Abstract
A new poroid wood-inhabiting fungal genus, Leifiporia, is proposed, based on morphological and molecular evidence, which is typified by L. rhizomorpha sp. nov. The genus is characterized by an annual growth habit, resupinate basidiocarps with white to cream pore surface, a dimitic hyphal system with generative hyphae bearing clamp connections and branching mostly at right angles, skeletal hyphae present in the subiculum only and distinctly thinner than generative hyphae, IKI–, CB–, and ellipsoid, hyaline, thin-walled, smooth, IKI–, CB– basidiospores. Sequences of ITS and LSU nrRNA gene regions of the studied samples were generated, and phylogenetic analyses were performed with maximum likelihood, maximum parsimony and Bayesian inference methods. The phylogenetic analysis based on molecular data of ITS + nLSU sequences showed that Leifiporia belonged to the core polyporoid clade and was closely related to Diplomitoporus overholtsii and Lopharia cinerascens, and then grouped with Pycnoporus and Trametes. Further investigation was obtained for more representative taxa in the Polyporaceae based on nLSU sequences, in which the results demonstrated that the genus Leifiporia formed a monophyletic lineage with a strong support (100 % BS, 100 % BP, 1.00 BPP). Both morphological and molecular evidence confirmed the placement of the new genus in the core polyporoid clade. In addition, a new combination, Leifiporia eucalypti, is proposed based on examination of its type material and phylogeny.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Polypores are a very important group of wood-inhabiting fungi which have been extensively studied Among them, the Polyporaceae is a diverse group of Polyporales, including species having annual to perennial, resupinate, pileate and stipitate basidiocarps, a monomitic to dimitic or trimitic hyphal structure with simple septa or clamp connections on generative hyphae, and thin- to thick-walled, smooth to ornamented, cyanophilous to acyanophilous basidiospores (Ryvarden and Johansen 1980; Gilbertson and Ryvarden 1986, 1987; Dai 2012; Ryvarden and Melo 2014).
Molecular systematics is a powerful tool to infer phylogenies within fungal groups including polypores (White et al. 1990; Binder et al. 2005, 2013; James et al. 2006; Hibbett et al. 2007). By using ribosomal DNA sequences, Binder et al. (2005) resolved the taxonomic structure of Polyporales and divided the polyporoid clade into three main groups—the core polyporoid clade, the antrodia clade, and the phlebioid clade—together with some ‘residual’ taxa not assigned to any group. Hibbett et al. (2007) analyzed a higher-level phylogenetic classification of the fungi, and showed that Polyporales formed a monophyletic lineage within Basidiomycota. Recently, molecular studies employing multi-gene (5.8S, nrLSU, nrSSU, rpb1, rpb2, tef1) datasets have helped to investigate the phylogenetic overview of the Polyporales, in which 35 families were recognized in Polyporales, and the Polyporaceae species were mainly nested in the core polyporoid clade (Binder et al. 2013; Zhao et al. 2015). Overviews of studies on fungal diversity worldwide have been carried out based on increasing specialized knowledge and new techniques (Hawksworth 2012; Dai et al. 2015), and new genera of polypores have recently been found (Miettinen and Larsson 2011; Cui 2013; Li et al. 2013, 2014; Sotome et al. 2013; Spirin et al. 2013; Chen et al. 2014, 2016; Ariyawansa et al. 2015; Ghobad-Nejhad 2015; Westphalen et al. 2015; Zhou et al. 2016).
During a study of Chinese polypores, two samples collected from eastern China were characterized by resupinate, white to cream basidiocarps with distinct rhizomorphic margins, a monomitic hyphal structure in tube trama, but few skeletal hyphae in the subiculum, and hyaline, thin-walled, ellipsoid basidiospores, which are negative in Melzer’s reagent and Cotton Blue, and causing a white rot. These characters distinguished them from all the known polypore taxa, and we have proposed a new genus for the two collections. To support our proposal, phylogenetic analyses on the position of the new genus and related taxa were carried out based on the internal transcribed spacer (ITS) regions and the large subunit nuclear ribosomal RNA gene (nLSU) sequences.
Materials and methods
Morphology
The studied specimens are deposited at the herbaria of Beijing Forestry University (BJFC) and the University of Oslo (O). Macro-morphological descriptions are based on field notes. Special color terms follow Petersen (1996). Micro-morphological data were obtained from the dried specimens, and observed under a light microscope following Dai (2010). The following abbreviations have been used: KOH = 5 % potassium hydroxide, CB = Cotton Blue, CB– = acyanophilous, IKI = Melzer’s reagent, IKI– = both inamyloid and indextrinoid, L = mean spore length (arithmetic average of all spores), W = mean spore width (arithmetic average of all spores), Q = variation in the L/W ratios between the specimens studied, n (a/b) = number of spores (a) measured from given number (b) of specimens.
Molecular phylogeny
CTAB rapid plant genome extraction kit-DN14 (Aidlab Biotechnologies, Beijing, China) was used to obtain PCR products from dried specimens, according to the manufacturer’s instructions with some modifications. ITS region was amplified with primer pairs ITS5 and ITS4 (White et al. 1990). The nuclear LSU region was amplified with primer pairs LR0R and LR7 (http://www.biology.duke.edu/fungi/mycolab/primers.htm). The PCR procedure for ITS was as follows: initial denaturation at 95 °C for 3 min, followed by 35 cycles at 94 °C for 40 s, 58 °C for 45 s and 72 °C for 1 min, and a final extension of 72 °C for 10 min. The PCR procedure for nLSU was as follows: initial denaturation at 94 °C for 1 min, followed by 35 cycles at 94 °C for 30 s, 48 °C 1 min and 72 °C for 1.5 min, and a final extension of 72 °C for 10 min. The PCR products were purified and directly sequenced at Beijing Genomics Institute. All newly generated sequences were deposited at GenBank (Table 1).
Sequencher 4.6 (GeneCodes, Ann Arbor, MI, USA) was used to edit the DNA sequence. Sequences were aligned in MAFFT 6 (Katoh and Toh 2008; http://mafft.cbrc.jp/alignment/server/) using the “G-INS-I” strategy and manually adjusted in BioEdit (Hall 1999). The sequence alignment was deposited in TreeBase (submission ID 18818). Sequences of Heterobasidion annosum (Fr.) Bref. and Stereum hirsutum (Willd.) Pers. obtained from GenBank were used as outgroups to root trees following Binder et al. (2013) in the ITS + nLSU analysis. Sequences of Ceriporiopsis gilvescens (Bres.) Domański and Phlebia radiata Fr., obtained from GenBank, were used as outgroups to root trees in nLSU tree.
Maximum parsimony analysis was applied to the nLSU and ITS + nLSU dataset sequences. Approaches to phylogenetic analysis followed Zhao et al. (2013, 2014), and the tree construction procedure was performed in PAUP* v.4.0b10 (Swofford 2002). All characters were equally weighted and gaps were treated as missing data. Trees were inferred using the heuristic search option with TBR branch swapping and 1000 random sequence additions. Max-trees were set to 5000, branches of zero length were collapsed and all parsimonious trees were saved. Clade robustness was assessed using a bootstrap (BT) analysis with 1,000 replicates (Felsenstein 1985). Descriptive tree statistics tree length (TL), consistency index (CI), retention index (RI), rescaled consistency index (RC), and homoplasy index (HI) were calculated for each Maximum Parsimonious Tree (MPT) generated. Sequences were also analyzed using Maximum Likelihood (ML) with RAxML-HPC2 on Abe through the Cipres Science Gateway (www.phylo.org; Miller et al. 2009). Branch support for ML analysis was determined by 1000 bootstrap replicate.
MrModeltest 2.3 (Posada and Crandall 1998; Nylander 2004) was used to determine the best-fit evolution model for each data set for Bayesian inference (BI). Bayesian inference was calculated with MrBayes3.1.2 with a general time reversible (GTR) model of DNA substitution and a gamma distribution rate variation across sites (Ronquist and Huelsenbeck 2003). Four Markov chains were run for 2 runs from random starting trees for 5 million generations (ITS + nLSU), and for 2 million generations (nLSU), and trees were sampled every 100 generations. The first one-quarter of the generations were discarded as burn-in. A majority rule consensus tree of all remaining trees was calculated. Branches that received bootstrap support for maximum likelihood (BS), maximum parsimony (BP) and Bayesian posterior probabilities (BPP) greater than or equal to 75 % (BP) and 0.95 (BPP) were considered as significantly supported, respectively.
Results
The ITS + nLSU dataset included sequences from 75 fungal specimens representing 65 species. The dataset had an aligned length of 2240 characters, of which 1326 characters are constant, 245 are variable and parsimony-uninformative, and 669 are parsimony-informative. Maximum parsimony analysis yielded four equally parsimonious trees (TL = 5510, CI = 0.287, HI = 0.713, RI = 0.529, RC = 0.152). The best model for the ITS + nLSU dataset estimated and applied in the Bayesian analysis was: GTR + I + G, lset nst = 6, rates = invgamma; prset statefreqpr = dirichlet (1,1,1,1). Bayesian analysis and ML analysis resulted in a similar topology as the MP analysis, with an average standard deviation of split frequencies = 0.007467 (BI).
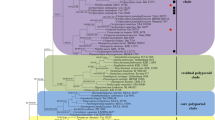
The phylogeny (Fig. 1) inferred from ITS + nLSU sequences demonstrated seven major clades for 65 species of the Polyporales. The new genus Leifiporia clustered in the core polyporoid clade and was closely related to Diplomitoporus overholtsii (Pilát) Gilb. & Ryvarden and Lopharia cinerascens (Schwein.) G. Cunn, and then grouped with Pycnoporus P. Karst. and Trametes Fr. with a high support (98 % BS, 87 % BP, 0.98 BPP).
Maximum Parsimony strict consensus tree illustrating the phylogeny of Leifiporia and related species in Polyporales based on ITS + nLSU sequences. Branches are labeled with maximum likelihood bootstrap higher than 70 %, parsimony bootstrap proportions higher than 50 % and Bayesian posterior probabilities more than 0.95, respectively. Clade names follow Binder et al. (2013)
The nLSU dataset included sequences from 35 fungal specimens representing 32 species. The dataset had an aligned length of 1370 characters, of which 1129 characters are constant, 76 are variable and parsimony-uninformative, and 165 are parsimony-informative. Maximum parsimony analysis yielded 12 equally parsimonious trees (TL = 693, CI = 0.414, HI = 0.586, RI = 0.418, RC = 0.173). The best model for the nLSU dataset estimated and applied in the Bayesian analysis was: GTR + I + G, lset nst = 6, rates = invgamma; prset statefreqpr = dirichlet (1,1,1,1). Bayesian analysis and ML analysis resulted in a similar topology as MP analysis, with an average standard deviation of split frequencies = 0.002783 (BI).
A further phylogeny (Fig. 2) inferred from nLSU sequences was obtained for more representative taxa in the Polyporaceae and showed that the new genus formed a monophyletic lineage and grouped with the related species Diplomitoporus overholtsii and Lopharia cinerascens with strong support (100 % BS, 100 % BP, 1.00 BPP).
Maximum Parsimony strict consensus tree illustrating the phylogeny of Leifiporia and related species obtained for more representative taxa in the Polyporaceae, based on the nLSU sequence datasets. Branches are labeled with maximum likelihood bootstrap higher than 70 %, parsimony bootstrap proportions higher than 50 % and Bayesian posterior probabilities more than 0.95 respectively. Clade names follow Binder et al. (2013)
Taxonomy
Leifiporia Y.C. Dai, F. Wu & C.L. Zhao, gen. nov.
MycoBank no.: MB 817362
Differs from other genera by its resupinate, brittle basidiocarps with poroid surface, a dimitic hyphal system with clamp connections on generative hyphae which are frequently branched at right angles, skeletal hyphae present in the subiculum only, distinctly thinner than generative hyphae, IKI–, CB–, and basidiospores ellipsoid, hyaline, thin-walled, smooth, usually bearing one or two guttules, IKI–, CB–.
Type species. Leifiporia rhizomorpha Y.C. Dai, F. Wu & C.L. Zhao.
Etymology. Leifiporia (Lat.): in honour of the Norway mycologist Prof. Leif Ryvarden.
Basidiocarps annual, resupinate, adnate, soft when fresh, brittle when dry. Pore surface white to cream. Pores angular; dissepiments thin, entire to slightly lacerate. Hyphal system dimitic, generative hyphae hyaline, thin-walled with clamp connections, frequently branched at right angles; skeletal hyphae present in subiculum only, interwoven, distinctly thinner than generative hyphae, IKI–, CB–; tissues unchanged in KOH. Basidia barrel-shaped to pyriform. Basidiospores ellipsoid, hyaline, thin-walled, smooth, usually bearing one or two guttules, IKI–, CB–.
Leifiporia rhizomorpha Y.C. Dai, F. Wu & C.L. Zhao, sp. nov. Figs. 3a-b and 4
MycoBank no.: MB 817363
Holotype. CHINA. Anhui Prov., Qimen County, Guniujiang Nature Reserve, on fallen angiosperm trunk, 9 August 2013, Dai 13380 (BJFC).
Etymology. Rhizomorpha (Lat.): referring to the species bearing a rhizomorphic margin.
Fruiting body. Basidiocarps annual, resupinate, soft, without odor or taste when fresh, brittle when dry, up to 20 cm long, 4 cm wide, 1.5 mm thick at center (measured in two hitherto available specimens). Pore surface white to cream when fresh, cream when dry; pores angular, 3–4 per mm; dissepiments thin, entire to slightly lacerate. Sterile margin distinctly rhizomorphic, white, up to 4 mm wide. Subiculum white, soft corky, up to 0.2 mm thick. Tubes concolorous with pore surface, brittle, up to 1.3 mm long.
Hyphal structure. Hyphal system dimitic; generative hyphae hyaline, thin-walled, bearing clamp connections; skeletal hyphae present in the subiculum only, IKI–, CB–; tissues unchanged in KOH.
Subiculum. Generative hyphae dominant, hyaline, thin-walled, frequently branched at right angles, interwoven, 2.5–4 μm in diameter; skeletal hyphae infrequent, hyaline, thick-walled with a narrow lumen or subsolid, occasionally branched, interwoven, distinctly thinner than generative hyphae, 1.5–2.5 μm in diameter.
Tubes. Generative hyphae hyaline, thin-walled, frequently branched at right angle, interwoven, 2–3.5 μm in diameter. Cystidia absent, but fusoid cystidioles occasionally present, hyaline, thin-walled, 11–13 × 5–6 μm; basidia barrel-shaped to pyriform, with four sterigmata and a basal clamp connection, 14–17 × 5.5–8 μm; basidioles dominant, in shape similar to basidia, but slightly smaller.
Spores. Basidiospores mostly ellipsoid, hyaline, thin-walled, smooth, usually bearing one or two guttules, IKI–, CB–, (4.5–)5–6(−6.5) × 2–3 μm, L = 5.68 μm, W = 2.51 μm, Q = 2.15–2.38 (n = 60/2).
Type of rot. White rot.
Additional specimen (paratype) examined: CHINA. Zhejiang Prov., Qingyun County, Baishanzu Nature Reserve, on fallen angiosperm branch, 14 August 2013, Dai 13432 (BJFC).
Leifiporia eucalypti (Ryvarden) Y.C. Dai, F. Wu & C.L. Zhao, comb. nov. Fig. 5
MycoBank no.: MB 817364
Basionym: Dichomitus eucalypti Ryvarden, Trans Br Mycol Soc 85: 539 (1985).
Fruiting body. Basidiocarps annual, resupinate, brittle when dry, up to 2 cm long, 1 cm wide, 1.5 mm thick at center. Pore surface white to pale cream when dry; pores angular, 2–3 per mm; dissepiments thin, entire to slightly lacerate. Sterile margin narrow, white, up to 1 mm wide. Subiculum white, soft corky, up to 0.3 mm thick. Tubes concolorous with pore surface, brittle, up to 1.2 mm long.
Hyphal structure. Hyphal system dimitic; generative hyphae hyaline, thin-walled, bearing clamp connections; skeletal hyphae present in subiculum only, IKI–, CB–; tissues unchanged in KOH.
Subiculum. Generative hyphae dominant, hyaline, thin-walled, frequently branched mostly at right angle, interwoven, 2.5–4 μm in diameter; skeletal hyphae infrequent, hyaline, thick-walled with a narrow lumen to subsolid, frequently branched, interwoven, distinctly thinner than generative hyphae, 1.5–2.5 μm in diameter.
Tubes. Generative hyphae hyaline, thin-walled, frequently branched at right angle, interwoven, 2–3.5 μm in diameter. Cystidia absent, but fusoid cystidioles present, hyaline, thin-walled, 10–13 × 4–5.5 μm; basidia barrel-shaped to pyriform, with four sterigmata and a basal clamp connection, 15–20 × 5–9 μm; basidioles dominant, in shape similar to basidia, but slightly smaller.
Spores. Basidiospores ellipsoid, hyaline, thin-walled, smooth, usually bearing one guttule, IKI–, CB–, (6.5–)7–8(−8.5) × (3–)3.5–4.5(−5) μm, L = 7.67 μm, W = 3.95 μm, Q = 1.94 (n = 30/1).
Type of rot. White rot.
Specimen (isotype) examined: AUSTRALIA, Northern Territory, Stokes Creek, George Gill Ranges, on Eucalyptus camaldulensis, 10 August 1981, leg. A. C. Kalotas, 910674 (O).
Discussion
In the present study, a new genus, Leifiporia, was described, based on phylogenetic analyses and morphological characters. The genus had unique morphological characters in Polyporaceae and formed a monophyletic lineage within core polyporoid clade.
Previously, seven clades were found in the Polyporales: antrodia clade, core polyporoid clade, fragiliporia clade, gelatoporia clade, phlebioid clade, residual polyporoid clade and tyromyces clade (Binder et al. 2013; Zhao et al. 2015). According to our results based on the combined ITS + nLSU sequence data (Fig. 1), the new genus is nested in the core polyporoid clade with strong support (100 % BS, 99 % BP, 1.00 BPP). In the nLSU analysis, Leifiporia grouped with Diplomitoporus overholtsii and Lopharia cinerascens based on phylogenetic analysis with high support (Fig. 1), which was mentioned in a previous study by Li and Cui (2013).
The species Dichomitus eucalypti Ryvarden was described by Ryvarden based on resupinate, brittle basidiocarps and a dimitic hyphal system with clamp connections (Ryvarden 1985). However, it grouped with Pycnoporus and Trametes in ITS analysis, and clustered with Diplomitoporus overholtsii and Lopharia cinerascens in nLSU analysis (Li and Cui 2013). In the present study, Dichomitus eucalypti was the sister species to L. rhizomorpha, and not related to the generic type of Dichomitus D.A. Reid, D. squalens (P. Karst.) D.A. Reid, which was mentioned by Li and Cui (2013). Binder et al. (2013) employed multi-gene datasets to investigate the phylogenetic overview of the Polyporales and showed that the type species of Dichomitus, D. squalens, was the sister species to Sparsitubus nelumbiformis L.W. Hsu & J.D. Zhao and then grouped with Perenniporia Murrill species. According to our inference with LSU sequences (Fig. 2), D. squalens also grouped with S. nelumbiformis and then was closely related to some species of Perenniporia s.l.; a similar result was indicated by Binder et al. (2013). Thus, the new combination, Leifiporia eucalypti, is proposed.
Morphologically, Dichomitus, Diplomitoporus Domański, Lopharia Kalchbr. & MacOwan, Pycnoporus and Trametes are different from Leifiporia (see the key to these genera, below). Dichomitus is separated from the new genus by hard and woody basidiocarps, and the presence of arboriform skeletal hyphae, which are occasionally cyanophilous and dominated in trama and context (Gilbertson and Ryvarden 1986; Núñez and Ryvarden 2001; Yuan 2013; Ryvarden and Melo 2014). Diplomitoporus differs from Leifiporia by its corky basidiocarps, weakly amyloid skeletal hyphae dominated in whole basidiocarps, and allantoid basidiospores (Ryvarden and Melo 2014). Lopharia differs from Leifiporia by a smooth or hydnoid hymenophore and the presence of thick-walled cystidia, cyanophilous skeletal hyphae and large basidiospores (10–16 × 6–7.5 μm; Hjortstam and Ryvarden 1989). Pycnoporus differs in pileate, hard corky and reddish basidiocarps and a trimitic hyphal system (Núñez and Ryvarden 2001). Trametes differs from Leifiporia by its pileate, corky basidiocarps and a trimitic hyphal system (Ryvarden and Melo 2014).
Fibroporia Parmasto is similar to Leifiporia in having infrequent skeletal hyphae in the subiculum only, but it has slightly thick-walled basidiospores, and causes a brown rot (Ryvarden and Melo 2014).
Having resupinate basidiocarps with brittle consistency is similar to two similar genera in the Polyporales: Fragiliporia Y.C. Dai, B.K. Cui & C.L. Zhao and Physisporinus P. Karst. The former differs from Leifiporia by a completely monomitic hyphal system with thick-walled generative hyphae and allantoid basidiospores (Zhao et al. 2015), while the latter is separated from Leifiporia by its ceraceous basidiocarps which change to red when bruised, a completely monomitic hyphal system with simple septa and globose basidiospores (Gilbertson and Ryvarden 1987; Núñez and Ryvarden 2001; Ryvarden and Melo 2014). In addition, the three genera were nested in different clades based on previous phylogenetic analyses (Binder et al. 2013).
Polypores are an extensively studied group of Basidiomycota (Gilbertson and Ryvarden 1987; Núñez and Ryvarden 2001; Ryvarden and Melo 2014), but the Chinese polypore diversity is still not well known, especially in the subtropics and tropics; many recently described genera of polypores have been from these areas (Cui 2013; Li et al. 2013, 2014; Chen et al. 2014, 2016; Zhao et al. 2015). The new genus in the present study, Leifiporia, is also from the subtropics. It is possible that new polypore taxa will be found after futher investigations and molecular analyses.
A key to Leifiporia and related genera
-
1 Hyphal structure trimitic, binding hyphae present 2
-
1* Hyphal structure dimitic, binding hyphae absent 3
-
2 Basidiocarps cinnabar red Pycnoporus
-
2* Basidiocarps white, gray, pale Trametes
-
3 Hymenophore smooth to irregularly irpicoid; cystidia present Lopharia
-
3* Hymenophore poroid; cystidia absent 4
-
4 Basidiocarps resupinate to pileate; arboriform skeletal hyphae present Dichomitus
-
4* Basidiocarps resupinate; arboriform skeletal hyphae absent 5
-
5 Hyphal structure monomitic· 6
-
5* Hyphal structure dimitic at least in subiculum 7
-
6 Clamp connection present Fragiliporia
-
6* Simple septa present Physisporinus
-
7 Skeletal hyphae present in both trama and subiculum, basidiospores allantoid Diplomitoporus
-
7* Skeletal hyphae present in subiculum only, basidiospores ellipsoid 8
-
8 Basidiospores thin-walled, skeletal hyphae thinner than generative hyphae Leifiporia
-
8* Basidiospores thick-walled, skeletal hyphae thicker than generative hyphae Fibroporia
References
Ariyawansa HA, Hyde KD, Jayasiri SC, Buyck B, Chethana KWT, Dai DQ, Dai YC, Daranagama DA, Jayawardena RS, Lücking R, Ghobad-Nejhad M, Niskanen T, Thambugala KM, Voigt K, Zhao RL, Li GJ, Doilom M, Boonmee S, Yang ZL, Cai Q, Cui YY, Bahkali AH, Chen J, Cui BK, Chen JJ, Dayarathne MC, Dissanayake AJ, Ekanayaka AH, Hashimoto A, Hongsanan S, Jones EBG, Larsson E, Li WJ, Li QR, Liu JK, Luo ZL, Maharachchikumbura SSN, Mapook A, McKenzie EHC, Norphanphoun C, Konta S, Pang KL, Perera RH, Phookamsak R, Phukhamsakda C, Pinruan U, Randrianjohany E, Singtripop C, Tanaka K, Tian CM, Tibpromma S, Abdel-Wahab MA, Wanasinghe DN, Wijayawardene NN, Zhang JF, Zhang H, Abdel-Aziz FA, Wedin M, Westberg M, Ammirati JF, Bulgakov TS, Lima DX, Callaghan TM, Callac P, Chang CH, Coca LF, Dal-Forno M, Dollhofer V, Fliegerová K, Greiner K, Griffith GW, Ho HM, Hofstetter V, Jeewon R, Kang JC, Wen TC, Kirk PM, Kytövuori I, Lawrey JD, Xing J, Li H, Liu ZY, Liu XZ, Liimatainen K, Lumbsch HT, Matsumura M, Moncada B, Nuankaew S, Parnmen S, Santiago ALCMA, Sommai S, Song Y, de Souza CAF, de Souza-Motta CM, Su HY, Suetrong S, Wang Y, Wei SF, Yuan HS, Zhou LW, Réblová M, Fournier J, Camporesi E, Luangsaard JJ, Tasanathai K, Khonsanit A, Thanakitpipattana D, Somrithipol S, Diederich P, Millanes AM, Common RS, Stadler M, Yan JY, Li XH, Hye LW, Nguyen TTT, Lee HB, Battistin E, Marsico O, Vizzini A, Vila J, Ercole E, Eberhardt U, Simonini G, Wen HA, Chen XH, Miettinen O, Spirin V, Hernawati (2015) Fungal diversity notes 111–252—taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers 75:27–274. doi:10.1007/s13225-015-0346-5
Binder M, Hibbett DS, Larsson KH, Larsson E, Langer E, Langer G (2005) The phylogenetic distribution of resupinate forms across the major clades of mushroom-forming fungi (Homobasidiomycetes). Syst Biodivers 3:113–157. doi:10.1017/S1477200005001623
Binder M, Justo A, Riley R, Salamov A, López-Giráldez F, Sjökvist E, Copeland A, Foster B, Sun H, Larsson E, Larsson KH, Townsend J, Grigoriev IV, Hibbett DS (2013) Phylogenetic and phylogenomic overview of the polyporales. Mycologia 105:1350–1373. doi:10.3852/13-003
Chen JJ, Cui BK (2014) Phlebiporia bubalina gen. et sp. nov. (Meruliaceae, Polyporales) from Southwest China with a preliminary phylogeny based on rDNA sequences. Mycol Prog 13:563–573. doi:10.1007/s11557-013-0940-4
Chen JJ, Cui BK, Dai YC (2016) Global diversity and molecular systematics of Wrightoporia s.l. (Russulales, Basidiomycota). Persoonia 37:21–36. doi:10.3767/003158516X689666
Cui BK (2013) Antrodia tropica sp nov. from southern China inferred from morphological characters and molecular data. Mycol Prog 12:223–230. doi:10.1007/s11557-012-0829-7
Dai YC (2010) Hymenochaetaceae (Basidiomycota) in China. Fungal Divers 45:131–343. doi:10.1007/s13225-010-0066-9
Dai YC (2012) Polypore diversity in China with an annotated checklist of Chinese polypores. Mycoscience 53:49–80. doi:10.1007/s10267-011-0134-3
Dai YC, Cui BK, Si J, He SH, Hyde KD, Yuan HS, Lui XY, Zhou LW (2015) Dynamics of the worldwide number of fungi with emphasis. Mycol Prog 14:62. doi:10.1007/s11557-015-1084-5
Felsenstein J (1985) Confidence intervals on phylogenetics: an approach using bootstrap. Evolution 39:783–791
Ghobad-Nejhad M (2015) Collections on Lonicera in Northwest Iran represent an undescribed species in the Inonotus linteus complex (Hymenochaetales). Mycol Prog 14:90. doi:10.1007/s11557-015-1100-9
Gilbertson RL, Ryvarden L (1986) North American polypores, vol 1. Abortiporus – Lindtneria. Fungiflora, Oslo, pp 1–433
Gilbertson RL, Ryvarden L (1987) North American polypores, vol 2. Megasporoporia – Wrightoporia. Fungiflora, Oslo, pp 434–885
Hall TA (1999) Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hawksworth DL (2012) Global species numbers of fungi: are tropical studies and molecular approaches contributing to a more robust estimate? Biodivers Conserv 21:2425–2433. doi:10.1007/s10531-012-0335-x
Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, Eriksson OE, Huhndorf S, James T, Kirk PM, Lücking R, Thorsten Lumbsch H, Lutzoni F, Matheny PB, McLaughlin DJ, Powell MJ, Redhead S, Schoch CL, Spatafora JW, Stalpers JA, Vilgalys R, Aime MC, Aptroot A, Bauer R, Begerow D, Benny GL, Castlebury LA, Crous PW, Dai YC, Gams W, Geiser DM, Griffith GW, Gueidan C, Hawksworth DL, Hestmark G, Hosaka K, Humber RA, Hyde KD, Ironside JE, Kõljalg U, Kurtzman CP, Larsson KH, Lichtwardt R, Longcore J, Miadlikowska J, Miller A, Moncalvo JM, Mozley-Standridge S, Oberwinkler F, Parmasto E, Reeb V, Rogers JD, Roux C, Ryvarden L, Sampaio JP, Schüssler A, Sugiyama J, Thorn RG, Tibell L, Untereiner WA, Walker C, Wang Z, Weir A, Weiss M, White MM, Winka K, Yao YJ, Zhang N (2007) A higher-level phylogenetic classification of the Fungi. Mycol Res 111:509–547. doi:10.1016/j.mycres.2007.03.004
Hjortstam K, Ryvarden L (1989) Lopharia and Porostereum (Basidiomycotina). Syn Fungal 4:1–68
James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J, Lumbsch HT, Rauhut A, Reeb V, Arnold AE, Amtoft A, Stajich JE, Hosaka K, Sung GH, Johnson D, O’Rourke B, Crockett M, Binder M, Curtis JM, Slot JC, Wang Z, Wilson AW, Schüssler A, Longcore JE, O’Donnell K, Mozley-Standridge S, Porter D, Letcher PM, Powell MJ, Taylor JW, White MM, Griffith GW, Davies DR, Humber RA, Morton JB, Sugiyama J, Rossman AY, Rogers JD, Pfister DH, Hewitt D, Hansen K, Hambleton S, Shoemaker RA, Kohlmeyer J, Volkmann-Kohlmeyer B, Spotts RA, Serdani M, Crous PW, Hughes KW, Matsuura K, Langer E, Langer G, Untereiner WA, Lücking R, Büdel B, Geiser DM, Aptroot A, Diederich P, Schmitt I, Schultz M, Yahr R, Hibbett DS, Lutzoni F, McLaughlin DJ, Spatafora JW, Vilgalys R (2006) Reconstructing the early evolution of fungi using a six-gene phylogeny. Nature 443:818–822. doi:10.1038/nature05110
Justo A, Hibbett DS (2011) Phylogenetic classification of Trametes (Basidiomycota, Polyporales) based on a five-marker dataset. Taxon 60:1567–1583
Katoh K, Toh H (2008) Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform 9:286–298
Kim KM, Lee JS, Jung HS (2007) Fomitopsis incarnatus sp. nov. based on generic evaluation of Fomitopsis and Rhodofomes. Mycologia 99:833–841. doi:10.3852/mycologia.99.6.833
Li HJ, Cui BK (2013) Taxonomy and phylogeny of the genus Megasporoporia and its related genera. Mycologia 105:368–383. doi:10.3852/12-114
Li HJ, Han ML, Cui BK (2013) Two new Fomitopsis species from southern China based on morphological and molecular characters. Mycol Prog 12:709–718. doi:10.1007/s11557-012-0882-2
Li HJ, Cui BK, Dai YC (2014) Taxonomy and multi-gene phylogeny of Datronia (Polyporales, Basidiomycota). Persoonia 32:170–182. doi:10.3767/003158514X681828
Miettinen O, Larsson KH (2011) Sidera, a new genus in Hymenochaetales with poroid and hydnoid species. Mycol Prog 10:131–141. doi:10.1007/s11557-010-0682-5
Miettinen O, Rajchenberg M (2012) Obba and Sebipora, new polypore genera related to Cinereomyces and Gelatoporia (Polyporales, Basidiomycota). Mycol Prog 11:131–147. doi:10.1007/s11557-010-0736-8
Miller MA, Holder MT, Vos R, Midford PE, Liebowitz T, Chan L, Hoover P, Warnow T (2009) The CIPRES portals. CIPRES. URL: http://www.phylo.org/sub_sections/portal. 2009-08-04. (Archived by WebCite(r) at http://www.webcitation.org/5imQlJeQa)
Núñez M, Ryvarden L (2001) East Asian polypores 2. Syn Fungal 14:165–522
Nylander JAA (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University
Petersen JH (1996) Farvekort. The Danish mycological society’s colour-chart. Foreningen til Svampekundskabens Fremme, Greve, pp 1–6
Posada D, Crandall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818
Robledo GL, Amalfi M, Castillo G, Rajchenberg M, Decock C (2009) Perenniporiella chaquenia sp. nov. and further notes on Perenniporiella and its relationships with Perenniporia (Poriales, Basidiomycota). Mycologia 101:657–673. doi:10.3852/08-040
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. doi:10.1093/bioinformatics/btg180
Ryvarden L (1985) Dichomitus eucalypti sp. nov. (Polyporaceae, Basidiomycota). Trans Br Mycol Soc 85:539–540
Ryvarden L, Johansen I (1980) A preliminary polypore flora of East Africa. Fungiflora, Oslo
Ryvarden L, Melo I (2014) Poroid fungi of Europe. Syn Fungal 31:1–455
Sotome K, Akagi Y, Lee SS, Ishikawa NK, Hattori T (2013) Taxonomic study of Favolus and Neofavolus gen. nov. segregated from Polyporus (Basidiomycota, Polyporales). Fungal Divers 58:245–266. doi:10.1007/s13225-012-0213-6
Spirin V, Miettinen O, Pennanen J, Kotiranta H, Niemela T (2013) Antrodia hyalina, a new polypore from Russia, and A. leucaena, new to Europe. Mycol Prog 12:53–61. doi:10.1007/s11557-012-0815-0
Swofford DL (2002) PAUP*: phylogenetic analysis using parsimony (*and other methods). Version 4.0b10. Sinauer, Sunderland
Tomšovský M, Menkis A, Vasaitis R (2010) Phylogenetic relationships in European Ceriporiopsis species inferred from nuclear and mitochondrial ribosomal DNA sequences. Fungal Biol 114:350–358. doi:10.1016/j.funbio.2010.02.004
Westphalen MC, Tomsovsky M, Kout J, Gugliotta AM (2015) Bjerkandera in the neotropics: phylogenetic and morphological relations of Tyromyces atroalbus and description of a new species. Mycol Prog 14:100. doi:10.1007/s11557-015-1124-1
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic, San Diego, pp 315–322
Yuan (2013) Dichomitus sinuolatus sp nov (Basidiomycota, Polyporales) from China and a key to the genus. Nova Hedwigia 97:495–501. doi:10.1127/0029-5035/2013/0098
Zhao CL, Cui BK, Dai YC (2013) New species and phylogeny of Perenniporia based on morphological and molecular characters. Fungal Divers 58:47–60. doi:10.1007/s13225-012-0177-6
Zhao CL, He XS, Wanghe KY, Cui BK, Dai YC (2014) Flammeopellis bambusicola gen. et. sp. nov. (Polyporales, Basidiomycota) evidenced by morphological characters and phylogenetic analysis. Mycol Prog 13:771–780. doi:10.1007/s11557-014-0960-8
Zhao CL, Cui BK, Song J, Dai YC (2015) Fragiliporiaceae, a new family of Polyporales (Basidiomycota). Fungal Divers 70:115–126. doi:10.1007/s13225-014-0299-0
Zhou LW, Vlasák J, Decock C, Assefa A, Stenlid J, Abate D, Wu SH, Dai YC (2016) Global diversity and taxonomy of the Inonotus linteus complex (Hymenochaetales, Basidiomycota): Sanghuangporus gen. nov., Tropicoporus excentrodendri and T. guanacastensis gen. et spp. nov., and 17 new combinations. Fungal Divers. doi:10.1007/s13225-015-0335-8
Acknowledgments
The research is supported by the National Natural Science Foundation of China (Project No. 31372115).
Author information
Authors and Affiliations
Corresponding author
Additional information
Section Editor: Franz Oberwinkler
Rights and permissions
About this article
Cite this article
Zhao, CL., Wu, F. & Dai, YC. Leifiporia rhizomorpha gen. et sp. nov. and L. eucalypti comb. nov. in Polyporaceae (Basidiomycota). Mycol Progress 15, 799–809 (2016). https://doi.org/10.1007/s11557-016-1210-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-016-1210-z