Abstract
A new species of whitish truffle, Tuber thailandicum, is described based on collections from northern Thailand. This species is characterized by whitish ascomata with dark brown gleba and subglobose spores with an alveolate reticulum. Tuber thailandicum is similar to T. castilloi, but differs in the thicker peridium and wider spores in one-spored asci. Molecular analysis of the internal transcribed spacer region and large subunit of ribosomal DNA also supports that T. thailandicum is clearly different from previously described whitish truffle species. It grows in mycorrhizal association with Betula alnoides, and the morphology and anatomy of mycorrhizae are described. Moreover, the identification of mycorrhizal status was confirmed by molecular methods.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Truffles (Tuber spp.) are edible fungi and belong to the order Pezizales, family Tuberaceae. Generally, truffle species produce hypogeous ascocarps and are the most expensive fungi in the world (Hall et al. 2007; Stobbe et al. 2012, 2013). Périgord black truffle (T. melanosporum Vittad.), Italian white truffle (T. magnatum Pico) and summer truffle (T. aestivum Vittad.) are a highly prized food in European countries (Hall et al. 2007; Bonito et al. 2010a; Stobbe et al. 2013). Oregon whitish truffles (T. gibbosum Harkn. and T. oregonense Trapp, Bonito & P. Rawl.) are commercially harvested in North America (Bonito et al. 2010b). Tuber indicum Cooke & Massee is one of the renowned commercial black truffles in China (García-Montero et al. 2010; Mortimer et al. 2012). Chinese truffles have been exported to Australia, Europe, Japan and America since the 1980s, and the popularity of truffles worldwide has increased (Rey 2001; Hall et al. 2003; Mortimer et al. 2012). Truffles form ectomycorrhizal symbiosis on a wide range of shrubs and trees including members of the families Betulaceae, Cistaceae, Corylaceae, Fagaceae and Pinaceae (Weden et al. 2004; Hall et al. 2007; Trappe et al. 2009; Bonito et al. 2011). Currently, more than half of the harvested truffle yield is produced in orchards (Hall et al. 2003). Traditional identification of Tuber species is based on morphological characteristics. There are 296 Tuber records in Index Fungorum (http://www.indexfungorum.org/ Names/Names.asp). This index might include many synonyms and misidentifications; therefore, it is difficult to estimate worldwide Tuber species diversity. Morphological variations associated with environmental conditions and developmental stage may make this manner of identification difficult. Closely related truffles are difficult to distinguish visually (Chen and Liu 2007; Kinoshita et al. 2011). Although Kirk et al. (2008) noted 86 Tuber species worldwide, this figure remains uncertain because of the limitations regarding information from Asia. Recently, molecular taxonomical analyses have provided powerful tools in the identification of Tuber species (Chen and Liu 2007; Karkouri et al. 2007; Bonito et al. 2010a, b; Kinoshita et al. 2011; Guevara et al. 2013).
Truffles have been researched in Asia for many decades (Trappe 1976; Liu 1985). Tuber indicum Cooke & Massee, found in India, was the first Asian truffle to be discovered (Cooke and Massee 1892). In the 1980s, four new Tuber species were reported, T. liaotongense Wang, T. taiyuanense Liu, T. tianshanense Tao and T. xizangense Xu from China (Liu and Liu 1994; Tao 1988; Wang et al. 1998; Xu 1999; Chen and Gong 2000). Tuber huidongense Wang and T. zhongdianense He, Hai, Li & Y. Wang were reported from China in 2002 and 2004, respectively (Wang and He 2002; He et al. 2004). In 2005, T. umbilicatum Chen & P.G. Liu and T. furfuraceum Hu & Y.I. Wang were reported from China and Taiwan, respectively (Chen et al. 2005; Hu and Wang 2005). Since 2010, more than ten known new species of Tuber were reported from China (García-Montero et al. 2010; Fan et al. 2011a, 2011b, 2012a, 2012b, 2012c, 2013; 2014; Deng et al. 2013; Su et al. 2013; Li et al. 2014). In addition, Kinoshita et al. (2011) reported that Tuber biodiversity in Asia is considered to be high and not well documented. However, no Tuber species have been reported from Thailand. During an investigation of ectomycorrhizal fungi associated with tree species in northern Thailand, we found an interesting species of Tuber under Betula alnoides Buch.-Ham. ex G. Don. (Betulaceae), which has white ascomata. It resembled several known whitish truffles, such as T. castilloi Guevara, Bonito & Trappe (Guevara et al. 2013) and T. oregonense Trappe, Bonito & P. Rawl. (Bonito et al. 2010b) from North America, T. magnatum and T. borchii Vittad. from Europe (Riousset et al. 2001) and T. latisporum Juan Chen & P.G. Liu from China (Chen and Liu 2007). However, detailed morphological observation and phylogenetic analysis of internal transcribed spacer (ITS) and large subunit (LSU) regions of ribosomal DNA revealed that it is a new species, which we describe in this present paper. Moreover, the ectomycorrhizal association with B. alnoides was confirmed and described by molecular techniques.
Material and methods
Sample collection
Ascomata of Tuber thailandicum were collected from Huay Kok Ma village (18°40′30″N, 98°54′25″E, elevation 1240 m), Doi Suthep-Pui National Park, Chiang Mai Province, Thailand in May 2014. Soil type in the field was sandy clay loam (52.5 % sand, 19.1 % silt and 28.4 % clay) with a pH of 4.5. Ascomata were wrapped in aluminum foil or kept in plastic specimen boxes until they were transported back to the laboratory, and photographs were taken within 24 h. The specimens were dried at 40–45 °C and deposited at the Research Laboratory for Excellence in Sustainable Development of Biological Resources, Faculty of Science, Chiang Mai University, Thailand (SDBR-CMU).
Morphological studies
Macromorphological data were derived from fresh specimens. Color names and codes follow Kornerup and Wanscher (1967). The micromorphological data were derived from dried specimens rehydrated in 95 % ethanol followed by distilled water, 5 % potassium hydroxide (KOH) or Melzer’s reagent. Size data of anatomical features are based on at least 50 measurements of each structure under a light microscope (Olympus CX51, Japan). For spore statistics, Q is the ratio of spore length divided by spore width and Q is the average Q of all specimens ± standard deviation. For scanning electron microscopy (SEM), ascospores were scraped from the dried specimen onto double-sided tape, which was mounted directly on an SEM stub, coated with gold, examined and photographed with a JEOL JSM-5910 LV SEM (JEOL, Japan).
Molecular studies
Genomic DNA of dried specimens (1–10 mg) was extracted using the fungal Genomic DNA Extraction Mini Kit (FAVORGEN, Taiwan). The internal transcribed spacer (ITS) region of ribosomal DNA (rDNA) was amplified by PCR using ITS1F and ITS4 primers (White et al. 1990) under the following thermal conditions: 95 °C for 2 min, 30 cycles of 95 °C for 30 s, 50 °C for 30 s, 72 °C for 1 min, and 72 °C for 10 min on a GeneAmp 9700 thermal cycler (Applied Biosystems, USA). In addition, the large subunit (LSU) region of rDNA was also amplified with LROR and LRO5 primers (Vilgalys and Hester 1990) under the following thermal conditions: 94 °C for 2 min, 30 cycles of 95 °C for 30 s, 52 °C for 30 s, 72 °C for 1 min, and 72 °C for 10 min. Negative controls lacking fungal DNA were run for each experiment to check for any contamination of the reagents. PCR products were checked on 1 % agarose gels stained with ethidium bromide under UV light and purified using NucleoSpin® Gel and PCR Clean-up Kit (Macherey-Nagel, Germany). The purified PCR products were directly sequenced. Sequencing reactions were preformed and the sequences were automatically determined in the genetic analyzer at the 1ST Base Company (Malaysia) using the same PCR primers mentioned above. Sequences were used to query GenBank via BLAST (http://blast.ddbj.nig.ac.jp/top-e.html).
For the phylogenetic analysis, a multiple sequence alignment was carried out using the alignment subroutines in Clustal X (Thompson et al. 1997), and the aligned ITS and LSU sequences deposited at TreeBASE under the numbers 16701 and 16704, respectively. A phylogenetic tree was constructed using the PAUP beta 10 software version 4.0 (Swofford 2002). In maximum parsimony analysis, all characters were equally weighted and gaps were treated as missing data. Heuristic searches with 100 random-addition sequence replicates and tree-bisection reconnection branch swapping were performed. Bootstrap analysis was conducted with 1,000 replicates using the same settings as above (Felsenstein 1985). The parsimony tree scores, including tree length and consistency, retention, rescaled consistency, and homoplasy indices (CI, RI, RC, and HI), were calculated. Bayensian phylogenetic analyses were carried out using the Metropolis-coupled Markov chain Monte Carlo (MCMCMC) method with MrBayes v3.2 (Ronquist et al. 2012), under a GRT + I + G model. Markov chains were run for 1,000,000 generations, with six chains and random starting trees. The chains were sampled every 100 generations. Among these, the first 2000 trees were discarded as the burn-in phase of each analysis and the resulting trees were used to calculate Bayesian posterior probabilities.
Characterization of Tuber thailandicum ectomycorrhizas
Morphological analysis
Ten ectomycorrhizal root samples were collected in the field point where the ascocarps were found, and separated from soil samples under the stereomicroscope (Olympus TL3, Japan). Ectomycorrhizal morphotypes were characterized based on anatomical and morphological characters following Agerer (1986, 1991, 2006). For anatomical characterization, sections of root tips mounted in 3 % KOH or 1 % Congo Red were observed under a light microscope.
Molecular analysis
The ectomycorrhizal roots having the macro-morphological characteristics of Tuber sp. were then analyzed molecularly (Rauscher et al. 1996; Kovács and Jakucs 2006; Boutahir et al. 2013). The ectomycorhizal roots were washed three times with sterilized water. Genomic DNA was extracted from single mycorrhizal root tips as described by Paolocci et al. (1999), using the Nucleospin Plant II kit (Macherey-Nagel, Germany) according to the manufacturer’s protocols. The ITS regions of the rDNA of the fungal symbiont were amplified by polymerase chain reaction with ITS1F and ITS4 primers following the thermal conditions described above. PCR products were checked, purified and sequenced. For phylogenetic analysis of the sequences, the same settings as above were used.
For plant identification, the ITS regions of rDNA were amplified from total genomic DNA extracted from ectomycorrhizal colonized root using the primers ITS4 and ITSLeu (Baum et al. 1998) under the following thermal conditions: 94 °C for 3 min, 35 cycles of 91 °C for 1 min, 55 °C for 2 min, 72 °C for 1 min, and 72 °C for 10 min. PCR products were checked, purified and sequenced. Sequences were used to query GenBank via BLAST.
Results
Taxonomy
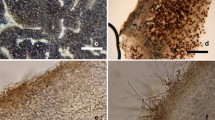
Tuber thailandicum N. Suwannarach, J. Kumla & S. Lumyong, sp. nov. Figure 1
Tuber thailandicum. (a) and (b), Ascomata. (c). A cross section of peridium. (d). Pseudoparenchymatous tissue of the outer layer of peridium. (e). The hyhae-like hairs on the surface of a peridium. (f) and (g). Asci and ascospores as observed under a compound microscope. (h). Ascospore as observed with a scanning electron microscope. Scale bar a and b = 1 cm, c = 50 μm, d and e = 10 μm, f = 25 μm, a and h = 10 μm
MycoBank No.: MB811502
Diagnosis: Differs from T. castilloi due to its thicker peridium and wider spores in one-spored asci.
Holotypus: Thailand, Chiang Mai Province, Muang Distric, Doi Suthep-Pui National Park, Huay Kok Ma village, 18°40′30″N, 98°54′25″E, elevation 1240 m, in a tropical deciduous forest, dominated by Betula alnoides and Castanopsis spp., 20 May 2014, Nakarin Suwannarach and Jaturong Kumla, SDBR-CMU-MTUF001 (holotypus), GenBank sequence KP196329.
Etymology: thailandicum referring to Thailand, where the new species was found.
Ascomata hypogeous, globose to subglobose or irregularly lobed, 2−3.5 cm in diameter, white (2A1) to pale yellow (4A3) when fresh, gradually becoming pale yellow (3A3) to brown (6D8) when dry (Fig. 1a, b). Odor slightly aromatic when mature. Peridium surface minutely verrucose, mostly 150 − 255 μm thick, composed of two layers (Fig. 1c). Outer cortical layer 75−112.5 μm thick, pseudoparenchymatous, composed of subglobose to subangular, pale yellow or hyaline cells 6−20 × 4−10.5 μm (Fig. 1d). The outer most cells giving rise the hyphae-like hairs, the hairs 20−63.5 μm long, septate, tapered, usually acute at the apex (Fig. 1e). Inner layer 70−170 μm thick, of intricately interwoven, hyaline hyphae 2−5 μm in diameter, the walls thin to somewhat thickened. Gleba solid, white when young, becoming dark brown at maturity, marbled with distinct, white and meandering veins. Asci globose to subglobose or ellipsoid, hyaline, with thin or slightly thickened walls, 37.5−80 × 50−87.5 μm, sessile with a short stalk 3−4 μm in diameter, one-spored to four-spored (Fig. 1f). Ascospores subglobose, sometimes globose or broadly ellipsoid, hyaline when young, becoming yellowish brown to brown at maturity, excluding their alveolate-recticulate ornamentation, in one-spored asci 40−65 × 40−62 μm, in two-spored asci 25−48 × 20−50 μm, in three-spored asci 22−33 × 20−29 μm, in four-spored asci 20−28 × 15−25 μm, Q = 1.00−1.28, Q = 1.09 ± 0.08, ornamented with a regular alveolate reticulum, 3−5 μm high, constituted of mostly hexagonal meshes 10−17.5 × 7.5−12.5 μm and 3−4 μm across the spore width (Fig. 1g, h).
Ecology and Distribution: Hypogeous, solitary or in groups in calcareous soils under B. alnoides, fruiting during the rainy season. Known only from Thailand.
Other Material Examined: THAILAND: Huay Kok Ma village, 20 May 2014, Nakarin Suwannarach (SDBR-CMU-MTUF001, SDBR-CMU-MTUF002), 5 June 2014, Jaturong Kumla and Santhiti Vadthanarat (SDBR-CMU-MTUF003).
Note: Tuber thailandicum resembles T. castilloi but T. castilloi has a smaller ascomata size (1.1−2.5 cm in diameter), thinner peridium (80−150 μm), and narrower spores in one-spored asci (40−65 × 40−62 μm). Tuber thailandicum and T. castilloi spores are subglobose to broadly ellipsoid, while those of T. castilloi are mostly broadly ellipsoid (Guevara et al. 2013). Tuber castilloi forms mycorrhizal associations with Quercus spp. in North America.
Phylogenetic analysis
The ITS sequences from ascomata of T. thailandicum SDBR-CMU-MTUF001, SDBR-CMU-MTUF002 and SDBR-CMU-MTUF003 were deposited at GenBank under accession numbers KP196328, KP196329 and KP196330, respectively. In addition, the LSU sequences of T. thailandicum SDBR-CMU-MTUF001, SDBR-CMU-MTUF002 and SDBR-CMU-MTUF003 were deposited under accession numbers KP196333, KP196334 and KP196335, respectively. The Tuber sequences for phylogenetic analysis were obtained in this study and from GenBank database (Table 1). Tuber magnatum was used as the outgroup. The aligned dataset of 55 sequences of the ITS consisted of 773 characters, of which 221 characters were constant, 131 variable characters were parsimony uninformative, and 431 characters were parsimony informative. Heuristic searches resulted in 22 equally parsimonious trees with a length of 1779 steps, CI = 0.567, RI = 0.816, RC = 0.462 and HI = 0.473. While the aligned data set of 46 sequences of LSU consisted of 733 characters of which 421 characters were constant, 137 variable characters were parsimony uninformative, and 175 characters were parsimony informative. Heuristic searches resulted in ten equally parsimonious trees with a length of 546 steps, CI = 0.703, RI = 0.861, RC = 0.606 and HI = 0.404. The phylogenetic dendrograms of ITS and LSU sequences are shown in Figs. 2 and 3. Both phylograms indicated that T. thailandicum clearly differed from other whitish truffle species and formed a monophyletic clade with a bootstrap support of 100 %. Based on the ITS analysis, the whitish truffles were separated into three main clades. Tuber thailandicum stands within the Gibbosum clade together with T. bellisporum, T. castellanoi, T. gibbosum and T. pseudospaerosporum, and forms a sister taxon to T. pseudospaerosporum with 98 % bootstrap support. The Maculatum clade comprises ten whitish truffle species (T. castilloi, T. foetidum, T. guevarai, T. lauryi, T. maculatum, T. mexiusanum, T. miquihuanense, T. pseudomagnatum, T. shearii and T. walkeri). The remaining clade includes 16 species, T. alboumbilicum, T. borchii, T. dryophilum, T. latisporum, T. liui, T. liyuanum, T. microspaerosporum, T. oligospermum, T. panzhihuanense, T. puberulum, T. sinosphaerosporum, T. sinopuberulum, T. spherosporum, T. sphaerospermum, T. vesicoperidium and T. zhongdianense. From the LSU region analysis, the number of operational taxonomic units used in phylogenetic analysis were fewer than in the ITS analysis because of limited availability of LSU sequence data in the database. Moreover, the ITS region is a more precise marker for the identification and species-level determination of Tuber than the LSU region (Chen and Liu 2007; Bonito et al. 2010a). Therefore, this study used only the ITS analysis to group the Tuber species corresponding to Bonito et al. (2010a).
One of 22 maximum parsimonious trees inferred from a heuristic search of the ITS region of rDNA of 55 sequences. Tuber magnatum was used as the outgroup. Numbers above branches identify the bootstrap statistics percentages (left) and Bayesian posterior probabilities (right). Branches with bootstrap values ≥ 50 % are shown at each branch and the bar represents ten shown substitutions per nucleotide position. The sequences from this study are in bold. a Sequence from ascomata and b Sequence from mycorrhizae
One of ten maximum parsimonious trees inferred from a heuristic search of the LSU region of rDNA of 46 sequences. Tuber magnatum was used as the outgroup. Number above branches identify the bootstrap statistics percentages (left) and Bayesian posterior probabilities (right). Branches with bootstrap values ≥ 50 % are shown at each branch and the bar represents ten shown substitutions per nucleotide position. The sequences from this study are in bold
Characterization of Tuber thailandicum ectomycorrhizas
Two ITS sequences of the mycorrhizae were obtained. Phylogenetic analysis confirmed that the fungal symbiont of the selected mycorrhizas was in the same clade as T. thailandicum fruiting bodies (Fig. 2). These sequences were deposited at GenBank under accession numbers KP196331 and KP196332, respectively and both sequences had 100 % similarity with T. thailandicum. Therefore, the fungal symbiont was T. thailandicum. For plant identification, the ITS sequence from ectomycorrhizal roots in field, containing 592 bp, was deposited in GenBank under accession number KP713780 and closely resembled the ITS sequence AY763114 and AY761101 from B. alnoides (Li and Shoup 2005), with a similarity of 99 % by BLAST. Therefore, the mycorrhizal root formed by the colonization of T. thailandicum was B. alnoides.
The mycorrhizae are simple or monopodial-pinnate, straight, cylindrical or sometimes club-shaped and always with rounded ends, 2.0–5.0 mm in length and 0.3–0.6 mm in diameter (Fig. 4a, b). Rhizomorphs were not observed. The color varied with different developmental stages; white when young then becoming pale grey, and dark grey when older. Two kinds of peritrophic elements, emanating hyphae and cystidia, were observed on T. thailandicum mycorrhizae. The emanating hyphae are hyaline, 2.0–3.0 μm in diameter, thin-walled, septate, ramified and tortuous (Fig. 4c). They bear abundant crystals and generally grow from a network on the surface of the mycorrhizae. The cystidia are hyaline, needle-shaped, septate, smooth, straight or slightly bent, 40–60 μm in length, 3.0–5.0 μm in diameter and have rounded tips (Fig. 4d). The cystidia grow radially from the surface of mantle and are arranged in a restricted area, often at the apex of the mycorrhizae. Mycorrhizae with cystidia are very rare and sometimes may lack these structures. The outer and inner mantle layers formed a pseudoparenchymatous mantle consisting of angular cells, 4.5–10.0 μm in diameter. The mantle type of hypha was designated as L type after Agerer’s (2006) categorization (Fig. 4e). A cross section of the mycorrhizal roots showed a 25–30 μm thick mantle arranged in four to six cell layers, with the Hartig net only reaching the epidermal layer and apparently only the first row of the cortical cells (Fig. 4f).
Morphological and anatomical traits of T. thailandicum mycorrhizae with Betula alnoides. a and b. Ectomycorrhizal root tips. c. Emanating hyphae with crystals. d. Cystidia. e. The angular cell of the outer mantle layer. f. Cross section of mycorrhizal root tips showing mantle sheath (M) and Hartig net (arrowed). Scale bars: a and b = 1 mm, c = 20 μm, d and e = 10 μm, f = 25 μm
Discussion
The present study is the first report to describe a new whitish truffle from Thailand and its ectomycorrhizal association. Tuber thailandicum is characterized by its white ascomata and subglobose spores with an alveolate reticulum; thus, it belongs to the gibbosum group. The morphological characteristics clearly separate the new species, T. thailandicum from other whitish truffle. It has a two-layered peridium that easily separated it from T. cistophilum, T. glabrum, T. gennadii, T. lacunosum, T. maculatum, T. oligospermum, T. panzhihuanense, T. sphaerospermum and T. vesicoperidium, which have a one-layered peridium (Bulman et al. 2010; Alvarado et al. 2012; Fan and Cao 2012c, 2014, Fan et al. 2014; Deng et al. 2013). The presence of hyphae-like hairs on peridial surface of T. thailandicum distinguishes it from T. alboumbilicum, T. bellisporum, T. castellanoi, T. gibbosum, T. microspiculatum, T. miquihuanense, T. oregonense, T. pseudomagnatum, T. pseudosphaerosporum, T. sinoalbidum and T. sinopuberulum (Guevara et al. 2013). Moreover, the spore shape of T. thailandicum differs from the broadly ellipsoidal, ellipsoid and long-ellipsoid spores of T. bellisporum, T. borchii, T. dryophilum, T. foetidum, T. gibbosum, T. glabrum, T. latisporum, T. liui, T. liyuanum, T. maculatum, T. oregonense, T. pseudomagnatum, T. rapaeodorum, T. sinoalbidum and T. zhongdianense (Xu 1999; He et al. 2004; Chen and Liu, 2007; Bonito et al. 2010a; Bulman et al. 2010; Fan and Cao 2012b; Fan et al. 2014). Tuber microsphaerosporum and T. sinosphaerosporum are distinguished from T. thailandicum by their reticulated globose spores (Fan and Yue 2013; Fan et al. 2012a). Tuber shearii has spores similar to T. thailandicum, but can be easily differentiated by its smaller reticulum size (9−10 × 5−6 μm) (Murill 1920). Tuber thailandicum shares some morphological characteristics with five North American truffles, T. castilloi, T. guevaria, T. lauryi, T. mexiusanum and T. walkeri, including a two-layered peridium with hyphae-like hair on its surface and subglobose, sometime globose or broadly ellipsoid reticulated spores (Table 2). However, the peridial thickness in T. thailandicum (150−225 μm) was different from T. castilloi (80−150 μm) and T. lauryi (300−1000 μm) (Guevara et al. 2013). The smaller ascomata size and 1−5 spores in asci of T. guevaria, T. mexiusanum and T. walkeri distinguished them from the new species reported here (Guevara et al. 2013). Interestingly, our ITS and LSU sequence analyses clearly separate T. thailandicum from the other whitish truffles. Based on the ITS sequence analysis, Tuber thailandicum forms a sister taxon to T. pseudosphaerosporum known from southwestern China with 87 % similarity, while T. pseudosphaerosporum has thicker outer peridium (150−250 μm) and regularly recticuted globose spores (Fan and Yue 2013). Therefore, the combination of morhphological and molecular characteristics strongly supported a new whitish truffle, T. thailandicum from Thailand.
Truffles grow in symbiosis with several trees such as beech (Fagus spp.), birch (Butula spp.), hazel (Corylus spp.), oaks (Quercus spp.), pine (Pinus spp.) and spruce (Picea spp.) (Weden et al. 2004; Hall 2007; Trappe et al. 2009; Bonito et al. 2011; Stobbe et al. 2012). The ectomycorrhzae of whitish truffle generally have a pseudoparenchymatous-epidermoid mantle (M type) and needle-shaped cystidia, as have also been detected in T. borchii (Rauscher et al. 1996; Benucci et al. 2012; Lancellotti et al. 2014), T. maculatum (Zambonelli et al. 1999), T. oligospermum (Boutahir et al. 2013), T. puberulum (Blaschke 1987), T. rapaeodorum (Kovács and Jakucs 2006) mycorrhizae. The shape and size of cystidia and the anatomy of mantle are characteristics used to identify the mycorrhizae of the most economically important Tuber species (Kovács and Jakucs 2006; García-Montero et al. 2008; Benucci et al. 2012). In this study, the pseudoparenchymatous, angular outer mantle layer (L type) of T. thailandicum mycorrhizae was a important characteristic in distinguishing it from other whitish truffle mycorrhizae. However, previous studies reported that the comparison of younger and older parts of the same mycorrhizal specimen showed variable feature overlap. Variable mantle patterns in the ectomycorrhizae of T. borchii were observed and these phenomena were explained as intraspecific and age-related variabilities of the strains (Giomaro et al. 2000). Those observations suggested that the anatomical characteristics of mycorrhizas are not sufficient for identification of the fungal symbiont species (Kovács and Jakucs 2006; Boutahir et al. 2013). Conversely, the molecular analyses have provided powerful tools to identify the genetic differences between fungal symbiont species. Therefore, combining molecular and anatomical approaches is suitable to identify ectomycorrhizae of truffle (Baciarelli-Falini et al. 2006; Kovács and Jakucs 2006; Rubini et al. 2001; Boutahir et al. 2013). In conclusion, our study presents a new whitish truffle, T. thailandicum from Thailand. This discovery is important to stimulate the investigations of Tuber in Thailand, and will help us understand more on the distribution and ecology of Tuber in Asia.
References
Agerer R (1986) Studies on ectomycorrhizae. II introducing remarks on characterization identification. Mycotaxon 26:473–492
Agerer R (1991) Studies on ectomycorrhizae XXXIV. Mycorrhizae of Gomphidius glutinosus and of G. roseus with some remarks on Gomphidiaceae (Basidiomycetes). Nova Hedwigia 53:127–170
Agerer R (2006) Fungal relationships and structural identity of their ectomycorrhizae. Mycol Prog 5:67–107. doi:10.1007/s11557-006-0505-x
Alvarado P, Moreno G, Manjón JL (2012) Comparison between Tuber gennadii and T. oligospermum lineages reveals the existence of the new species T. cistophilum (Tuberaceae, Pezizales). Mycologia 104:894–910. doi:10.3852/ 11-254
Baciarelli-Falini L, Rubini A, Riccioni C, Paolocci F (2006) Morphological and molecular analyses of ectomycorrhizal diversity in a man-made T. melanosporum plantation: description of novel truffle-like morphotypes. Mycorrhiza 16:475–484. doi:10.1007/s00572-006-0066-5
Baum DA, Small RL, Wendel JF (1998) Biogeography and floral evolution of Baobabs (Adansonia, Bombacaceae) as inferred from multiple data sets. Syst Biol 47:181–207
Benucci GMN, Bonito G, Falini LB, Bencivenga M (2012) Mycorrhization of Pecan trees (Carya illinoinensis) with commercial truffle species: Tuber aestivum Vittad. and Tuber borchii Vittad. Mycorrhiza 22:383–392. doi:10.1007/s00572-011-0413-z
Blaschke H (1987) Vorkommen und charakterisierung der ektomykorrrhizaassoziation Tuber puberulum mit Picea abies. Z Mykol 53:283–288
Bonito G, Grygansyi AP, Trappe JM, Vilgalys R (2010a) A global meta-analysis of Tuber ITS rDNA sequences: species diversity, host associations and long-distance dispersal. Mol Ecol 19:4994–5008. doi:10.1111/j.1365-294X.2010.04855.x
Bonito G, Trappe JM, Donovan S, Vilgalys R (2011) The Asian black truffle Tuber indicum can form ectomycorrhizas with North American host plants and complete its life cycle in non-native soils. Fungal Ecol 4:83–93. doi:10.1016/j.funeco.2010.08.003
Bonito G, Trappe JM, Rawlimson P, Vilgalys R (2010b) Improved resolution of major clades whitish Tuber and taxonomy of species within the Tuber gibbosum complex. Mycologia 102:1042–1057. doi:10.3852/09-213
Boutahir S, Iotti M, Piattoni F, Zambonelli A (2013) Morphological and molecular characterization of Tuber oligospermum mycorrhizas. Afr J Agric Res 8:4081–4087. doi:10.5897/AJAR2013.7354
Bulman SR, Visnovsky SB, Hall IR, Guerin-Laguette A, Wang Y (2010) Molecular and morphological identification of truffle-producing Tuber species in New Zealand. Mycol Prog 9:205–214. doi:10.1007/s11557-009-0626-0
Chen J, Liu PG (2007) Tuber latisporum sp. nov. and related taxa, based on morphology and DNA sequence data. Mycologia 99:475–481. doi:10.3852/mycologia.99.3.475
Chen J, Liu PG, Wang Y (2005) Tuber umbilicatum, a new species from China, with a key to the spinose-reticulate spored Tuber species. Mycotaxon 94:1–6
Chen YL, Gong MQ (2000) Truffles Tuber spp.: biodiversity and geological distribution. Edible Fungi of China 19:25–26
Cooke MC, Massee G (1892) Himalayan truffles. Grevellea 20:67
Deng XJ, Liu PG, Liu CY, Wang Y (2013) A new white truffle species, Tuber panzhihuanense from China. Mycol Prog 12:557–561. doi:10.1007/s11557-012-0862-6
Fan L, Cao JZ (2012) Two new species of white truffle from China. Mycotaxon 121:297–304. doi:10.5248/121.297
Fan L, Cao JZ, Liu YY, Li Y (2011a) Two new species of tuber from China. Mycotaxon 116:349–354. doi:10.5248/116.349
Fan L, Cao JZ, Yu J (2012a) Tuber in China: T. sinopuberulum and T. vesicoperidium spp. nov. Mycotaxon 121:255–263. doi:10.5248/121.255
Fan L, Cao JZ, Zheng ZH, Li Y (2012b) Tuber in China: T. microspermum and T. microspiculatum spp. nov. Mycotaxon 199:391–395. doi:10.5248/119.391
Fan L, Feng S, Cao JZ (2014) The phylogenetic position of Tuber glabrum sp. nov. and T. sinomonosporum nom. nov., two Paradoxa-like truffle species from China. Mycol Prog 13:241–246. doi:10.1007/s11557-013-0908-4
Fan L, Hou CL, Cao JZ (2011b) Tuber sinoalbidum and T. polyspermum – new species from China. Mycotaxon 118:403–410. doi:10.5248/118.403
Fan L, Yue SF (2013) Phylogenetic divergence of three morphologically similar truffles: Tuber sphaerosporum, T. sinosphaerosporum, and T. pseudospaerosporum sp. nov. Mycotaxon 125:283–288. doi:10.5248/125.283
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
García-Montero LG, Díaz P, Martín-Fernández S, Casermeiro MA (2008) Soil factors that favour the production of Tuber melanosporum carpophores over other truffle species: a multivariate statistical approach. Acta Agric Scand B 58:322–329. doi:10.1080/09064710701743286
García-Montero LG, Díaz P, Massimo GD, García-Abril A (2010) A review of research on Chinese Tuber species. Mycol Prog 9:315–335. doi:10.1007/s11557-009-0647-8
Giomaro G, Zambonelli A, Sisti D, Cecchini M, Evangelista, Stocchi V (2000) Anatomical and morphological characterization of mycorrhizas of five strains of Tuber borchii Vittad. Mycorrhiza 10:107–114. doi:10.1007/s005720000065
Guevara G, Bonito G, Trappe JM, Cázares E, Williams G, Healy RA, Schadt C, Vilgalys G (2013) New north american truffles (Tuber spp.) and their ectomycorrhizal associations. Mycologia 105:194–209. doi:10.3852/12-087
Hall IR, Brown GT, Zambonelli A (2007) Taming the truffle, the history, lore and science of the ultimate mushroom. Timber Press, Portland
Hall IR, Yun W, Amicucci A (2003) Cultivation of edible ectomycorrhizal mushrooms. Trends Biotechnol 21:433–438. doi:10.1139/B04-051
He XY, Li HM, Wang Y (2004) Tuber zhongdianense sp. nov. from China. Mycotaxon 90:213–216. doi:10.5248/121.255
Hu HT, Wang Y (2005) Tuber furfuraceum sp. nov. from Taiwan. Mycotaxon 93:155–157
Karkouri KE, Murat C, Zampieri E, Bonfante P (2007) Identification of internal transcribed spacer sequence motifs in truffles: a first step toward their DNA bar cording. Appl Environ Microbiol 73:5320–5330. doi:10.1128/AEM.00530-07
Kinoshita A, Sasaki H, Nara K (2011) Phylogeny and diversity of Japanese truffles (Tuber spp.) inferred from sequences of four nuclear loci. Mycologia 103:779–794. doi:10.3852/10-138
Kirk PM, Cannon PF, Minter DW, Stalpers JA (2008) Ainsworth and Bisby’s dictionary of the fungi, 10th edn. Oxon, UK
Kornerup A, Wanscher JH (1967) Methuen handbook of colour, 2nd edn. Eyre Methuen, London
Kovács GM, Jakucs E (2006) Morphological and molecular comparison of white truffle ectomycorrhizae. Mycorrhiza 16:567–574. doi:10.1007/s005720060071-8
Lancellotti E, Iotti M, Zambonelli A, Franceschini A (2014) Characterization of Tuber borchii and Arbutus unedo mycorrhizas. Mycorrhiza 24:481–486. doi:10.1007/s00572-014-0564-9
Li J, Shoup S (2005) Phylogenetic of Betula (Betulaceae) inferred from sequences of nuclear ribosomal DNA. Rhodora 107:69–86. doi:10.3119/04-14.1
Li SH, Zheng LY, Liu CY, Wang Y, Li L, Zhao YC, Zhang XL, Yang M, Xiong HK, Qing Y, Wang L, Zhou DQ (2014) Two new truffles species, Tuber alboumbilicum and Tuber pseudobrumale from China. Mycol Prog 13:1157–1163. doi:10.1007/s11557-014-1004-0
Liu B (1985) New species and new records of hypogeous fungi from China I. Acta Mycol Sin 4:84–89
Liu B, Liu YH (1994) Study on hypogeous fungi in China. Chinese Fungi 9:157–165
Mortimer PE, Karunarathna SC, Li Q, Gui H, Yang X, Yang X, He J, Ye L, Guo J, Li H, Sysouphanthong P, Zhou D, Xu J, Hyde KD (2012) Prized edible Asian mushrooms: ecology, conservation and sustainability. Fungal Divers 56:31–47. doi:10.1007/s13225-012-0196-3
Murrill WA (1920) Another new truffle. Mycologia 12:157–158
Paolocci F, Rubini A, Granetti B, Arcioni S (1999) Rapid molecular approach for a reliable identification of Tuber spp. ectomycorrhizae. FEMS Microbiol Ecol 28:23–30. doi:10.1016/S0168-6496(98)00088-9
Rauscher T, Müller WR, Chevalier G, Agerer R (1996) Tuber borchii Vitt. In: Agerer R (ed) Colour atlas of ectomycorrhizae. Einhorn Verlag, Schwäbisch Gmüad, p 113
Rey JF (2001) Creation et origine de la commercialisation de la truffe de Chine en France et dans le monde. In: Corvoisier M, Olivier JM, Chevalier G (eds) Federation Française des Trufficulteurs, Proceedings du Ve Congrès International Science et Culture de la Truffe, Paris, pp 517–518
Riousset G, Riousset L, Chevalier G, Bardet MC (2001) Truffe d’Europe et de Chine. INRA, Paris
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and modelchoice across a large model space. Syst Biol 61:539–542. doi:10.1093/sysbio/sys029
Rubini A, Paolocci F, Granetti B, Arcioni S (2001) Morphological characterization of molecular-typed Tuber magnatum ectomycorrhizae. Mycorrhiza 11:179–185. doi:10.1007/s005720100116
Stobbe U, Büntgen U, Sproll L, Tegel W, Egli S, Fink S (2012) Spatial distribution and ecological variation of re-discovered German truffle habitats. Fungal Ecol 5:591–599. doi:10.1016/j.funeco.2012.02.001
Stobbe U, Egli S, Tegel W, Peter M, Sproll L, Büntgen U (2013) Potential and limitation of Burgundy truffle cultivation. Appl Microbiol Biotechnol 97:5215–5224. doi:10.1007/s00253-013-4956-0
Su KM, Xiong WP, Wang Y, Li SH, Xie R, Baima D (2013) Tuber bomiense, a new truffle species from Tibet, China. Mycotaxon 126:127–132. doi:10.5248/126.127
Swofford DL (2002) PAUP*: phylogenetic analysis using parsimony (*and other methods), beta version 4.0b10. Sinauer Associates, Sunderland, Massachusetts
Tao K (1988) Taxonomic study on hypogeous fungi of China. The University of Shanxi, Taiyuan, Dissertation
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higguns DG (1997) The CLUSTAL_X windows interface: flexible stratgies for multiple sequene alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Trappe JM (1976) Notes on Japanese hypogeous ascomycetes. Trans Mycol Soc Jpn 17:209–217
Trappe JM, Molina R, Luoma DL, Cázares E, Pilz D, Smith JE, Castellano MA, Miller SL, Trappe MJ (2009) Diversity, ecology, and conservation of truffle fungi in forests of the pacific northwest. Portland, Oregon
Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172:4239–4246
Wang Y, He XY (2002) Tuber huidongense sp. nov. from China. Mycotaxon 83:191–194
Wang Y, Moreno G, Riousset L, Manjón JL, Riousset G, Fourré G, Di Massimo G, García-Montero LG, Díez J (1998) Tuber pseudoexcavatum sp. nov. a Chinese species commercialised in Spain, France and Italy with comments on Chinese truffles. Cryptogam Mycolog 19:113–120
Weden C, Chevalier G, Danell E (2004) Tuber aestivum (syn. T. uncinatum) biotopes and their history on Gotland, Sweden. Mycol Res 108:304–310. doi:10.1017/S0953756204009256
White TJ, Burns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, Whitish TJ (eds) PCR protocols, a guide to methods and applications. Academic Press, San Diego, pp 315–322
Xu AS (1999) A taxonomic study of the genus Tuber in Xizang. Mycosystema 18:361–365
Zambonelli A, Iotti M, Amicucci A, Pisi A (1999) Caratterizzazione anatomo-morfologica delle micorrize di Tuber maculatum Vittad. su Ostrya carpinifolia Scop. Micol Ital 28:29–35
Acknowledgments
This work was supported by grants from Chiang Mai University and TRF Research-Team Association Grant (RTA5880006). We are grateful to Prof. Dr. Robert J. McGovern and Mr. Keegan Kennedy for English proof reading.
Author information
Authors and Affiliations
Corresponding author
Additional information
Section Editor: Franz Oberwinkler
Rights and permissions
About this article
Cite this article
Suwannarach, N., Kumla, J. & Lumyong, S. A new whitish truffle, Tuber thailandicum from northern Thailand and its ectomycorrhizal association. Mycol Progress 14, 83 (2015). https://doi.org/10.1007/s11557-015-1107-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11557-015-1107-2