade B), while the pedological analysis was based on 80 sites (and 84 soil samples: 54 of clade A, 30 of clade B). Non-metric multidimensional scaling analysis (NMDS) was used to detect differences between the species. The use of Bray–Curtis distances (1000 iterations and 3 reduced dimensions) resulted in a convenient representation of the multidimensional space (stress values between 0.12 and 0.17). Soils, herbs, and woody plants were analysed separately. In the case of the pedological data, some pairs of variables were highly correlated (Pearson correlation, R 2 > 0.5), thus we excluded 3 characters from the NMDS: lime-dissolving carbonic acid, zinc and sulfate. The two species’ fruiting periods were recorded at a resolution of months (n = 46 for clade A, n = 28 for clade B), and were compared with the Kolmogorov–Smirnov test.
After the results of the above-mentioned analysis, it proved interesting to compare the soil copper content of T. brumale agg. with other Tuber species nests. We consulted the FHTA’s database concerning the copper content spectrum of 12 Tuber species (or species complexes), and found data on 975 soil samples (n ≥ 18 per species). We checked the equality of their variances with Bartlett’s test, and used the nonparametric Neményi post hoc test (PMCMR package; Pohlert 2014) to compare the copper content of the 12 species.
Predictive distribution modelling
In order to predict the potential distribution area of T. brumale agg. in Europe and the Middle East, we used the dedicated software MaxEnt (v.3.3.3 k; Phillips et al. 2006). MaxEnt combines presence-only data with spatially explicit environmental variables to predict species’ environmental envelope (realized ecological niche), and, based on that, their potential distribution (Phillips et al. 2006). We chose this software because it is the most capable of producing useful results even with as few as 5 occurrences (Hernandez et al. 2006).
We compiled 225 instances of T. brumale agg. location data (GPS coordinates), including all specimens from this study and Merényi et al. 2014. Clade A’s 182 datapoints occupied 124 grid cells of ∼4 × 4 km (2.5 arcmin), while clade B’s 43 datapoints occupied 32 grid cells. The grid cell data were randomly partitioned into training and test sets, used to build the distribution model and to validate it, respectively. The training set had 93 datapoints for clade A and 24 datapoints for clade B, while the test set had 31 datapoints for clade A and 8 datapoints for clade B.
The MaxEnt model’s present conditions data was based on the elevation and 19 bioclimatic layers, downloaded from the WorldClim database (www.worldclim.org, 13 Apr. 2015; Hijmans et al. 2005). These layers were converted to ESRI ASCII raster format with Quantum GIS (QGIS Wroclaw 1.7.3 software; QGIS Development Team 2009). MaxEnt selected the most relevant layers in determining species distributions. We used the following parameters: 25% random test, 20 replicates, max. 5000 iterations, ‘subsample’ type replicate, and a ‘logistic’ modelling output; all other settings were left as default. The performance of the species distribution model was evaluated by the Area Under the Curve (AUC) statistic of the Receiver Operating Characteristic (ROC) plots. Jackknife resampling was used to assess the importance of the variables. The final potential species distribution map was made with QGIS.
Results
Our 119 independent T. brumale agg. specimens, originating from all over Europe, show great similarity across geographic regions (Online Resource 1). Twenty eight of these 119 specimens were newly examined, as part of this study, by use of the ITS-RFLP method. We could not detect any differences between clades A and B based on macromorphology and odour descriptions. Their spore size and ornamentation did not differ significantly (Online Resource 3). After adding further characters, we found a few traits which showed strong significant difference between the two species, e.g. the size of peridial cells, wart size and dry weight (Online Resource 3). From the number of spores per asci (1–8) we derived additional characters: their mode (“dominant number”), and their ratios (relative to the sum total). The dominant number of spores per asci is 4 in both species; also, the ratio of four-spored asci shows no significant difference. In contrast, the ratio of three- and five-spored asci differed significantly between the two species: clade A has a higher ratio of five-spored asci and a lower ratio of three-spored asci than clade B. Accordingly, a strong correlation occured between the ratios of three- and five-spored asci (Pearson correlation: R 2 = −0.806, p < 0.001).
Multicharacter species delimitation
None of the 16 independent or derived characters could separate the two species without overlap. Variable selection indicated that the most useful characters for distinguishing the two species are ASPC, Warts and R5. We arrived at the following delimitation formula, capable of efficiently delimiting the two species:
If D < 0 for a given specimen, it is expected to be of clade A, otherwise it is of clade B. This formula misclassifies only four of the training set’s 83 specimens: its “internal error” is 4.8%.
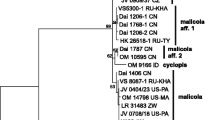
We also performed variable selection to delimit, beyond the two clades, the two haplogroups of clade A (AI and AII; Fig. 1). The resulting most useful characters turned out to be the same as above (ASPC, Warts and R5); however, they could not effectively separate the haplogroups (43.1% misclassification).
Linear discriminant analyses (LDA) of clades based on morphological data. Point colours represent the three phylogenetic ITS lineages. Red triangle Tuber brumale s.str. (clade A haplogrop I.); blue square Tuber brumale s.str. (clade A haplogrop II); green circle Tuber cryptobrumale sp. nov. (clade B)
For a combined analysis of the morphology and the soil, we selected those specimens for measurement of all their respective characters (39 specimens). On this set of specimens, a delimitation using only morphological characters resulted in 5.1% misclassification (selected variables: ASPC, R3); use of solely pedological characters led to 20.5% misclassification (selected variables: Cu, pH(KCl)). Using both sets of characters, we found a mere 2.6% “internal error” with the character combination ASPC, Cu and R3. As yet, this is the most effective separation of the two species.
The morphology-based classification rule (formula 1) was also validated externally. A total of 13 newly measured specimens could all be correctly classified on the level of clades (Online Resource 4), corresponding to no “external error” (100% efficiency). It is interesting to note, however, in this validation dataset, that none of the 3 characters alone would meaningfully delimit the clades. Half of the character values are in the overlapping ranges of their respective characters. And, apart from a single exception, all specimens have such “overlapping” character values (i.e. at least one out of three).
Ecological descriptions of T. brumale aggr.
Of the 12 soil variables measured, 7 differed significantly between the two species (p < 0.05; Online Resource 5). The most remarkable difference was found in the copper content of the soil: clade A had 3.65 ± 1.75 mg/kg while clade B had 6.71 ± 3.89 mg/kg. Copper levels similar to the soil of clade B were found in T. borchii Vittad., T. macrosporum Vittad. and T. rapaeodorum Tul. and C. Tul. All other examined Tuber species had a significantly lower copper level (Online Resource 6).
Clade A and B were also found to be separable using NMDS plots (Online Resource 7). The most separating dimension was composed of phosphate (P2O5, R 2 = 0.36, p < 0.001), potassium (K2O, R 2 = 0.60, p < 0.001) and copper (Cu, R 2 = 0.60, p < 0.001). Also, zinc is highly correlated with potassium (Pearson correlation R 2 = 0.61, p < 0.01).
Most of the explored habitats of both species had brown forest soil (> 70%; different subtypes), but T. brumale s.str. was also collected from rendzina (21%), while T. cryptobrumale sp.nov. was occasionally found in meadow soils (28%).
Based on the coenological surveys, both species were often associated with mountainous oak–hornbeam forests (Carici pilosae–Carpinetum; A: 50%; B: 33%). Clade B was found similarly often in lowland oak–hornbeam forests (Circaeo–Carpinetum; 33%). Of the ectomycorrhizae-forming woody plants, Carpinus betulus L. (A: 72%; B: 87%), Quercus robur L. (A: 31%; B: 48%), and Quercus cerris L. (A: 39%; B: 26%) were found to be most frequently present in the vicinity of T. brumale agg.
According to the NMDS (Online Resources 8–9), trees and herbs showed no substantial difference in their association with the two species. However, weak effects were attributable to Acer campestre L. seedlings (R 2 = 0.32, p < 0.01), Fraxinus angustifolia subsp. pannonica Soó & Simon seedlings (R 2 = 0.3, p < 0.01), Poa pratensis agg. (R 2 = 0.40, p < 0.001), Rubus caesius L. (R 2 = 0.34, p < 0.001) and Tilia cordata Mill. seedlings (R 2 = 0.30, p < 0.1).
The distribution of the harvesting periods have shown no significant difference between the two species (p = 0.9). More than 90% of the collections of both species dated from October to February.
Distribution modelling
The potential area reconstructions of both T. brumale s.str. and T. cryptobrumale sp. nov. (Online Resource 10) were found to be accurate by cross-validation, as indicated by their high AUC scores (0.935 ± 0.022 and 0.957 ± 0.031, respectively).
T. brumale s.str.’s potential area was based mostly on temperature seasonality (BIO4: 33.4%), max. temperature of the warmest month (BIO5: 17.9%), and precipitation of the driest quarter (BIO17: 13.2%). Based on jackknife testing, temperature seasonality (BIO4) provides the most useful information by itself. T. cryptobrumale sp. nov.’s potential area was based mostly on the mean temperature of the driest quarter (BIO9: 37,5%), precipitation of the driest month (BIO14: 25,5%) and isothermality (BIO3: 12,1%). BIO9 was the most useful variable according to jackknife tests.
T. cryptobrumale’s distribution, both realized and potential, is restricted to a considerably smaller region than that of T. brumale s.str. The former’s realized distribution is in eastern–central Europe, and mainly in the Carpathian Basin. Its predicted potential area is in a much wider range (from northeastern Spain to the Caucasus), but is confined to small patches.
Taxonomy
Tuber brumale Vittad., Monographia Tuberacearum: 37, Tab, fig. VI (1831) [MB#228019].
≡ Oogaster brumalis (Vittad.) Corda (1854), Icones fungorum hucusque cognitorum 6: 72, 73, t. 17:127
= Tuber cibarium Sibthorp (1794) Flora Oxoniensis 398
= Tuber hiemalbum Chatin (1869) La Truffe 46
= Tuber renatii Bonnet (1884) Rev. Myc. 6(23): 137
= Tuber moschatum Bonnet (1869), La Truffe 44 [non T. moschatum Bull., Herb Fr. (Paris): tab. 479 (1790), nec. T. moschatum Sowerby, Col. fig. Engl. Fung. Mushr. (London) 3: tab. 426 (1803), = Melanogaster variegatus (Vittad.) Tul. & C. Tul., Fungi hypog.: 92 (1851)]
= Tuber montanum Chatin (1891), Bull. Soc. Bot. France 38: 58
≡ Tuber brumale Vittadini subsp. montanum (Chatin) E. Fischer (1897) Ascomyceten: Tuberaceen und Hemiasceen 44
Description: Ascomata hypogeous, globose to flattened-subglobose, often with a basal depression or cavity. 6–60 mm in diam.; dried weight 0.06–7.05 g; warted, black (36) (RBGE, 1969), sometimes reddish at the base of warts. Warts 1.8 ± 0.4 mm (0.9–3.3 mm) (this is mean and standard deviation value pair and min–max range in parentheses) across, angular, usually flattened but sometimes pyramidal, usually 5- to 6-sided, often pointed or depressed at the apex, radially fissured. Warts sometimes separated by fissures in the surface of the ascomata, easily detaching. Gleba firm, solid, whitish at first, becoming grey-brown, and most frequently sepia (26) and drab (33) (RBGE, 1969), dark-brown at maturity, marbled with a few, infrequently or widely spaced, white veins. Odour pleasant, slightly chocolate-like or rarely strong, acetone-like. Peridium 324 ± 70 μm (210–475 μm) thick in total, the external layer 80 ± 29.5 μm (33.7–126.3 μm) thick, of cells as a hyaline or pale yellow pseudoparenchyma. The uppermost cells (layer: 89 ± 42.8 μm) are highly pigmented, orange-brown and often have extremely thick cell walls. The size of the largest peridial cells (ASPC) are 15.65 ± 2.62 μm (10.93–23.4 μm). The internal peridial layer is 156.7 ± 53.8 μm (50.5–244.1 μm) thick, this prosenchyma is intricately interwoven with hyaline hyphae (3–6 μm). Asci globose to subglobose or broadly ellipsoid, stemless, 50–70 × 60–80 μm. The distribution of spore numbers per asci is 1: 3.1 ± 3.1%; 2: 9.3 ± 8.6%; 3: 23.7 ± 9.3%; 4: 37.0 ± 10.7%; 5: 22.6 ± 11.1%; 6: 4.0 ± 6.4%;7: 0.2 ± 0.7%; and 8: 0.0 ± 0.2%. Thus, the 3- to 5-spored asci are the most common. Ascospores ellipsoid Q = 1.62 (1.44–1.82), sometimes plum seed-shaped, yellow to light golden brown, 28.1 × 17.4 μm ((25.3–33.7) × (15.7–19.1) μm), in four-spored asci excluding ornamentation. Spore volume is 4487 ± 541 μm3 (3392–6469 μm3). Spores are ornamented with slender flexuous spines, spicule height is 2.32 ± 0.49 μm (0.63–3.73 μm). In Melzer’s reagent: immature asci orange, spores orange-brown, peridium deep orange brown, other tissue yellowish. In ammonium hydroxide, spores could became greenish in a low rate of asci.
Distribution and habitat: The fruiting period is mainly in late autumn-winter (October–January, occasionally August–March). Ascomata can be found under a variety of potential host plants (eg. Carpinus betulus, Quercus robur, Quercus cerris, Quercus petraea, Corylus avellana, Tilia cordata and Fagus sylvatica), mostly in Carici pilosae–Carpinetum phytocoenoses. Nearly neutral pH (pHwater = 6.79 ± 0.51, pHKCl = 6.56 ± 0.49) humus-rich (6.04 ± 1.47%) heavy soil (‘sticky point according to Arany’ is 61.5 ± 9.9). Occurs in the nonmountaineous areas of Europe.
Conservation status: T. brumale is common in the Mediterranean and continental regions of Europe. In addition, its existence was detected with environmental DNA sampling from outside Europe (e.g. Iran and New Zealand).
Additional specimens examined: See Online Resource 11.
Notes: T. brumale and its sister taxon, T. cryptobrumale, share notable morphological similarities in their colour, shape, ascomata size, and spore size. Their separating features are: the size of the largest peridial cells (ASPC), the average size of warts (Warts), and the ratio of five-spored asci (R5). The formula D = 0.23 × ASPC − 1.07 × Warts − 3.64 × R5–2.06 efficiently (95.2%) delimits the two species. A negative D value is indicative of T. brumale. T. brumale also differs from T. cryptobrumale in some unique fixed alleles on LSU and ITS loci, based on the alignments of loci deposited in TreeBASE (S18611). T. brumale unique fixed alleles are: ITS positions: 11(T), 18(C), 38(C), 39(T), 41(C), 44(T), 106(G), 111(C), 115(G), 120(A), 121(G), 123(C), 149(T), 158(A), 176(C), 196(C), 201(G), 205(A), 208(T), 213(T), 216(T), 229(C), 242(A), 251(A), 258(A), 263(A), 266(G), 269(C), 275(A), 283(G), 285(T), 286(G), 290(T), 291(G), 296(A), 297(A), 309(T), 317(A), 326(T), 329(C), 338(T), 340(T), 341(T), 346(C), 350(A), 351(A), 364(T), 420(G), 435(G), 444(T), 470(A), 637(T), 666(T), 680(G), 688(T), 689(T), 690(A), 693(A), 694(C), 701(T), 705(G), 712(T), 713(C), 718(A), 727(C), 728(A), 732(G), 748(A), 779(G), 793(G), 794(T), 802(A), 804(A); LSU positions: 971(T), 972(A), 975(C), 1026(A), 1042(T), 1084(G), 1105(C), 1300(T), 1362(A), 1368(A), 1405(T), 1407(T), 1415(T), 1433(C).
Tuber cryptobrumale Z. Merényi, T. Varga & Z. Bratek, sp. nov. – Holotype: HUNGARY, Bükk mountains, Felsőtárkány (48°0.091′N, 20°26.517′E), 290 m asl, 09 December 2012, Leg: Zs. Merényi, É. Bordás, I. Merényi ZB4690A, BP107922 (BP!). GenBank: KU203777 MycoBank: MB 815399; Fig. 2
Tuber cryptobrumale sp. nov. a Ascomata of the holotype (BP107922). Bar 1 cm. b Ascospores of the holotype (BP107922) Bar 25 μm. c Cross-section of the peridium. Below the pigmented layer, there lies the ectal excipulum with pseudoparenchymatous big roundish, isodiametric cells, and beneath that, the inner excipulum of textura intricata from paratype ZB4705H, Bar 50 μm
Diagnosis: Tuber cryptobrumale morphologically resembles T. brumale, but differs in the size of the largest peridial cells (ASPC) and the average size of warts (Warts). ASPC of T. cryptobrumale is larger, 24.0 ± 4.98 μm (in the range of 16.6–38.73 μm) [vs. 15.65 ± 2.62 μm (10.93–23.4)]; Warts of T. cryptobrumale are smaller 1.3 ± 0.3 mm (0.9–2.0 mm) [vs. 1.8 ± 0.4 mm (0.9–3.3 mm)]. In addition, the ratios of three- and five-spored asci (R3 and R5, respectively) are different in the two species (31.4 ± 7.7 and 13.2 ± 6.8% in T. cryptobrumale vs. 23.7 ± 9.3 and 22.6% ± 11.1% in T. brumale, respectively). In addition, the two species differ in some unique fixed alleles on LSU and ITS loci, based on the alignments of loci deposited in TreeBASE (S18611). T. cryptobrumale’s unique fixed alleles are: ITS positions: 11(C), 18(T), 38(T), 39(C), 41(A), 44(C), 106(A), 111(T), 115(A), 120(G), 121(A), 123(T), 149(C), 158(G), 163(C), 176(T), 196(T), 201(A), 205(C), 208(A), 213(A), 216(G), 229(T), 242(T), 251(G), 258(G), 263(G), 266(T), 269(T), 275(G), 283(A), 285(C), 286(T), 290(G), 291(A), 296(gap), 297(C), 309(C), 317(G), 326(C), 329(T), 338(G), 340(C), 341(A), 346(T), 350(G), 351(T), 364(C), 420(C), 435(C), 444(C), 470(T), 637(C), 666(C), 680(A), 688(gap), 689(gap), 690(gap), 693(G), 694(A), 701(C), 705(A), 712(C), 713(G), 718(G), 727(T), 728(G), 732(A), 748(G), 779(A), 793(C), 794(C), 802(G), 804(T); LSU positions: 971(C), 972(T), 975(G), 1026(G), 1042(C), 1084(A), 1105(T), 1300(C), 1362(G), 1368(G), 1405(C), 1407(C), 1415(C), 1433(T).
Etymology: crypto (ancient greek for hidden) refers to the pseudocryptic nature of this species and brumale refers to its high similarity to its sibling species, T. brumale.
Description: Ascomata hypogeous globose to flattened subglobose, kidney-like, almost always with a basal depression or cavity. 5–27 mm in diam.; dry weight 0.06–1.62 g; warted, black (36) (RBGE, 1969), sometimes reddish at the base of warts. Warts 1.3 ± 0.3 mm (0.9–2.0 mm) across, angular, much flattened, usually 5–6-sided, often depressed at the apex, radially fissured. Warts sometimes separate from each other, forming fissures in the surface of the ascomata, also warts easily detaching. Gleba firmly solid, whitish at first, becoming grey-brown, and most frequently sepia (26), cigar-brown (16) and drab (33) (RBGE, 1969), dark-brown at maturity, marbled with a few, infrequently or widely spaced, white veins. Odour similar to T. brumale, pleasant, slightly chocolate-like or rarely strong, acetone-like. Peridium 381 ± 103 μm (244–522 μm) thick in total, with an external layer of 92 ± 26.3 μm (51.5–134.7 μm), composed of cells arranged as a hyaline or pale yellow pseudoparenchyma. The uppermost cells (layer: 108 ± 46.9 μm) are highly pigmented and often have extremely thick cell walls. The size of the largest peridial cells (ASPC) are 24.0 ± 4.98 μm (16.6–38.73 μm). The internal peridial layer is 180.7 ± 49.5 μm (101–261 μm) thick, intricately interwoven with the hyaline hyphae. Asci globose to subglobose, ellipsoid or irregular. The distribution of spore numbers per asci is 1: 4.0 ± 2.3%; 2: 11.7 ± 5.4%; 3: 31.4 ± 7.7%; 4: 38.2 ± 5.7%; 5: 13.2 ± 6.8%; 6: 1.3 ± 2.0%; 7: 0.0 ± 0.2%; and 8: 0.0 ± 0.0%. Thus, the 3- and the 4-spored asci are the most common. Ascospores ellipsoid Q = 1.57 (1.49–1.67), sometimes plum seed-shaped, yellow to brown, 27.6 × 17.5 μm [(25.3–30.5) × (16.1–18.9) μm], in four-spored asci excluding ornamentation. Spore volume is 4452 ± 587 μm3 (3598–5598 μm3). Spores are spiny, spicule height is 3.51 ± 0.51 μm (2.72–4.76 μm). In ammonium hydroxide, spores often became greenish in 5–40% of asci.
Distribution and habitat: The fruiting period is mainly in late autumn-winter (October–January, occasionally August–February). Ascomata can be found under a variety of potential host plants (e.g. Carpinus betulus, Quercus robur, Q. cerris, Q. petraea, Corylus avellana, Tilia cordata and Fagus sylvatica), mostly in Carici pilosae–Carpinetum and Circaeo–Carpinetum phytocoenoses. Nearly neutral pH (pHwater = 6.79 ± 0.57, pHKCl = 6.16 ± 0.71) humus-rich (6.85 ± 2.01%) heavy soil (‘sticky point according to Arany’ is 6.,8 ± 9.5). Occurs in the nonmountaineous areas of the Carpathian Basin; also, a single specimen has been found in Istria.
Conservation status: In the FHTA’s mycotheca, approximately 30% of the specimens assigned to T. brumale s.l. belong to T. cryptobrumale. T. brumale agg. is common in the Mediterranean and continental regions of Europe, therefore T. cryptobrumale does not seem rare. According to our current knowledge, its distribution is limited to the Carpathian Basin and its immediate vicinity. It is most probably a locally common species.
Additional specimens examined: See Online Resource 11.
Notes: T. cryptobrumale and its sister taxon, T. brumale, share notable morphological similarities in their colour, shape, ascomata size, and spore size. Their separating features are: the size of the largest peridial cells (ASPC), the average size of warts (Warts), and the ratio of five-spored asci (R5). The formula D = 0.23 × ASPC − 1.07 × Warts − 3.64 × R5–2.06 efficiently (95.2%) delimits the two species. A positive D value is indicative of T. cryptobrumale. Our 83 specimens of the two species had D values between −2.6958 and 5.5162. The 4 erroneously classified specimens had near zero values of D (−0.5221 ≤ D ≤ 0.6430; in this range, we also had 7 correctly classified specimens).
Testing of species-specific primers
According to our trials (e.g. gradient PCR,) the following PCR protocol can be recommended: DreamTaq Green Buffer (Fermentas) (20 mM MgCl2, 1.0 μl), dNTPmix (Fermentas) (2 mM, 1.0 μl), primers TbrA, TbrB and ITS4 (White et al. 1990) (0.01 mM, 0.2 μl), Milli-Q water (2.35 μl), Dream Taq polymerase (Fermentas) (5 unit/μl, 0.05 μl) and template DNA dissolved in Milli-Q water (5.0 μl). Thermocycling was carried out under the following conditions: 94 °C for 4.5 min, 30 cycles of 94 °C for 20 s, 56 °C for 30 s and 72 °C (elongation) for 40 s, and finally a 72 °C for 3 min. Our designed primers proved to be effective in multiplex PCR conditions. Primers with the above-mentioned conditions are not able to amplify product in other Tuber species (tested with T. excavatum, T. macrosporum and T. rufum). In the case of T. brumale s.str. (clade A), amplified PCR product (owing to TbrA primer) was 627 bp, while from T. cryptobrumale (clade B) the PCR product was 750 bp. This 123-bp difference is easily distinguishable in agarose gel electrophoresis (Online Resource 12).
Discussion
A new species in the genus Tuber
For decades, it was common knowledge that T. brumale and T. moschatum were distinct species pairs, but both Gandeboeuf et al. (1994, 1997) and our molecular results (Merényi et al. 2014) found no basis for such differentiation. A recent, PSR-based species delimitation of T. brumale agg. found no morphological difference, indicative of a cryptic species complex (Merényi et al. 2014). To verify T. brumale agg.’s cryptic nature, we analysed more than 100 specimens for this article. We found all examined characters to be overlapping, but still identified a (linear) combination of characters capable of efficient species delimitation; it is based on exoperidial cell size (ASPC), wart diameter (Warts) and the ratio of five-spored asci (R5). Used as a classifier, this formula (1) correctly identified the clade of 95.2% of the specimens, demonstrating that there is fundamental morphological difference between the two monophyletic lineages. These lineages shall thus be treated as being pseudocryptic rather than cryptic. The suggested species names are T. brumale s.str. (clade A) and T. cryptobrumale sp. nov. (clade B). Concerning the two haplogroups of T. brumale s.str., we found that none of the characters could support their efficient separation. This is in accordance with our earlier hypothesis that these haplogroups form a single species (Merényi et al. 2014).
The ecological study of T. brumale and T. cryptobrumale focuses on the question whether or not the two species share the same niche. Their climatic demands are similar. The flora in the habitat of the two species are almost identical. Moreover, in some forests, the two species were found to coexist ( e.g. KF551031–KF551034; see Merényi et al. 2014). However, this might only be due to their host-generalism—which has been proven by independent observations (Merényi et al. 2016). The two species have the same harvesting period, which might affect the range of their consumers and vectors (see below). But the two species’ habitats show significant differences in their soil characteristics. The main difference is in the copper content of the soil, which is higher for T. cryptobrumale than for most other Tuber species. This can either mean that T. brumale and T. cryptobrumale do not share the same niche or that the mycelia of T. cryptobrumale (and similar species) accumulate copper—this has been hypothesised earlier in the case of T. brumale s.l. (Bratek 1991). The high level of copper in their fruiting bodies supports this hypothesis (Granetti et al. 2005; Orczán et al. 2012). But the two species also differ in their preference of genetic soil types, and the radical transformation of soil types is certainly beyond the capabilities of fungi.
The harvesting period of the Melanosporum group, including T. brumale agg. (also called winter truffle), is uncommon: it lasts from November to March (Riousset et al. 2001; Hall et al. 2007). This is in sharp contrast with the majority of hypogeous fungi which have a summer to early winter harvesting period. Having a harvesting period unlike most concurrent species must provide certain benefits: their fruiting bodies have a better chance of being consumed, and thus the spores contained within are helped in being spread.
Contrary to intuition, a winter harvesting period does not indicate a greater temperature tolerance of a species (i.e. its whole life cycle). Winter truffles, similarly to T. melanosporum Vittad., avoid northern Europe (Ławrynowicz 1992; Ławrynowicz et al. 2008; Stobbe et al. 2012), as confirmed by potential area reconstruction (Online Resource 10). They are also unlikely to be found in high mountains (the Pyrenees, Alps or Carpathian Mountains).
The reconstructed potential areas of T. brumale s.str. and T. cryptobrumale show similar patterns of environmental preference in the Carpathian Basin, from where they have both been collected, implying the two species’ similar climatic demands. The difference in the extent of the observed and potential distributions of T. cryptobrumale suggest that historical circumstances (climatic changes or past barriers) have contributed to its present, limited range. It has already been hypothesised that in the last glacial the Carpathian Basin and the Balkans, and the Iberian or Italian peninsulas, provided refugia for different winter truffle species and haplogroups (Merényi et al. 2014). The Balkans might also have been the refugia of a recently described T. brumale agg. relative, T. petrophilum Milenković, P. Jovan., Grebenc, Ivančević & Marković.
The area reconstruction of T. brumale aggr. also shows favourable conditions on the eastern shore of the Black Sea and the Caucasus, where as yet no winter truffles have been observed. However, T. brumale has been detected even farther to the east, in Iran, by environmental sampling (Bahram et al. 2012). Also, other truffle species have been reported from Armenia (Badalyan et al. 2005). It would be helpful if these localities received more attention from the truffle research community, as they may hold the key to the past interactions of the truffle mycota of Europe and Asia.
Problems and bias in the course of species delimitation
Sampling
This study showcases the advantages of broad geographical sampling and a large sample size. These features have enabled the discovery of a new species, and, hopefully, the reliable exploration of the aggregate’s genetic diversity. Until the 1990s, T. brumale agg. has only been sampled in western Europe and thus could not reveal the existence of T. cryptobrumale. This warns us that, if molecular sampling does not cover a presumed species’ complete distribution, morphologically similar (cryptic/pseudocryptic) species can evade discovery, no matter how thorough the respective studies are in other respects.
Extensive sampling coverage does not just expose genetic diversity but also uncovers morphological variability. According to our literature search in peer-reviewed journals, 75.5% of the species descriptions in the genus Tuber were based on fewer than five specimens (Online Resource 13). Moreover, 14.3% of the reviewed 49 species were described without any molecular examinations (between 2007 and 2016). In the case of T. brumale agg., preliminary measurements on only 5–10 specimens (and a focus on classical traits, i.e. macromorphology and spore features) did not show any morphological differences between the two species, suggesting that they were cryptic (Merényi et al. 2014). By increasing the sample size to n > 30 per clades (and including non-traditional characters), we succeeded in uncovering a significant difference between the two species—even if not without character overlap.
Characters
Among both epi- and hypogeous mushrooms, the most frequently examined identification characters are related to spores, e.g. their shape, diameter or ornamentation; this is also true for the genus Tuber (e.g. Montecchi and Sarasini 2000; Chen et al. 2011). But spore characters do not play a significant role in the differentiation of the winter truffle species, with the notable exception of a derived character: the distribution of the number of spores per asci. Separation benefitted from the use of peridial features (wart size and exoperidial cell size)—differences in these may be due to selection on the peridium, which acts as a barrier protecting the spore-bearing structures from their underground habitat. Researchers of other Tuber species have also found peridial characters to be useful identifiers (Chen et al. 2005; Halász et al. 2005; Chen and Liu 2007; Bonito et al. 2010b).
The earlier erroneous description of T. moschatum demonstrates the remarkable phenotypic variability (both in colour and odour) of T. brumale s.str. haplogroup I. This might have been due to the non-representative sampling of high-variability continuous morphological characters (often the symptom of quantitative genetic traits and/or environmental effects). Non-representative sampling can blur the difference between a single wide distribution and several narrow ones, potentially leading to the discription of non-existent species (Verbruggen et al. 2005b) or deeming two species morphologically identical, thus cryptic. Several studies caution against underestimating the variability of characters between closely related species (Verbruggen et al. 2005a, b; Chen et al. 2011, Szurdoki et al. 2014). When characters covary with the environment or the geographic location, it is even more difficult to use them for species delimitation and identification (Verbruggen 2014).
Multicharacter analyses
When trying to delimit two or more species, it is sufficient to find a single character whose distributions (for the given species) do not overlap. In the absence of such characters, multivariate analyses may offer a solution (for a general overview, see Podani 2000; James 1990), e.g. principle component analysis (cf. Rivera et al. 2011; Schwarzfeld and Sperling 2014), canonical variate analysis (cf. Florio et al. 2012; Barrett and Freudenstein 2011), non-metric multidimensional scaling (cf. Jiménez-Pérez and Lorea-Hernández 2009), and linear discriminant analysis (cf. Lumley et al. 2010; Szurdoki et al. 2014). These tools can integrate data coming from morphological, ecological and other measurements—as such, they are instruments of integrative taxonomy.
We chose LDA, and it provided us with a linear classifier in the form of the weighted sum of a small subset of our characters. This classifier, specific to our purposes, can identify whether a given winter truffle specimen belongs to T. brumale s.str. or to T. cryptobrumale. This approach, however, is probabilistic, and thus cannot offer absolute certainty. But for those able to tolerate a mere 95% reliability, this classifier can prove useful in practice. And even if some of the characters are inconvenient for species identification (e.g. soil variables), their role in species delimitation—the real focus of integrative taxonomy (Pante et al. 2015)—is undeniable.
The advantages of integrative taxonomy are particularly evident in the case of apparently cryptic species: it is most often in these cases that only a combination of diverse characters will prove suitable for species delimitation. The cross-validation of different methods is also recommended when delimiting morphological similar species (Jörger and Schrödl 2013).
References
Agapow PM, Bininda-Emonds OR, Crandall KA, Gittleman JL, Mace GM, Marshall JC, Purvis A (2004) The impact of species concept on biodiversity studies. Q Rev Biol 79:161–179
Arribas P, Andújar C, Sánchez-Fernández D, Abellán P, Millán A (2013) Integrative taxonomy and conservation of cryptic beetles in the Mediterranean region (Hydrophilidae). Zool Scr 42:182–200. doi:10.1111/zsc.12000
Badalyan SM, Hovsepyan RA, Iotti M, Zambonelli A (2005) On the presence of truffles in Armenia. Flora Mediterr 15:683–692
Bahram M, Põlme S, Kõljalg U, Zarre S, Tedersoo L (2012) Regional and local patterns of ectomycorrhizal fungal diversity and community structure along an altitudinal gradient in the Hyrcanian forests of northern Iran. New Phytol 193:465–473. doi:10.1111/j.1469-8137.2011.03927.x
Balasundaram SV, Engh IB, Skrede I, Kauserud H (2015) How many DNA markers are needed to reveal cryptic fungal species? Fungal Biol 9:940–945. doi:10.1016/j.funbio.2015.07.006
Barrett CF, Freudenstein JV (2011) An integrative approach to delimiting species in a rare but widespread mycoheterotrophic orchid. Mol Ecol 20:2771–2786. doi:10.1111/j.1365-294X.2011.05124.x
Bonito GM, Gryganskyi AP, Trappe JM, Vilgalys R (2010a) A global meta-analysis of Tuber ITS rDNA sequences: species diversity, host associations and long-distance dispersal. Mol Ecol 19:4994–5008. doi:10.1111/j.1365-294X.2010.04855.x
Bonito G, Trappe JM, Rawlinson P, Vilgalys R (2010b) Improved resolution of major clades within Tuber and taxonomy of species within the Tuber gibbosum complex. Mycologia 102:1042–1057. doi:10.3852/09-213
Bonuso E, Zambonelli A, Bergemann SE, Iotti M, Garbelotto M (2009) Multilocus phylogenetic and coalescent analyses identify two cryptic species in the Italian bianchetto truffle, Tuber borchii Vittad. Conserv Genet 11:1453–1466. doi:10.1007/s10592-009-9972-3
Borhidi A (2003) Magyarország növénytársulásai. Akadémiai Kiadó, Budapest
Bratek Z (1991) A Tuber brumale és egy hazai élőhelye. Thesis, Eötvös University.
Bratek Z, Merényi ZS, Varga T (2013) Changes of hypogeous funga in the Carpathian-Pannonian region in the past centuries. Acta Mycol 48:1–8. doi:10.5586/am.2013.005
Cai L, Giraud T, Zhang N, Begerow D, Cai G, Shivas RG (2011) The evolution of species concepts and species recognition criteria in plant pathogenic fungi. Fungal Divers 50:121–133. doi:10.1007/s13225-011-0127-8
Ceruti A, Fontana A, Nosenzo C (2003) Le specie europee del genere Tuber. Regione Piemonte Museo Regionale de Scienze Naturali. Monografie XXXVII, Torino
Chen J, Guo SX, Liu PG (2011) Species recognition and cryptic species in the Tuber indicum complex. PLoS ONE 6:e14625. doi:10.1371/journal.pone.0014625
Chen J, Liu PG (2007) Tuber latisporum sp. nov. and related taxa, based on morphology and DNA sequence data. Mycologia 99:475–481
Chen J, Liu P, Wang Y (2005) Tuber umbilicatum, a new species from China, with a key to the spinose-reticulate spored Tuber species. Mycotaxon 94:1–6
Deng XJ, Liu PG, Liu CY, Wang Y (2012) A new white truffle species, Tuber panzhihuanense from China. Mycol Prog 12:557–561. doi:10.1007/s11557-012-0862-6
Fan L, Feng S, Cao JZ (2013) The phylogenetic position of Tuber glabrum sp. nov. and T. sinomonosporum nom. nov., two Paradoxa-like truffle species from China. Mycol Prog 13:241–246. doi:10.1007/s11557-013-0908-4
Fan L, Hou C, Li Y (2012) Tuber microverrucosum and T. huizeanum- two new species from China with reticulate ascospores. Mycotaxon 122:161–169
Fan L, Liu X, Cao J (2015) Tuber turmericum sp. nov., a Chinese truffle species based on morphological and molecular data. Mycol Prog. 14: 111. doi:10.1007/s11557-015-1134-z
Florio AM, Ingram CM, Rakotondravony HA, Louis EE, Raxworthy CJ (2012) Detecting cryptic speciation in the widespread and morphologically conservative carpet chameleon (Furcifer lateralis) of Madagascar. J Evol Biol 25:1399–1414. doi:10.1111/j.1420-9101.2012.02528.x
Gandeboeuf D, Dupré C, Chevalier G (1994) Différenciation des truffes européennes par l’analyse des isoenzymes. Acta Bot Gall 141:455–463. doi:10.1080/12538078.1994.10515183
Gandeboeuf D, Dupre C, Nicolas P, Chevalier G (1997) Grouping and identification of Tuber species using RAPD markers. Can J Bot 75:36–45. doi:10.1139/b97-005
Granetti B, Angelis A, Materozzi G (2005) Umbria terra di tartufi. Regione Umbria
Halász K, Bratek Z, Szegő D, Rudnóy S, Rácz I, Lásztity DL, Trappe JM (2005) Tests of species concepts of the small, white, European group of Tuber spp. based on morphology and rDNA ITS sequences with special reference to Tuber rapaeodorum. Mycol Prog 4:281–290
Hall IR, Brown G, Zambonelli A (2007) Taming the truffle: the history, Lore, and science of the ultimate mushroom. Timber, Portland.
Hall IR, Zambonelli A (2012) Laying the foundations. pp 3–16. In: Edible ectomycorrhizal mushrooms soil biology 34. Springer, Berlin
Hernandez PA, Graham CH, Master LL, Albert DL, Albert DL (2006) The effect of sample size and species characteristics on performance of different species distribution modeling methods. Ecography 29:773–785
Hibbett DS, Ohman A, Glotzer D, Nuhn M, Kirk P, Nilsson RH (2011) Progress in molecular and morphological taxon discovery in fungi and options for formal classification of environmental sequences. Fungal Biol Rev 25:38–47. doi:10.1016/j.fbr.2011.01.001
Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A (2005) Very high resolution interpolated climate surfaces for global land areas. Int J Climatol 25:1965–1978. doi:10.1002/joc.1276
James F (1990) Multivariate analysis in ecology and systematics: panacea or Pandora box. Annu Rev Ecol Syst 21:129–166. doi:10.1146/annurev.ecolsys.21.1.129
Jiménez-Pérez N, Lorea-Hernández FG (2009) Identity and delimitation of the American species of Litsea Lam. (Lauraceae): A morphological approach. Plant Syst Evol 283:19–32. doi:10.1007/s00606-009-0218-0
Jörger KM, Schrödl M (2013) How to describe a cryptic species? Practical challenges of molecular taxonomy. Front Zool 10:1
Kirk PM, Cannon RF, David JC, Stalpers JA (2003) Dictionary of the fungi, ed. 9 edn. CAB International, Wallingford
Ławrynowicz M (1992) Distributional limits of truffles in the northern Europe. Micol Veg Mediterr 7:31–38
Ławrynowicz M, Krzyszczyk T, Fałdziński M (2008) Occurrence of black truffles in Poland. Acta Mycol 43:143–151
Lumley LM, Sperling FAH (2010) Integrating morphology and mitochondrial DNA for species delimitation within the spruce budworm (Choristoneura fumiferana) cryptic species complex (Lepidoptera: Tortricidae). Syst Entomol 35:416–428. doi:10.1111/j.1365-3113.2009.00514.x
Lundholm N, Bates SS, Baugh KA, Bill BD, Connell LB, Léger C, Trainer VL (2012) Cryptic and pseudo-cryptic diversity in diatoms–with descriptions of Pseudo-nitzschia hasleana sp. nov. and P. fryxelliana sp. nov. J Phycol 48:436–454. doi:10.1111/j.1529-8817.2012.01132.x
Mattirolo O (1907) Gli autoptici di Carlo Vittadini e la loro importanza nello studio della Idnologia. Atti del Congresso del Naturalisti Italiani, Milano 15–19 Settembre 1906, pp 396–402
Medina R, Lara F, Goffinet B, Garilleti R, Mazimpaka V (2012) Integrative taxonomy successfully resolves the pseudo-cryptic complex of the disjunct epiphytic moss Orthotrichum consimile s. l. (Orthotrichaceae). Taxonomy 61:1180–1198
Merényi Z (2014) Molecular biological and ecological studies on hypogeous fungi. Dissertation, Eötvös University. Retrieved from http://www.doktori.hu/index.php?menuid=193&vid=14379
Merényi Z, Illyés Z, Völcz G, Bratek Z (2010) Creation database application for development on truffle cultivation methods. In: Proceedings of the First Conference on the "European" Truffle Tuber aestivum/uncinatum. Austrian J Mycol 19: 239–244
Merényi Z, Varga T, Bratek Z (2016) Tuber brumale: the most controversial Tuber species. In: Zambonelli A, Iotti M, Murat C (eds) True Truffle (Tuber spp.) in the World: Soil Ecology, Systematics and Biochemistry. Soil Biology Series 47. Springer, Berlin, pp 49–68. doi:10.1007/978-3-319-31436-5
Merényi Z, Varga T, Geml J, Orczán ÁK, Chevalier G, Bratek Z (2014) Phylogeny and phylogeography of the Tuber brumale aggr. Mycorrhiza 24 Suppl 1: S101–S113. doi:10.1007/s00572-014-0566-7
Montecchi A, Sarasini M (2000) Funghi ipogei d’Europa. Associazione Micologica Bresadola, Fondacione Centro Studi Micologici, Trento, Vicenza
Orczán Á, Vetter J, Merényi Z, Bonifert É, Bratek Z (2012) Mineral composition of hypogeous fungi in Hungary. J Appl Bot Food Qual 104:100–104
Orive E, Pérez-Aicua L, David H, García-Etxebarria K, Laza-Martínez A, Seoane S, Miguel I (2013) The genus Pseudo-nitzschia (Bacillariophyceae) in a temperate estuary with description of two new species: Pseudo-nitzschia plurisecta sp. nov. and Pseudo-nitzschia abrensis sp. nov. J Phycol 49:1192–1206. doi:10.1111/jpy.12130
Pante E, Puillandre N, Viricel A, Arnaud-Haond S, Aurelle D, Castelin M, Chenuil A, Destombe C, Forcioli D, Valero M, Viard F, Samadi S (2015) Species are hypotheses: avoid connectivity assessments based on pillars of sand. Mol Ecol 24: 525–544. doi:10.1111/mec.13048
Phillips SJ, Anderson RP, Schapire RE (2006) Maximum entropy modeling of species geographic distributions. Ecol Model 190: 231–259. doi:10.1016/j.ecolmodel.2005.03.026
Podani J (2000) Introduction to the exploration of multivariate biological data. Backhuys, Leiden
Pohlert T (2014) The Pairwise Multiple Comparison of Mean Ranks Package (PMCMR) Retrieved from https://cran.r-project.org/web/packages/PMCMR/vignettes/PMCMR.pdf
QGIS Development Team 2009. QGIS Geographic information system. Open Source Geospatial Foundation. URL http://qgis.osgeo.org
R Core Team 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/
Reynolds HT (2011) Systematics, Phylogeography and Ecology of Elaphomycetaceae. Dissertation, Duke University
Riousset L, Riousset G, Chevalier G, Bardet M (2001) Truffes d’Europe et de Chine. INRA, Paris
Rivera PC, Di Cola V, Martínez JJ, Gardenal CN, Chiaraviglio M (2011) Species delimitation in the continental forms of the genus epicrates (serpentes, boidae) integrating phylogenetics and environmental niche models. PLoS ONE 6:e22199. doi:10.1371/journal.pone.0022199
Sáez AG, Probert I, Geisen M, Quinn P, Young JR, Medlin LK (2003) Pseudo-cryptic speciation in coccolithophores. Proc Natl Acad Sci U S A 100:7163–7168. doi:10.1073/pnas.1132069100
Schwarzfeld M, Sperling F (2014) Species delimitation using morphology, morphometrics, and molecules: definition of the Ophion scutellaris Thomson species group, with descriptions of six new species (hymenoptera, Ichneumonidae). Zookeys 462:59–114. doi:10.3897/zookeys.462.8229
Stobbe U, Büntgen U, Sproll L, Tegel W, Egli S, Fink S (2012) Spatial distribution and ecological variation of re-discovered German truffle habitats. Fungal Ecol 5:591–599. doi:10.1016/j.funeco.2012.02.001
Szurdoki E, Márton O, Szövényi P (2014) Genetic and morphological diversity of Sphagnum angustifolium, S. flexuosum and S. fallax in Europe. Taxon 63:237–248
Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC (2000) Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol 31:21–32. doi:10.1006/fgbi.2000.1228
Urban A, Stielow B, Orczán A, Varga T, Tamaskó G, Bratek Z (2013) High phylogenetic diversity in the Tuber excavatum group. p. 73, in: First International Congress of Trufficulture, Teruel, Spain
Venables W, Ripley B (2002) Modern applied statistics with S. Springer, New York
Verbruggen H (2014) Morphological complexity, plasticity, and species diagnosability in the application of old species names in DNA-based taxonomies. J Phycol 50:26–31. doi:10.1111/jpy.12155
Verbruggen H, De Clerck O, Cocquyt E, Kooistra WHCF, Coppejans E (2005b) Morphometric taxonomy of Siphonous green algae: a methodological study within the genus Halimeda (Bryopsidales). J Phycol 41:126–139. doi:10.1111/j.1529-8817.2005.04080.x
Verbruggen H, De Clerck O, Kooistra WHCF, Coppejans E (2005a) Molecular and morphometric data pinpoint species boundaries in Halimeda section Rhipsalis (Bryopsidales, Chlorophyta). J Phycol 41: 606–621. doi:10.1111/j.1529-8817.2005.00083.x
Vittadini C (1831) Monographia Tuberacearum. Milano
Weihs C, Ligges U, Luebke K, Raabe N (2005) klaR analyzing German business cycles. In: Baier D, Decker R, Schmidt-Thieme L (eds) Data analysis and decision support. Springer, Berlin, pp 335–343
White TJ, Bruns T, Lee S, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic, New York, pp 315–322
Acknowledgements
We are obliged to Giovanni Pacioni for advice on taxonomic issues. We are grateful to the people who provided us with specimens, namely Alessandra Zambonelli, Amer Montecchi, Chriss Chrysopoulos, Diamandis Stephanos, Gérard Chevalier, Joseph Maria Vidal, Marjanović Zaklina, Stanislav Glejdura, Tine Grebenc and the truffle hunters of the FHTA. We would like to thank Judit Vikor, Péter László and Tibor Simon for their help in the classification of soil types and plant coenological associations. We are also grateful to Ádám Solti, Annamária Tóth, Lilla Bóna, Sára Pólya, and Zsófia Gáti for their various help and technical assistance. ZsM was supported by NTP-NFTÖ-16-0216 and GINOP- 2.1.1-15- 2015-00115 (Széchenyi 2020 Programme), while TV was supported by NTP-EFÖ-P-15-0295, the National Talent Program of the Ministry of Human Capacities (EMMI) and the Human Capacities Grant Management Office (EMET). This study was supported by the MIKOQUAL project under the Ányos Jedlik Programme and by the QUTAOMEL project under the National Technology Programme.
Author information
Authors and Affiliations
Corresponding author
Additional information
Section Editor: Zhu-Liang Yang
Rights and permissions
About this article
Cite this article
Merényi, Z., Varga, T., Hubai, A.G. et al. Challenges in the delimitation of morphologically similar species: a case study of Tuber brumale agg. (Ascomycota, Pezizales). Mycol Progress 16, 613–624 (2017). https://doi.org/10.1007/s11557-017-1296-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-017-1296-y