Abstract
The Golovinomyces biocellatus complex consists of powdery mildew (Erysiphales) species restricted to hosts of the family Lamiaceae. Previous authors used minor morphological features of the sexual state and host range data to split the complex. The data, however, were not sufficient to define a convincing species concept. Our taxonomic study is based on molecular phylogenetic and asexual state morphology data. For morphological studies, mainly features of the asexual morph (conidiophores, conidia, germination patterns) were studied using light and scanning electron microscopy. Detailed line drawings of asexual state features are provided. For phylogenetic analyses, two markers (rDNA: ITS, LSU) of 64 specimens were applied. The phylogeny resulted in two major clades. Clade I consists of specimens with Lamiaceae hosts and three specimens of Verbena. Clade II consists of two sister groups, the first (IIa) with Salvia spp. and the second (IIb) with Lycopus europaeus (the type host G. biocellatus) and Glechoma. Clades I and IIb and two subclades of IIa with Salvia hosts are characterized by specific morphological traits (differences in conidiophore length, conidial shape, width, and germination patterns). Based on these data, we suggest to consider specimens of clades I (including specimens on Verbena) and IIb and the two subclades of IIa as distinct species, namely G. monardae, G. biocellatus, G. salviae, and G. neosalviae sp. nov. A key for the identification of species based on asexual state features is provided. The results are discussed with respect to host range, jumps, co-evolutionary aspects, and distribution patterns.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Golovinomyces biocellatus (Ehrenb.) Heluta (Erysiphales) as described by Braun and Cook (2012) is a species with worldwide distribution and broad host range. Hosts are mainly species and genera of the subfamily Nepetoideae (tribe Mentheae) within the family Lamiaceae, but some host genera, namely Ajuga, Teucrium (Ajugoideae), Scutellaria (Scutellarioidae), Stachyopsis (Lamieae), and Stachys (Stachydaeae), are members of other subfamilies (Braun and Cook 2012; Lamiaceae classification after Bendiksby et al. 2011; Walker et al. 2004). This species is the only member of the genus Golovinomyces on the Lamiaceae. Several authors (e.g., Jaczewski 1927; Blumer 1933; Nagy 1977) tried to split the species because of many biologically specialized strains and based on differences in fruiting body size: according to Blumer (1933), the fungus on Salvia has smaller fruiting bodies (chasmothecia) than strains on other host genera (91–110 μm vs. 118–135 μm).
The latter feature, however, could not be confirmed by us in this study. We measured fruiting bodies on Salvia officinalis and S. lavandulifolia with up to 135 μm diam. (unpublished). Jaczewski’s (1927) measurements did not confirm Blumer’s (l.c.) results either. The sexual state was described well (although often rarely formed), whereas previous descriptions of the asexual morph were generally scarce. Most authors, including Ehrenberg (1821) in his protologue, did not provide any information on conidiophores and conidia.
Major information on the G. biocellatus complex was provided by Takamatsu et al. (2013) in their comprehensive phylogenetic analysis of the genus Golovinomyces. In this study using ITS and 28S rDNA sequences as markers, the authors confirmed close evolutionary relationships of Golovinomyces species and their host plants, with one clade containing G. biocellatus on various species of the Lamiaceae and on Verbena, a species of the family Verbenaceae which is closely related to Lamiaceae. The authors, however, could not confirm a special position of Salvia specimens within the complex. In fact, Salvia specimens were placed in two different subclades of the G. biocellatus clade (Takamatsu et al. 2013).
In order to decide whether the G. biocellatus complex has to be maintained or needs to be split into several taxa, additional taxonomically relevant information is required. In the following, we focused on more phylogenetic information from rDNA sequence data and on morphological descriptions of the asexual morph, since the sexual morphs (chasmothecia) are little differentiated and barely diagnostic (in Golovinomyces, they are rather uniform and useful for taxonomic purposes only to a very limited extent).
Materials and methods
Molecular analyses
Whole-cell DNA was extracted from mycelia using the chelex method (Walsh et al. 1991), as described by Hirata and Takamatsu (1996). The 5′-end of the 28S rDNA (including the domains D1 and D2) and internal transcribed spacer (ITS) regions were amplified by polymerase chain reaction (PCR) using the respective primer pairs: PM3 (Takamatsu and Kano 2001) and TW14 (Mori et al. 2000) for 28S, PM5/ITS4 for ITS fragment 1, and ITS5/PM6 (Takamatsu and Kano 2001) for ITS fragment 2. For some specimens, Golovinomyces-specific primers PM5G (5′-GACCCTCCACCCGTGT-3′) and PM6G (5′-CGAGCCCCAACACCAA-3′) were used instead of PM5 and PM6, respectively. KOD FX Neo DNA polymerase (Toyobo, Japan) was used in the PCR according to the manufacturer’s protocol. The amplicons of the 28S rDNA and ITS regions were sent to Solgent Co. Ltd. (Daejeon, South Korea) for direct sequencing using primer pairs of NL1 (Mori et al. 2000) and NLP2 (Hirose et al. 2005) for the 28S region, and ITS1 and ITS4 (White et al. 1990) for the ITS region. New sequences determined in this study were deposited in the DNA Data Bank of Japan (DDBJ) under the accession numbers LC076800–LC076842.
These sequences were aligned with other sequences of Golovinomyces retrieved from DNA databases using MUSCLE (Edgar 2004) implemented in MEGA6 (Tamura et al. 2013). Alignments were further manually refined using the MEGA6 program and were deposited in TreeBASE (http://www.treebase.org/) under the accession number S18141. Phylogenetic trees were obtained from the data using the maximum parsimony (MP) and maximum likelihood (ML) methods. MP analysis was performed in PAUP 4.0a144 (Swofford 2002) with heuristic search option using the tree bisection reconnection (TBR) algorithm with 100 random sequence additions to find the global optimum tree. All sites were treated as unordered and unweighted, with gaps treated as missing data. The strength of internal branches of the resulting trees was tested with bootstrap (BS) analysis using 1000 replications with the step-wise addition option set as simple (Felsenstein 1985). Tree scores, including tree length, consistency index (CI), retention index (RI), and rescaled consistency index (RC), were also calculated. The ML analysis was done using raxmlGUI (Silvestro and Michalak 2012), under a GTRGAMMA model. The BS supports and trees were obtained by running rapid bootstrap analysis of 1000 pseudo-replicates followed by a search for the tree with the highest likelihood.
Morphology
Specimens of mainly German origin (and a few others from Armenia, Hungary, and the USA) from KR, HAL, and PUL were studied. For light microscopy, both fresh (most specimens from KR, PUL) and dried (specimens from HAL, M, ZT, specimens KR-M-0043412 and KR-M-0043413 on Verbena, specimen KR-M-0018634 on Salvia pratensis) asexual structures of specimens were examined in tap water mounts using an Olympus BH2 and Zeiss Axioskop 2 Plus. Pertinent features were measured at magnifications of 400× and 1000× and documented by line drawings of fresh material. From each specimen, up to 25 conidia were measured. To induce conidial germination, the method of Schmidt and Scholler (2002) was applied. Fresh conidia were spread on a microscope slide and placed in a Petri dish with moist cellulose tissue. The closed Petri dishes were incubated at room temperature and exposed to daylight through a north-facing window for 24 h. For scanning electron microscopy (SEM), fresh fungus material with host tissue was submerged for two hours in 2 % glutaraldehyde buffered in a 0.1 M K3PO4 and 0.1 M Na3PO4 solution at pH 6.8. After 3× rinsing in the same buffer, samples were submerged in 1 % osmium tetroxide prepared with the phosphate buffer as well. After briefly rinsing in buffer, the material was dehydrated in an ethanol column of 30 %, 50 %, 70 % up to 100 % and dried with a critical point dryer. Sori were mounted with coloidal silver on metal blocks and coated with gold-palladium in a Hummer 1 sputter coater. Samples were studied with a JEOL JSM-840 scanning electron microscope with digital image acquisition at 5 kV accelerating voltage.
Results
Phylogenetic analyses
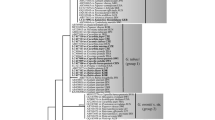
A total of 44 ITS and 28S rDNA sequences, including 40 from lamiaceous hosts and four from Verbena spp., were determined in this study. These sequences were aligned with 20 Golovinomyces sequences retrieved from DNA databases. The ITS + 28S rDNA combined dataset consists of 64 sequences and 1277 characters, of which 69 (5.4 %) characters were variable and 66 (5.2 %) characters were informative for parsimony analysis. A total of ca. 3 × 105 equally parsimonious trees with 84 steps (CI = 0.9524, RI = 0.9896, RC = 0.9425) were constructed by the MP analysis. The tree topologies were mostly consistent among the trees, except for branching orders of the terminal branches and branch length. One of the trees is shown in Fig. 1. ML analysis generated a tree topology almost identical to the MP tree. Thus, only BS values of the ML analysis are shown in Fig. 1. The 59 sequences from the G. biocellatus complex are divided into two distinct large clades. Clade I consists of 37 sequences from the specimens on Melissa, Mentha, Monarda, Origanum, Rosmarinus, Thymus (Lamiaceae), and Verbena (Verbenaceae), and one sequence from a specimen on Salvia collected in Australia. This clade is homogeneous, and supported by strong BS supports (MP = 99 %, ML 96 %). Clade II is also supported by high BS values (MP = 99 %, ML = 98 %) and divided into two subgroups, IIa and IIb. Subgroup IIa is a paraphyletic group composed of specimens on Salvia spp. This group further splits into four small groups. Subgroup IIb consists of specimens on Lycopus and Glechoma and forms a distinct clade (MP = 75 %, ML = 81 %). Two sequences from the materials on Verbena did not belong to the G. biocellatus complex, and form a clade with G. magnicellulatus with high BS supports (MP = 100 %, ML = 100 %).
A phylogenetic tree of the Golovinomyces biocellatus complex inferred from the combined dataset of the rDNA ITS regions and the divergent domains D1 and D2 sequences of the 28S rDNA. This tree is a phylogram of one of the 485,103 most parsimony trees with 84 steps, which was found using a heuristic search by PAUP*. Horizontal branch lengths are proportional to the number of substitutions that were inferred to have occurred along a particular branch of the tree. Bootstrap (BS) (≧70 %) values by the maximum parsimony (MP) and maximum likelihood (ML) methods are shown on the respective branch
Taxonomy
Clades and subgroups resulting from our phylogeny (Fig. 1) are supported by features of the asexual morphs. Therefore, we suggest to place species on Melissa, Mentha, Monarda, Origanum, Rosmarinus, Thymus (Lamiaceae), and Verbena (Verbenaceae) of clade I in a separate species, G. monardae. Specimens of subgroup IIb with host species of the genera Lycopus and Glechoma represent G. biocellatus s. str. Subgroup IIa consists of Old World specimens on Salvia. Major morphological differences (conidiophore length) between specimens on S. fruticosa, S. lavandulifolia, S. officinalis, and those on S. pratensis and S. nemorosa are supported by the taxonomy of the hosts. Both groups also form a distinct clade; however, the two clades have no strong bootstrap support. Despite low bootstrap support, but based on obvious morphological and host differences, we propose two species on Salvia, G. salviae comb. nov. and G. neosalviae sp. nov.
The following descriptions refer to asexual and partly sexual morphs of accepted species within the G. biocellatus complex:
Golovinomyces monardae (G.S. Nagy) M. Scholler, U. Braun & Anke Schmidt, comb. nov. (Figs. 2 and 3)
MycoBank, MB815612
Basionym: Erysiphe monardae G.S. Nagy, Phytopathol. Z. 88: 285 (1977); holotype: Hungary, Budapest, garden of the University of Horticulture, on Monarda didyma L., 26 Aug. 1976, G.S. Nagy (HAL 2919 F).
≡ E. biocellata var. monardae (Nagy) U. Braun, Zentralbl. Mikrobiol. 137: 316 (1982).
= E. cichoracearum f. menthae Jacz., Karmany opredelitel’ gribov. Vyp. 2: 217 (1927).
Mycelium more (e.g., on Monarda) or less (e.g., on Origanum) conspicuous, on stems and leaves, amphigenous, but more evident on the upper leaf surface, initially in patches, later effuse, often around leaf veins, whitish; hyphae hyaline, 4–8 μm wide; hyphal appressoria slightly or distinctly nipple-shaped, developing rarely in May to June, more frequently from July to August; conidiophores 50–145 μm long and 9–14 μm wide, partly widening from base to top, rarely about central on mother cell, mostly arising about 1/3–2/3 towards one end of the mother cell, no difference between epiphyllous and hypophyllous conidiophores, foot-cell 40–100(−140) μm long, mostly curved at the base, basal septum of the foot-cell partly raised above the junction with the hyphal mother cell, foot-cell followed by 0–3 shorter cells, forming catenescent conidia; conidia mostly ellipsoid, partly doliiform, swelling up in water, particularly in width, (24.4–)30–38(−46) × (16.4–)18–22(−24) μm, length/width ratio 1.5–2.0 (average 1.7); germ tubes subapically inserted, mostly without septum, rarely one-septate in short germ tubes, germ tubes mostly short to moderately long, sometimes also long, 11–84(−192) × 3.5–8.5 μm, terminating simply or in a club-shaped appressorium.
Sexual morph: see Braun and Cook (2012: 305), under Golovinomyces biocellatus (chasmothecia of G. monardae indistinguishable).
Specimens studied
On Lamiaceae: On Melissa officinalis L.: Germany, Schleswig-Holstein, Lübeck, St. Gertrud, Grootkoppel 6, Flora nursery, flower-pot, 10 Jun. 2010, leg. A. Schmidt (KR-M-0027770) (GenBank LC076811); Baden-Württemberg, Karlsruhe, Grünwinkel, Silcherstr. 25, garden, 25 Jul. 2011, leg. M. Scholler (KR-M-0033268) (GenBank LC076821). On Mentha aquatica Ehrenb.: Germany, Schleswig-Holstein, Lübeck, Lauerholz, Wesloer Moor, 18 Sep. 1999, leg. A. Schmidt (KR-M-0020808) (GenBank LC076804); Schleswig-Holstein, Ostholstein near Malente between Kellersee and Krummsee, lakeside, 18 Sep. 2012, leg. A. Schmidt (KR-M-0035015) (GenBank LC076826); Mecklenburg-Vorpommern, Klein Schmölen, Schmölener Brack, bank, 20 Oct. 2012, leg. A. Schmidt (KR-M-0035011) (GenBank LC076822); Mecklenburg-Vorpommern, Klein Schmölen, Schmölener Brack, bank, 20 Oct. 2012, leg. A. Schmidt (KR-M-0035016). On Mentha × piperita L. ‘chocolate’: Germany, Schleswig-Holstein, Lübeck, St. Gertrud, Grootkoppel 6, Flora nursery, two flower-pots, 17 Jun. 2009, leg. A. Schmidt (KR-M-0024166) (GenBank LC076810); Schleswig-Holstein, Lübeck, St. Gertrud, Grootkoppel 6, Flora nursery, flower-pot, 22 May 2014 (asexual state, KR-M-0039096) (GenBank LC076835) and 19 Jun. 2014 (teleom.; KR-M-0039097) (GenBank LC076836), leg. A. Schmidt. On Monarda citriodora Cerv. ex Lag.: Germany, Baden-Württemberg, Konstanz, botanical garden, 25 Jul. 2000, leg. H. Jage (KR-M-0022183) (GenBank LC076807); Sachsen-Anhalt, Halle (Saale), botanical garden, 02 Aug. 2004, leg. U. Braun (KR-M-0024044) (GenBank LC076809). On Monarda didyma L.: Armenia, Yerevan, botanical garden, 03 Sep. 1952, leg. S. Simonyan (HAL 1040 F); USA, Wisconsin, Sauke County, Parfrey’s Glen, 03 Oct. 1956, H.C. Greene (HAL 1105 F); Germany, Bezirk Rostock, 1980, Neukloster, leg. H. Henker (HAL 1106 F); Baden-Württemberg, Isny, Großholzleute, Gasthof Adler, flowerbed, 28 Jun. 2008, leg. M. Scholler (KR-M-0002831) (GenBank LC076800); Baden-Württemberg, Konstanz, botanical garden, 25 Jul. 2000, leg. H. Jage (KR-M-0022187) (GenBank LC076808); Baden-Württemberg, Isny, Großholzleute, Gasthof Adler, flowerbed, 28 Jun. 2008, leg. M. Scholler (KR-M-004090) (GenBank LC076802). On Monarda-hybrid ‘Prärienacht’: Germany, Hamburg, Klein-Flottbek, botanical garden, 16 Jul. 2011, leg. A. Schmidt (KR-M-0033267) (GenBank LC076820). On Monarda fistulosa L. ‘Blaustrumpf’: Germany, Schleswig-Holstein, Herzogtum Lauenburg, Groß Grönau, An der Gärtnerei 1, Aeschlimann nursery, flower-pot, 27 Jun. 2012 (asexual state; KR-M-0035018) (GenBank LC076827) and 09 Sep. 2012 (teleom.; KR-M-0035019) (GenBank LC076828), leg. A. Schmidt, on Monarda fistulosa L. ‘Schneewittchen’: Schleswig-Holstein, Herzogtum Lauenburg, Groß Grönau, An der Gärtnerei 1, Aeschlimann nursery, flower-pot, 27 Jun. 2012 (asexual state; KR-M-0035022) (GenBank LC076829) and 11 Sep. 2012 (teleom.; KR-M-0035023) (GenBank LC076830), leg. A. Schmidt. On Origanum majorana Moench: Germany, Schleswig-Holstein, Lübeck, St. Gertrud, Grootkoppel 6, Flora nursery, flower-pot, 12 Jul. 2011, leg. A. Schmidt (KR-M-0033265) (GenBank LC076818). On Origanum vulgare L.: Germany, Schleswig-Holstein, Lübeck, St. Gertrud, Grootkoppel 6, Flora nursery, flower-pot, 29 Jun. 2010, leg. A. Schmidt (KR-M-0027771) (GenBank LC076812). On Thymus × citriodorus (Pers.) Schreb. ex Schweigg.: Germany, Schleswig-Holstein, Ostholstein, Schürsdorf, Sandenredder 13, Rahlf nursery, flower-pot, 2 Aug. 2011 (asexual state; KR-M-0033259) (GenBank LC076815) and 25 Sep. 2011 (teleom.; KR-M-0033260) (GenBank LC076816), leg. A. Schmidt. On Thymus × citriodorus (Pers.) Schreb. ex Schweigg. ‘Variegata’: Germany, Schleswig-Holstein, Lübeck, St. Gertrud, Grootkoppel 6, Flora nursery, flower-pot, 2 Aug. 2011, leg. A. Schmidt (KR-M-0033261) (GenBank LC076817).
On Verbenaceae: On Verbena bonariensis L.: Germany, Brandenburg, Oranienburg, Schloßgarten, 26 Aug. 2009, leg. V. Kummer (KR-M-0043413) (GenBank LC076842). On Verbena sp.: Germany, Brandenburg, Potsdam, botanical garden, 27 Sep. 1996, leg. V. Kummer (KR-M-0043412).
The following specimens on Verbena cannot be assigned to a species known to occur on Verbena according to Braun and Cook 2012 (Golovinomyces monardae, G. verbenae (Schwein.) Heluta) and do not belong to the G. biocellatus complex (see outgroup in Fig. 1). Morphologically, these differ from the latter two species by a higher number of conidiophore cells (4–6 vs. 1–3):
On Verbenaceae: On Verbena hastata L.: Germany, Brandenburg, Potsdam, Botanical Garden, 25 Sep. 2003, leg. V. Kummer (KR-M-0043410) (GenBank LC076839). On Verbena sp.: Germany, Brandenburg, Oberspreewald-Lausitz, market place, 9 Sep. 2002, leg. V. Kummer (KR-M-0043411) (GenBank LC076840).
Golovinomyces biocellatus (Ehrenb.) Heluta, Ukrayins’k. Bot. Zhurn. 45: 62 (1988) emend. M. Scholler, U. Braun & Anke Schmidt (Figs. 4, 5, and 6)
≡ Erysiphe biocellata Ehrenb., Nova Acta Phys.-Med. Acad. Caes. Leop.-Carol. Nat. Cur. 10: 211 (1821); lectotype [designated by Braun (1987)]: Ehrenberg (1821: Pl. 13, original drawing).
≡ Erysibe biocellata (“biocellaris”) (Ehrenb.) Link, Sp. pl. 4, 6(1): 109 (1824).
≡ Erysiphe communis f. biocellata (Ehrenb.) Fr., Syst. Mycol. 3: 239 (1829).
= Oidium erysiphoides Fr., Syst. Mycol. 3: 432 (1832).
≡ Euoidium erysiphoides (Fr.) Y.S. Paul & J.N. Kapoor, Indian Phytopathol. 38: 762 (1985), nom. inval. (ICN, Art. 33.3).
≡ Euoidium erysiphoides (Fr.) Y.S. Paul & J.N. Kapoor, Indian J. Mycol. Pl. Pathol. 17: 302 (1987), nom. inval. (ICN, Art. 33.3).
= E. cichoracearum f. lycopi Jacz. (Jaczewski 1927: 216).
Mycelium not conspicuous, on stems and leaves, amphigenous, rarely in patches, mostly effuse, often around leaf veins, thin, whitish to grayish; hyphae hyaline, 4–6 μm wide; hyphal appressoria not frequently formed, nipple-shaped; conidiophores 70–145 μm long and (8–) 10–12(−14) μm wide, partly increasing from base to top, erect or curved at the base, rarely about central on mother cell, mostly arising about 1/3–2/3 or 1/4–3/4 towards one end of the mother cell, no difference between epiphyllous and hypophyllous conidiophores, foot-cell, longest cell of conidiophore, 55–100(−130) μm long, basal septum of the foot-cell partly raised above the junction with the hyphal mother cell, foot-cell followed by 0–3 shorter cells, forming catenescent conidia; conidia predominantly doliiform to limoniform, occasionally ellipsoid, 28–36(−43) × (18–)19–24(−26) μm, length/width ratio 1.3–2.0 (average 1.6), surface (SEM) polygonal, almost reticulate ridges; germ tubes subapically inserted, non- to one-septate (in relation about 1:1), mostly short to moderately long, sometimes also long, 24–72(−156) × 2.4–6.0 μm, terminating simply or in a club-shaped appressorium.
Sexual morph: see Braun and Cook (2012: 305).
On Glechoma hederacea L.: USA., Indiana, West Lafayette, Purdue University, greenhouse near Lilly Hall, 17 Sep. 2001, leg. M. Scholler (PUL F1563) (GenBank KX229701); Germany, Schleswig-Holstein, Lübeck, Israelsdorf, Holunderweg 5, garden, 04 Jul. 2007, leg. A. Schmidt (KR-M-21890) (GenBank LC076805); Niedersachsen, near Wulmstorf, Röttiger-barracks, way to the former military training area, 25 Aug. 2007, leg. A. Schmidt (KR-M-0021901) (GenBank LC076806). On Lycopus europaeus L.: Germany, Baden-Württemberg, Karlsruhe, Schloßpark, edge of the pond, 09 Jul. 2010, leg. A. Schmidt (KR-M-0027775) (GenBank LC076814); Schleswig-Holstein, Lübeck, St. Gertrud, city park, lakeside, 26 Aug. 2012, leg. A. Schmidt (KR-M-0035014) (GenBank LC076825); Schleswig-Holstein, Lübeck, St. Gertrud, city park, lakeside, 26 Aug. 2012 (asexual state; KR-M-0035026) (GenBank LC076831) and 14 Oct. 2012 (teleom.; KR-M-0035027) (GenBank LC076832), leg. A. Schmidt.
Golovinomyces salviae (Jacz.) M. Scholler, U. Braun & Anke Schmidt, comb. nov. (Figs. 7, 8, and 9)
MycoBank, MB815613
Basionym: Erysiphe labiatarum f. salviae Jacz., Karmanyi opredelitel’ gribov. Vyp. 2. Muchnisto-rosyanye griby: 162 (1927) (type hosts: Salvia glutinosa L., S. nutans L. S. pratensis L., S. silvestris, S. verticillata, S. virgata, S. sp.); lectotype* (designated here, MycoBank, MBT203828): On S. pratensis, Switzerland, Valais (Wallis), Riddes, Ecône, Sep. 1893, 500 m, leg. Abbé Besse (LEP 130001 ex herb. A. de Jaczewski 2214). *Note: Jaczewski (1927) introduced f. salvia for collections on several Salvia species, but without specification of the type material. He cited particular hosts and localities in Russia and mentioned that this forma also occurs in west Europe. The lectotype on Salvia pratensis is a collection from Jaczewski’s herbarium collected before 1927, which is accordingly qualified for a lectotypification.
≡ E. salviae (Jacz.) S. Blumer, Beitr. Krypt.-Fl. Schweiz 7(1): 273 (1933).
? = Oidium hormini Farneti, Atti Ist. Bot. Univ. Pavia 7: 255 (1902).
= Oidium verbenacea Pass., in Thüm., Mycoth. Univ., Cent. VIII, No. 789, Klosterneuburg 1877; lectotype (designated here, MycoBank, MBT203829): On Salvia verbenaca, Italy, Parma, May 1876, G. Passerini, Thüm., Mycoth. Univ. 789 (M-0014092).
? = E. cichoracearum f. salviae Koshk., Bot. Mater. Otd. Sporov. Rast. Bot. Inst. Komarova Akad. Nauk S. S. S. R. 14: 123 (1961) (type host S. virgata).
= E. simplex Heluta, Ukrayins’k. Bot. Zhurn. 43(5): 53 (1986), nom. illeg. (nom. superfl.).
≡ Golovinomyces simplex (Heluta) Heluta, Ukrayins’k. Bot. Zhurn. 45(5): 63 (1988).
Mycelium conspicuous, on leaves, hypophyllous or amphigenous, sometimes causing reddish spots epiphyllously, in distinct patches (later effuse), whitish to grayish; hyphae hyaline, 4–7(−10) μm wide; hyphal appressoria nipple-shaped to slightly lobed; conidiophores (50–)60–140 μm long and 8–15 μm wide (sometimes basal cell swelling up to 20 μm in the middle), erect or curved at the base, formed about 1/3–2/3 or 1/4–3/4 towards one end of the mother cell, basal septum of the foot-cell mostly raised 5–15 μm above junction with mother cell, 1–3(−4) cells, basal cell longest, 30–100(−120) μm, followed by 0–3 shorter cells, forming catenescent conidia; conidia doliiform, rarely cylindrical, (25–)30–36(−45) × (15–)20–24 μm, length/width ratio 1.3–1.7 (average 1.5); germ tube terminating in a club-shaped appressorium.
Sexual morph: Chasmothecia scattered to gregarious, subglobose, (75–)90–125 μm diam., peridium cells irregularly polygonal to daedaleoid, 5–25 μm diam., appendages numerous, arising from the lower half, mycelioid, simple, rarely branched, 0.5–2 times as long as the chasmothecial diam., 3–7 μm wide, septate, hyaline to pale brown, thin-walled, smooth or almost so, asci 5–13 per chasmothecium, broadly ovoid-saccate, 40–80 × 25–45 μm, subsessile to short-stalked, thin-walled, without distinct terminal oculus, 2-spored, ascospores ellipsoid, 17–28 × 10–17 μm, colorless.
On Salvia pratensis L.: Germany, Baden-Württemberg, Owingen, NSG an der Eyach N Weilerkirche, dry grassland, south slope, 19 Aug. 2005, leg. M. Scholler (KR-M-0018634) (GenBank LC076803). On Salvia nemorosa L.: Germany, Berlin, Kreuzberg, entrance to Marheinikeplatz 13, flower-pot, 04 Sep. 2015, leg. M. Scholler (KR-M-0046020) (GenBank LC100001).
Further specimens studied with light microscopy
On Salvia pratensis L.: Germany, Baden-Württemberg. Kreis Aalen, Goldberg near Goldburghausen, 23 Sep. 1967, H. & H. Doppelbaur (asexual state; M-0014107); Baden-Württemberg. Kreis Wangen, Großholzleute, 3 Sep. 1964, O. Klement (asexual state; M-0014106); Bavaria, Nördlingen, 19 Sep. 1955, K. Ruttmann (asexual state; M-14120); Bavaria, Schwaben, Landkreis Günzburg, Offingen, 25 Sep. 1968, H. & H. Doppelbaur (asexual state; M-0014118); Saarland, Habkirchen, Bliestal, 1 Sep. 2012, U. Richter (asexual state; HAL 2977 F).
On Salvia nutans L.: Ukraine, Melitopol, Altagir, 2 June 1984, leg. U. Braun (asexual and sexual state; HAL 1049 F).
On Salvia verbenaca L. Italy, Parma, May 1876, Passerini, Thüm., Mycoth. Univ. 8789 (asexual state; M-0014092).
On Salvia verticillata L.: Armenia, Stepanavan, Dendropark “Sosnyaki”, 10 Sep. 1949, D. N. Babayan (asexual and sexual state; HAL 1048 F). Germany, Bavaria, Oberbayern, Landkreis Altöttingen, Burghausen, Alter Burgberg, 2 Sep. 1969, H. & H. Doppelbaur (asexual state; M-0014119). Romania, distr. Craiova, Oltenia, garden, 17 Oct. 1965, I. Comes et al., Flora Olteniae Exsiccata 542 (asexual and sexual state; M-0014108, ZT Myc 55370). Russia, Bashkortostan, Ufa, slope on the river Belaya with steppe vegetation, 5 Jul. 1977, U. Braun 514 (asexual and sexual state; HAL 1341 F). Switzerland, Cantoin de Vaud, between Montcherand and Orbe, 17 Sep. 1943, E. Mayor (ZT Myc 55369); Canton Neuchâtel, between Frensens and Vernéaz, 11 Jul. 1951, E. Mayor (ZT Myc 55365).
Golovinomyces neosalviae M. Scholler, U. Braun & Anke Schmidt, sp. nov. (Figs. 10 and 11)
MycoBank, MB815614
Mycelium conspicuous, on stems and leaves, amphigenous, but mainly epiphyllous, initially in distinct patches, later effuse, covering the entire surface of the leaves, whitish to grayish, infected leaves soon become yellowish and falling off already in July or August, after that fresh leaves growing without any infection; hyphae hyaline, 4–7 μm wide; hyphal appressoria not frequently formed, nipple-shaped to slightly lobed; conidiophores 140–400 μm long and (8–)9–13(−14.5) μm wide, often increasing from base to top, erect or curved at the base, central on mother cell or formed about 1/3–2/3 or 1/4–3/4 towards one end of the mother cells, no difference between epiphyllous and hypophyllous conidiophores, basal septum of the foot-cell always raised 7–25 μm above junction with mother cell, size and arrangement of cells very variable, mostly at the base with 1–3 cells between 45 and 75(−115) μm long, followed by the longest cell of 105–205 μm, followed by 0–3 shorter cells, or, rarely, foot-cell, longest cell of conidiophore, followed by 1–3 shorter cells, forming catenescent conidia; conidia distinctly doliiform to limoniform, rarely ellipsoid, swelling up in water, particularly in width, (28.5–)33–44(−47.5) × (20–)22–26(−28) μm, length/width ratio 1.2–2.2 (average 1.7); germ tubes subapically inserted, non- to one-septate (in relation about 1:2), mostly short to moderately long, sometimes also long, 18–96(−170) × 2.4–10.8 μm, terminating simply or in a club-shaped appressorium.
Sexual morph: Chasmothecia scattered to subgregarious, subglobose, 100–135 μm diam., peridium cells irregularly polygonal to daedaleoid, 7–30 μm diam., appendages numerous, arising from the lower half, mycelioid, simple, rarely branched, about as long as the chasmothecial diameter or shorter, 3–8 μm wide, septate, hyaline to pale brown towards the base, thin-walled, smooth or almost so, asci 5–10 per chasmothecium, broadly ovoid-saccate, subcylindrical or somewhat asymmetric, 50–80 × 25–45 μm, subsessile to distinctly stalked, thin-walled (to 1 μm), without distinct terminal oculus, 2-spored, ascospores ellipsoid to somewhat ellipsoid-ovoid, 18–35 × 12–18 μm, colorless.
Holotype: On S. officinalis L. ‘Purpurascens’: Germany, Schleswig-Holstein, Lübeck, St. Gertrud, Grootkoppel 6, Flora nursery, two flower-pots, 04 Jun. 2012, leg. A. Schmidt (KR-M-0035013) (GenBank LC076824).
Further specimens studied
On S. fruticosa Miller: Germany, Baden-Württemberg, Karlsruhe, botanical garden, 06 Jul. 2011, leg. A. Schmidt (KR-M-33266) (GenBank LC076819). On S. lavandulifolia Vahl: Germany, Schleswig-Holstein, Ostholstein, Schürsdorf, Sandenredder 13, Rahlf nursery, two flower-pots, 07 Jun. 2014 (asexual state; KR-M-39098) (GenBank LC076837) and 03 Jul. 2014 (teleom.; KR-M-39099) (GenBank LC076838), leg. A. Schmidt. On Salvia cf. officinalis L.: Germany, Schleswig-Holstein, Lübeck, St. Gertrud, Grootkoppel 6, Flora nursery, flower-pot, 10 Jun. 2010, leg. A. Schmidt (KR-M-27772) (GenBank LC076813). On S. officinalis L. ‘Icterina’: Germany, Schleswig-Holstein, Lübeck, Grootkoppel 6, Flora nursery, three flower-pots, 04 Jun. 2012, leg. A. Schmidt (KR-M-35012) (GenBank LC076823). On S. officinalis L.: Germany, Schleswig-Holstein, Lübeck, St. Gertrud, Grootkoppel 6, Flora nursery, flower-pot, 01 Jun. 2013, leg. A. Schmidt (KR-M-38207) (GenBank LC076834); Schleswig-Holstein, Herzogtum Lauenburg, Groß Grönau, An der Gärtnerei 1, Aeschlimann nursery, flower-pot, 04 Jun. 2013, leg. A. Schmidt (KR-M-0038206); Schleswig-Holstein, Lübeck, St. Gertrud, Wesloer Landstraße 72–74, OBI, flower-pot, 05 Jul. 2013 (asexual state; KR-M-0038203) (GenBank LC076833) and 03 Aug. 2013 (teleom.; KR-M-0038204), leg. A. Schmidt. Switzerland, Neuchâtel, garden, 15 Aug. 1944, leg. E. Mayor (ZT Myc 55367. 55369).
Key to the recognized species of the Golovinomyces biocellatus complex and other Golovinomyces spp. on Verbenaceae and Lamiaceae based on features of the asexual morph:
On Verbenaceae (Verbena spp.):
1 Conidiophores curved at base2
1* Conidiophores not curved at baseGolovinomyces verbenae
2 Conidiophore cells 4–6G. sp.
2* Conidiophore cells 1–33
3 Conidia sometimes below 16 μm wideG. orontii s. lat. (= G. monardae?)
3* Conidia almost never below 16 μm wideG. monardae
On Mentheae
1 On Salvia s. str. (Salvia clade 1 species, Walker et al. 2004), conidiophores either short (60–140 μm) or long (140–400 μm)2
1* Not on Salvia s. str., conidiophores 70–145 μm3
2 Conidiophores 60–140 μm longGolovinomyces salviae
2* Conidiophores 140–400 μm longG. neosalviae
3 Conidia mostly ellipsoid, conidial germ tubes non-septate, very rarely one-septate
G. monardae
3*Conidia mostly doliiform to limoniform, conidial germ tubes often one-septate
G. biocellatus
Discussion
Powdery mildew on lamiaceous hosts, nowadays belonging to Golovinomyces, were previously assigned to Erysiphe cichoracearum sensu latissimo (Salmon 1900, Jaczewski 1927) comprising most species of the current genus Golovinomyces (“Erysiphe sect. Euerysiphe” sensu Blumer 1967). Blumer (1933, 1967) split E. cichoracearum s. lat. into several species, recognized the corresponding powdery mildews on Lamiaceae as a distinct species, and reintroduced the name Erysiphe biocellata. However, Blumer (1933) excluded the powdery mildew on Salvia species, treated it as a separate species, and introduced the combination Erysiphe salviae, based on E. labiatarum f. salviae (Jaczewski 1927). Braun (1987) used a broad concept for the name Erysiphe biocellata, encompassing all corresponding collections on lamiaceous hosts, including those on Salvia spp., i.e., E. salviae was reduced to synonymy with E. biocellata, but without any discussion. Braun (1995) followed the broad concept of E. biocellata, but added a brief discussion. Braun and Cook (2012) maintained this broad concept, provided a more detailed discussion, and emphasized the necessity of comprehensive biological and molecular examinations for the expected further splitting of G. biocellatus into smaller taxonomic units. Hence, the present investigations aimed at clarifying the unresolved phylogeny and taxonomy of the G. biocellatus complex. The results are clear and convincing with respect to host range and morphology. Concerning the host range, we could not confirm G. biocellatus s. lat. on Ajuga, Scutellaria, Stachys, or Teucrium, which belong to host subfamilies other than Nepetoideae. The powdery mildews on these plants were often named Golovinomyces biocellatus (and listed accordingly in the recent literature, e.g., in Braun and Cook 2012; Klenke and Scholler 2015), but turned out to be Neoerysiphe galeopsidis (DC.) U. Braun. We think that species of the G. biocellatus complex are restricted to members of the Lamiaceae subfamily Nepetoideae tribe Mentheae, although occasional infections of plants of other Lamiaceae subfamilies, tribes, or related families such as Verbenaceae (see below) may occur. The phylogenetic data are unambiguously in favor of two well-supported clades, I and II (Fig. 1). Clade I including Monarda spp. and numerous other host plant species and genera is considered an own species for which the name Erysiphe monardae is available. This species differs from G. biocellatus in morphological traits of its asexual morphs. The sexual morphs (chasmothecia) of the two species are little differentiated and barely diagnostic, which is, however, not surprising since fruiting bodies in Golovinomyces are rather uniform and, for taxonomic purposes, only of very limited use. Taking these results into account, the new combination Golovinomyces monardae is introduced. This plurivorous species occurs, as far as known and examined, on diverse hosts of genera belonging to the Nepetoideae and is obviously able to infect Verbena spp., at least accidentally. Braun and Cook (2012) recognized two species of the genus Golovinomyces on Verbena spp., viz. G. verbenae (Schwein.) Heluta and the plurivorous G. orontii (Castagne) Heluta. Previous records of G. orontii (= Erysiphe polyphaga) on Verbena spp. possibly refer to G. monardae. An additional, possibly undescribed species characterized by up to six conidiophore cells was found within the framework of this study. It clusters outside the G. biocellatus complex in our phylogeny (Fig. 1). Provided that G. monardae is different from G. orontii, four species of Golovinomyces occur on Verbena (see key).
Golovinomyces on Salvia spp. represents a particular case. All sequences retrieved from Salvia powdery mildew included in the present analysis cluster adjacent to the G. biocellatus s. str. clade (clade IIa), but in any case in separate groups, i.e., not mixed with G. biocellatus sequences. There is no bootstrap support for the Salvia powdery mildew at the species level, at least by ITS and LSU data, but the conidiophores and conidia are quite distinct from those of G. biocellatus (see descriptions and key) and due to rather broad, doliiform–limoniform conidia, they are rather reminiscent of the asexual state of species belonging to Golovinomyces sect. Depressi (U. Braun) U. Braun (Braun and Cook 2012), which is, however, a non-phylogenetic section just based on morphology. Two groups within clade IIa may be distinguished, one with three related host plants (S. officinalis, S. lavandulifolia, S. fruticosa) and very long conidiophores, and another one with two related host plants (S. pratensis, S. nemorosa) with very short conidiophores. Although the two features are not supported by sequence data, we think that the morphological and the host range data suggest that powdery mildews on Salvia spp. should be better considered as two species of their own, which we call G. salviae (lectotype host S. pratensis) and G. neosalviae (type host S. officinalis). All host species belong to clade I in the Salvia phylogeny performed by Walker et al. (2004). ITS sequences are often not sufficient for a reliable resolution at the species level (Kovács et al. 2011; Kiss 2012). Additional markers are probably necessary. Furthermore, there are additional Salvia species reported to be hosts of G. salviae that have not yet been included in the phylogenetic analyses. Characters of the chasmothecia, above all the size of the fruiting bodies, are rather variable and were previously used for the differentiation between particular species in the G. biocellatus complex. Blumer (1933) and Heluta (1989) confined E. salviae (E. simplex) to collections with relatively small chasmothecia. Blumer (l.c.) included Salvia glutinosa, S. pratensis, and S. verticillata as hosts, whereas Heluta (1989) cited only S. glutinosa and S. verticillata, and assigned collections on S. pratensis and other hosts to G. biocellatus. However, based on Braun and Cook (2012) and our own examinations, the chasmothecial size is rather variable and barely applicable for taxonomic purposes. The key to solve the taxonomic problems around Salvia powdery mildew lies in the asexual morph, which has been little and insufficiently examined until now. The biological specialization of Salvia powdery mildews is barely known. Laibach (1930) carried out inoculation experiments and stated that powdery mildew from Salvia verticillata was not able to infect S. pratensis, and with powdery mildew from S. pratensis, he could successfully infect S. dumetorum, S. pratensis × sylvestris, but not S. glutinosa, S. verticillata, S. officinalis, and other hosts, suggesting that biological specialization may occur among powdery mildew on Salvia spp., but modern examinations under verifiable conditions are not available. The study of the Golovinomyces species on S. glutinosa, which we could not examine molecularly, will be of particular interest because this plant belongs to Salvia clade III species and not clade I, as demonstrated in the phylogeny of Walker et al. (2004). Several herbarium specimens on S. glutinosa (only asexual morphs) have been examined [Germany, Baden-Württemberg, Geißkopf, 3 Sept. 1964, O. Klement 12993 (M-0014104); Bavaria, Rosenheim, between Gattern and Kraimoos, 25 Aug. 1934, H. Poeverlein (M-0014115); Bavaria, Rosenheim, near Aschau, 20 Aug. 1934, H. Poeverlein (M-0014116); Bavaria, Rosenheim, above Hohenaschau, 24 Aug. 1934, H. Poeverlein (M-0014117). Romania, Baile Herculeane, Valea Cernei, 8 Sep. 1962, E. Eliade (M-0014105). Switzerland, Canton Neuchâtel, Neuchâtel, Candolles, 21 Sep. 1945, E. Mayor (ZT Myc 55366)]. Conidiophores and conidia of all samples agree well with G. salviae, but since sequence data retrieved from S. glutinosa powdery mildew are not yet available, we refrain from a final taxonomic conclusion. Also, Salvia sp. from Australia hosting G. monardae (AF154323, see Fig. 1) may be a Salvia clade III species.
Due to the fact that we could not confirm Ajuga, Stachys, or Teucrium as hosts of G. biocellatus s. lat., there is evidence that species of the G. biocellatus complex are host-specific and restricted to species of the Mentheae within the Lamiaceae (apart from occasional infections of Verbena spp.). The present analyses are, of course, just a first step in resolving problems and open questions around the G. biocellatus complex. Although the results are unequivocal, the morphological examinations of asexual morphs and phylogenetic data were mainly focused on Central European collections. A broader sampling is necessary for a more detailed verification of the distribution of the particular species.
References
Bendiksby M, Thorbek L, Scheen AC, Lindqvist C, Ryding O (2011) An updated phylogeny and classification of Lamiaceae subfamily Lamioideae. Taxon 60:471–484
Blumer S (1933) Die Erysiphaceen Mitteleuropas unter besonderer Berücksichtigung der Schweiz. Beiträge zur Kryptogamenflora der Schweiz 7:1–483
Blumer S (1967) Echte Mehltaupilz (Erysiphaceae). G. Fischer Verlag, Jena
Braun U (1987) A monograph of the Erysiphales (powdery mildews). Beihefte zur Nova Hedwigia 89:1–700
Braun U (1995) The powdery mildews (Erysiphales) of Europe. G. Fischer, Jena, 337 pp
Braun U, Cook RTA (2012) Taxonomic manual of the Erysiphales (powdery mildews). CBS Biodiversity Series 11. CBS, Utrecht, 707 pp
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Ehrenberg CG (1821) De Mycetogenesi ad Acad. C.L.C.N.C. Praesidem Epistola. Nova Acta Phys-Med Acad Caesareae Leopoldino-Carolinae Naturae Curiosorum 10(1):160–222
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Heluta VP (1989) Flora Gribov Ukrainy. Muchnistorosyanye griby. Naukova Dumka, Kiev
Hirata T, Takamatsu S (1996) Nucleotide sequence diversity of rDNA internal transcribed spacers extracted from conidia and cleistothecia of several powdery mildew fungi. Mycoscience 37:283–288
Hirose S, Tanda S, Levente KI, Grigaliunaite B, Havrylenko M, Takamatsu S (2005) Molecular phylogeny and evolution of the maple powdery mildew (Sawadaea, Erysiphaceae) inferred from nuclear rDNA sequences. Mycol Res 109:912–922
Jaczewski AA (1927) Karmany opredelitel’ gribov. Vyp. 2. Muchnisto-rosyanye griby. Mikologicheskaya Laboratoriya Imeni Professora A.A. Jaczewskogo, Gosudarstvennogo Instituta Opytnoy Agronomii, Leningrad
Kiss L (2012) Limits of nuclear ribosomal DNA internal transcribed spacer (ITS) sequences as species barcodes for Fungi. Proc Natl Acad Sci U S A 109(27):E1811
Klenke F, Scholler M (2015) Pflanzenparasitische Kleinpilze. Springer, Berlin, 1172 pp
Kovács GM, Jankovics T, Kiss L (2011) Variation in the nrDNA ITS sequences of some powdery mildew species: do routine molecular identification procedures hide valuable information? Eur J Plant Pathol 131:135–141
Laibach F (1930) Über die Bedingungen der Perithezienbildung bei den Erysipheen. Jahrb Wiss Bot 72:106–136
Mori Y, Sato Y, Takamatsu S (2000) Evolutionary analysis of the powdery mildew fungi using nucleotide sequences of the nuclear ribosomal DNA. Mycologia 92:74–93
Nagy GS (1977) Erysiphe monardae sp. nov. Phytopathol Z 88:285–286
Salmon E (1900) A monograph of the Erysiphaceae. Memoirs of the Torrey Botanical Club 9:1–292
Schmidt A, Scholler M (2002) Studies in Erysiphales anamorphs (II): Colutea arborescens, a new host for Erysiphe palczewskii. Feddes Repertorium 113:107–111
Silvestro D, Michalak I (2012) raxmlGUI: a graphical front-end for RAxML. Org Divers Evol 12:335–337
Swofford DL (2002) PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4.0b10. Sinauer, Sunderland
Takamatsu S, Kano Y (2001) PCR primers useful for nucleotide sequencing of rDNA of the powdery mildew fungi. Mycoscience 42:135–139
Takamatsu S, Matsuda S, Grigaliunaite B (2013) Comprehensive phylogenetic analysis of the genus Golovinomyces (Ascomycota: Erysiphales) reveals close evolutionary relationships with its host plants. Mycologia 105:1135–1152. doi:10.3852/13-046
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729
Walker JB, Sytsma KJ, Treutlein J, Wink M (2004) Salvia (Lamiaceae) is not monophyletic: implications for the systematics, radiation, and ecological specializations of Salvia and tribe Mentheae. Am J Bot 91:1115–1125
Walsh PS, Metzger DA, Higuchi R (1991) Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 10:506–513
White TJ, Bruns TD, Lee S, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, San Diego
Acknowledgments
Volker Kummer provided specimens on Verbena and Jessica Cavaletto and Steve Goodwin provided the sequence of PUL F1563 (G. biocellata on Glechoma hederacea). Jörg Böllmann helped to prepare specimens for SEM, Philipp Gannibal took light microscopy pictures of the holotype of G. salviae, and H. Jage identified Salvia nemorosa and confirmed Salvia fruticosa. We are much obliged to Walter Gams for reading the manuscript. Finally, we thank the curators of the herbaria M and ZT for providing specimens.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Scholler, M., Schmidt, A., Siahaan, S.A.S. et al. A taxonomic and phylogenetic study of the Golovinomyces biocellatus complex (Erysiphales, Ascomycota) using asexual state morphology and rDNA sequence data. Mycol Progress 15, 56 (2016). https://doi.org/10.1007/s11557-016-1197-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11557-016-1197-5