Abstract
Cytospora species are cosmopolitan, and commonly associated with dieback and canker diseases of numerous hosts. In the present study, isolates were collected and identified from diseased branches or twigs of Ulmus pumila in northern China. The morphological characteristics and multilocus phylogeny (act1, ITS, LSU, tefA and tubB) indicate four distinct lineages with high branch support, i.e., C. carbonacea, C. chrysosperma, C. ribis and C. pruinopsis sp. nov. Cytospora pruinopsis is distinguishable from the other Cytospora spp. on Ulmus by its single conidiomatal locule with one ostiole per disc, and its smaller conidia. This study represents the first attempt to clarify the taxonomy of Cytospora spp. associated with canker and dieback symptoms of Ulmus pumila in northern China.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Cytospora (Sordariomycetes, Diaporthales, Valsaceae), which was introduced by Ehrenberg (1818), is presently regarded as a major causal agent of canker diseases in dicots and monocots (Adams et al. 2005). Cytospora has been regarded as the asexual morph of Valsa Fr., Valsella Fuckel, Valseutypella Höhn. and Leucostoma (Nitschke) Höhn. (Fries 1823; Saccardo 1884; Spielman 1985; Adams et al. 2005). Following the end of dual nomenclature for pleomorphic fungi (Wingfield et al. 2012; Crous et al. 2015), the older and more commonly encountered genus Cytospora (1818) was chosen over that of its sexual morph, Valsa (1849), for placement on the list of protected fungi (McNeill et al. 2012; Fan et al. 2015a, b; Rossman et al. 2015). Morphologically, the Cytospora sexual morph is characterized by a diaporthalean-like perithecial ascoma, clavate to elongate obovoid asci, and allantoid, hyaline, aseptate ascospores (Spielman 1983, 1985; Adams et al. 2005). The asexual morph is characterized by single or labyrinthine locules, filamentous conidiophores, and allantoid, hyaline, aseptate conidia (Spielman 1983, 1985; Adams et al. 2005). Species identification in Cytospora was previously based on host affiliations and morphology (Deng 1963; Tai 1979; Wei 1979), but because these characters have limited value, molecular phylogenetic data were needed to accurately distinguish Cytospora spp. (Adams et al. 2002, 2005). Several recent papers have subsequently provided updated phylograms for the genus based on multigene phylogenies using ex-type or reference strains (Zhang et al. 2014a, b; Fan et al. 2015a, b; Wang et al. 2015).
Elms (Ulmus L.) are temperate plants with worldwide distribution. In China, Ulmus pumila L. is a component of natural forests, with some specimens also planted as ornamental trees in streets, gardens and parks. Although Cytospora spp. have been recorded from a range of disease symptoms on Ulmus spp., the identities of the species involved have never been confirmed based on molecular techniques (Spaulding 1961; Conway and Morrison 1983; Chen 2002; Dudka et al. 2004; Zhuang 2005; Fotouhifar et al. 2010; Zhang et al. 2014a, b).
Previous studies have reported several Cytospora spp. from Ulmus spp. worldwide, i.e., C. ambiens Pers., C. carbonacea Fr., C. chrysosperma (Pers.) Fr., C. leucostoma (Pers.) Sacc., C. pulchella Sacc., and C. sacculus (Schwein.) Gvrit. (Gilman et al. 1957; Conway and Morrison 1983; Zhuang 2005; Mulenko et al. 2008; Fotouhifar et al. 2010). Thus far, three Cytospora spp. have been recorded from this host in China: C. ambiens, C. carbonacea and C. pulchella (Zhuang 2005; Zhang et al. 2014a, b). However, these species were recorded without detailed morphological or multi-gene phylogenetic analyses, and the phytopathogenic taxa causing Cytospora canker disease of Ulmus have not been clarified. In order to confirm the identities of Cytospora species that occur on Ulmus spp. in China, a total of 37 specimens were collected from symptomatic trees in six provinces in northern China. The objectives of this study were to 1) isolate the Cytospora spp. from symptomatic twigs and branches of Ulmus spp. and compare them to reference strains; and to 2) generate a multi-gene DNA phylogeny of the taxa concerned, and describe, illustrate and compare the new Cytospora species from Ulmus pumila.
Materials and methods
Isolates
Fresh specimens of Cytospora spp. were collected from infected branches or twigs of Ulmus pumila during collecting trips in Heilongjiang, Jilin, Ningxia, Qinghai, Shaanxi and Tibet Provinces in China (Table 1). Single conidial isolates were established from fruiting bodies by removing a mucoid conidial mass from pycnidial ostioles, and spreading the suspension on the surface of 1.8 % potato dextrose agar (PDA), incubated at 25 °C for up to 24 h. Single germinating conidia were removed and plated onto fresh PDA plates. Ten representative strains (selected based on morphology and cultural characteristics) were used in the phylogenetic analysis (Table 1). Specimens are deposited in the Museum of Beijing Forestry University (BJFC). Axenic cultures are maintained in the China Forestry Culture Collection Center (CFCC). Isolates of the new species are maintained in the China Center for Type Culture Collection (CCTCC).
DNA extraction, PCR amplification, and sequencing
Genomic DNA was extracted from colonies grown on cellophane-covered PDA using a modified CTAB [cetyltrimethylammonium bromide] method (Doyle and Doyle 1990). DNA was estimated by electrophoresis in 1 % agarose gel, and the quality was measured using the NanoDrop™ 2000 (Thermo Scientific, Waltham, MA, USA), following the user manual (Desjardins et al. 2009). PCR amplifications were performed in a DNA Engine Peltier Thermal Cycler (PTC-200; Bio-Rad Laboratories, Hercules, CA, USA). The ITS region was amplified using primers ITS1 and ITS4 (White et al. 1990). The partial large nuclear ribosomal RNA subunit (LSU) region was amplified using primers NL1 and NL4 (O’Donnell 1993). The partial actin (act1) region was amplified using primers ACT512F and ACT783R (Carbone and Kohn 1999). The partial translation elongation factor 1-alpha (tefA) gene region was amplified using primers EF1-728F and EF1-986R (Carbone and Kohn 1999), and tubB was amplified using primers Bt2a and Bt2b (Glass and Donaldson 1995). The PCR amplification products were estimated visually by electrophoresis in 2 % agarose gel. DNA sequencing was performed using an ABI PRISM® 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA, USA) with BigDye® Terminator Kit v.3.1 (Invitrogen, Carlsbad, CA, USA) at the Shanghai Invitrogen Biological Technology Company Limited (Beijing, China).
DNA sequence analysis
DNA sequences generated by forward and reverse primers were used to obtain consensus sequences using SeqMan v.7.1.0 in the DNASTAR Lasergene Core Suite software program (DNASTAR Inc., Madison, WI, USA). Sequences were aligned using MAFFT v.6 (Katoh and Toh 2010) and edited manually using MEGA5 (Tamura et al. 2011). Phylogenetic analysis was performed using PAUP* v.4.0b10 for maximum parsimony (MP) analysis (Swofford 2003), MrBayes v.3.1.2 for Bayesian analysis (Ronquist and Huelsenbeck 2003), and PhyML v.7.2.8 for maximum likelihood (ML) analysis (Guindon et al. 2010). The first analysis was performed on the combined multi-gene dataset (act1, ITS, LSU, tefA and tubB). A second analysis using ITS sequence data was performed to compare Cytospora species from the current study with other ex-type strains in GenBank (Supplementary Table 1). Phomopsis vaccinii Shear was selected as outgroup (Adams et al. 2005). Trees were shown using FigTree v.1.3.1 (Rambaut and Drummond 2010).
MP analysis was performed by a heuristic search option of 1000 random-addition sequences with a tree bisection and reconnection (TBR) algorithm. Maxtrees were set to 5000, branches of zero length were collapsed, and all equally parsimonious trees were saved. Clade stability was assessed with bootstrap analysis of 1000 replicates (Hillis and Bull 1993). Other calculated parsimony scores included tree length (TL), consistency index (CI), retention index (RI) and rescaled consistency (RC). ML analysis was also performed with a generalised time-reversible (GTR) site substitution model, according to previous studies (Guindon et al. 2010; Fan et al. 2015a). The branch support was evaluated with a bootstrapping (BS) method of 1000 replicates (Hillis and Bull 1993).
Bayesian inference (BI) analysis employing a Markov chain Monte Carlo (MCMC) algorithm was performed (Rannala and Yang 1996). A nucleotide substitution model was estimated by MrModeltest v.2.3 (Posada and Crandall 1998). Two MCMC chains were run from random trees for 1000,000 generations, and trees were sampled every 100th generation, resulting in a total of 10,000 trees. The first 25 % of trees were discarded as the burn-in phase of each analysis. Branches with significant Bayesian posterior probabilities (BPP) were estimated in the remaining 7500 trees.
Sequence data were deposited in GenBank (Table 1). The multilocus and ITS sequence alignment files were deposited in TreeBASE (www.treebase.org) as accession S16893. The taxonomic novelty was deposited in MycoBank (Crous et al. 2004).
Morphology
Species identification was based on morphological features of the fruiting bodies produced on infected plant tissues, supplemented by cultural characteristics. Hence, thin cross-sections were prepared by hand using a double-edged blade. Morphological characteristics of the fruiting bodies including size and arrangement of stromata, presence or absence of conceptacle in stromata, number and diameter of ostioles per disc, shape and size of discs, arrangement of locules, and size and shape of conidiophores and conidia (asci and ascospores) were determined under a light microscope. More than 20 fruiting bodies were sectioned, and 50 spores were randomly selected for measurement using a Leica compound microscope (DM2500; Leica Microsystems, Wetzlar, Germany). Cultural characteristics of isolates incubated on PDA in the dark at 25 °C were recorded, including colony colour and pycnidium structure.
Results
Molecular phylogeny
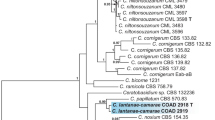
A total of 37 specimens associated with Cytospora infections were collected from Ulmus pumila in six provinces in China. Of these, ten strains representing four species were added to 12 reference sequences for further phylogenetic analyses. The phylogram (Fig. 1) generated here indicated 22 ingroup taxa, including 2863 characters, of which 1827 characters were constant; 138 variable characters were parsimony-uninformative and 898 were parsimony-informative. The heuristic search using maximum parsimony (MP) generated two parsimonious trees (TL = 2332, CI = 0.673, RI = 0.804, RC = 0.541), from which one was selected and is shown in Fig. 1.
Phylogram of combined act1, ITS, LSU, tefA, and tubB genes generated from maximum parsimony (MP) analyses. Values above the branches indicate maximum parsimony bootstrap (MP BP ≥ 50 %) and maximum likelihood bootstrap (ML BP ≥ 50 %). Thickened branches represent posterior probabilities (BI PP ≥ 0.90) from Bayesian inference. Scale bar = 70 nucleotide substitutions. Ex-type strains are in bold
Phylogenetic analysis of ITS sequence data of representative Cytospora sequences, including sequences from available ex-type strains, comprised 101 sequences. The Cytospora dataset included a total of 584 base pairs used for analyses after alignment. Of these, 352 characters were constant; 62 variable characters were parsimony-uninformative and 170 were parsimony-informative. A heuristic MP search generated 500 parsimonious trees, with the best tree with short tree length (TL = 820, CI = 450, RI = 0.829, RC = 0.373) shown in Fig. 2. All trees of ML and Bayesian analysis are in agreement, and do not significantly differ from the MP tree.
Phylogram of ITS regions generated from maximum parsimony (MP) analyses. Values above the branches indicate maximum parsimony bootstrap (MP BP ≥ 50 %) and maximum likelihood bootstrap (ML BP ≥ 50 %). Values below branches represent posterior probabilities (BI PP ≥ 0.90) from Bayesian inference. Scale bar = 20 nucleotide substitutions. The new sequences resulting from the current study are in blue. Ex-type strains are in bold
Taxonomy
Cytospora pruinopsis C.M. Tian & X.L. Fan, sp. nov. (Fig. 3)
Cytospora pruinopsis from Ulmus pumila (BJFC-S1073, holotype). a, b Habit of conidiomata on a twig in vivo. c Transverse sections through conidiomata. d Longitudinal sections through conidiomata. e Conidia. f Conidiophores. g Colonies on PDA at 3 days (left) and 30 days (right). Scale bars: A = 1 mm; B = 500 μm; C, D = 200 μm; E, F = 5 μm
MycoBank 813225
Etymology: Named after its morphological similarity to C. pruinosa.
Differs from C. pruinosa in wing-like ectostroma around ostiole and smaller conidial size, 2–4 × 1 μm.
Holotype: BJFC-S1073. CHINA, Shaanxi Province: Yulin City, Yuyang, 37°26′50.46″N, 116°20′39.44″E, 1120 m.a.s.l., on twigs and branches of U. pumila, coll. X.L. Fan, 14 May 2013 (BJFC-S1073, holotype; living ex-type culture, CFCC50034, CCTCC AF2014026);
Host/Distribution: Pathogen on twigs and branches of Ulmus pumila in China.
Conidiomata immersed in bark, slightly erumpent through the bark surface, discoid, with a single locule; superficially resembling cytophomoid conidiomata. Disc grey to black, nearly flat, circular to ovoid, (280–)290–310(−330) μm (av. = 300 μm, n = 20) diam, with one ostiole per disc. Ostiole medium grey to black, prominent, (106–)120.5–145.5(−152.5) μm (av. = 130.5 μm, n = 20) diam. Locule undivided, irregular, (470–)520–720(−790) μm (av. = 600 μm, n = 20) diam. Conidiophores hyaline, unbranched or occasionally branched at base and commonly branched above the base, (13–)14–20(−21) μm (av. = 18 μm, n = 20). Conidia hyaline, eguttulate, elongate-allantoid, aseptate, (2–)2.5–3.5(−4) × 1 μm (av. = 3 × 1 μm, n = 50).
Cultures: Colony growth on PDA initially white, becoming grey after 7–10 days. Colonies flat, felt-like, with a uniform texture. Conidiomata irregularly dispersed over agar surface.
Materials examined: CHINA, Jilin Province: Tonghua City, 41°73′56.12″N, 125°96′85.84″E, 225 m.a.s.l., on twigs and branches of U. pumila, coll. X.L. Fan, 7 June 2012 (BJFC-S334, paratype; ex-paratype culture, CFCC50035).
Cytospora chrysosperma (Pers.) Fr., Syst. Mycol. 2: 542, 1823. (Fig. 4),
Morphology of Cytospora chrysosperma from Ulmus pumila (BJFC-S788). a, b Habit of ascomata on twig. c Transverse sections through ascomata. d, e Habit of conidiomata on twig. f Transverse sections through conidiomata. g Longitudinal sections through ascomata. h Longitudinal sections through conidiomata. i Asci. j ascospores. k Conidiophores. l Conidia. m Colonies on PDA at 3 days (left) and 30 days (right). Scale bars: A, D = 1 mm; B, E = 500 μm; C, F – H = 200 μm; I – L = 5 μm
Host/Distribution: Pathogen on twigs and branches of U. pumila. Known from Armeniaca vulgaris, Crataegus azarolus, Ficus carica, Ligustrum latifolium, Malus pumila, Morus alba, Olea sativa, Persica vulgaris, Prunus domestica, Robinia pseudoacacia and Thuja orientalis in Iran; Fraxinus in Europe and Iran; Juglans regia in China and Iran; Salicaceae in China, Iran, Netherlands, South Africa, Switzerland, UK and USA; Sophora japonica in China; Triticum in Germany; and Ulmus in USA. (Saccardo 1884; Adams et al. 2005; Gadgil 2005; Fotouhifar et al. 2010; Fan et al. 2014a, 2015a).
Ascostromata immersed in the bark, erumpent, scattered, circular to ovoid, (800–)870–1090(−1120) μm (av. = 900 μm, n = 20) diam. Disc usually obscured by tightly ostiolar necks, when apparent pale brown to black, nearly hemispherical, circular to ovoid, (300–)330–370(−400) μm (av. = 350 μm, n = 20) diam, with 4–8 ostioles arranged circinately in a disc, brown to black, (65–)68.5–81(−89.5) μm (av. = 75.5 μm, n = 20) diam; 4–8 perithecia arranged circinately in black entostromata, flask-shaped to spherical, (420–)460–550(−610) μm (av. = 500 μm, n = 20) diam. Asci free, clavate to elongate obovoid, 25–33.5(−34) × 4–4.5 μm (av. = 31 × 4 μm, n = 20), 8-spored. Ascospores elongate-allantoid, thin-walled, hyaline, aseptate, (9–)9.5–11.5(−12) × (2–)2.5–3 μm (av. = 10 × 2.5 μm, n = 50). Conidiomata immersed in bark, erumpent, discoid, flask-shaped to conical, with a large multiple locules. Disc grey to black, nearly flat, circular to ovoid, (200–)230–310(−340) μm (av. = 300 μm, n = 20) diam, with one ostiole per disc. Ostiole medium grey, prominent, (61.5–)65–81(−86.5) μm (av. = 75.5 μm, n = 20) diam. Locules complex multi-loculed, subdivided frequently by invaginations, sharing common walls, (620–)650–1250(−1280) μm (av. = 1110 μm, n = 20) diam. Conidiophores hyaline, unbranched or occasionally branched at the base, (15.5–)16–23 μm (av. = 17.5 μm, n = 20). Conidia hyaline, eguttulate, elongate-allantoid, aseptate, (4–)4.5–5(−5.5) × 1–1.5 μm (av. = 4.5 × 1.5 μm, n = 50).
Cultures: Culture initially white, becoming partially pale yellow after 6–7 days, chiefly white. Colonies flat, felt-like, with uniform texture. Conidiomata formed randomly over agar surface.
Materials examined: CHINA, Tibet Province: Shigatse City, 29°27′34.53″N, 89°90′23.08″E, elev. 3976 m.a.s.l., on twigs and branches of U. pumila, coll. X.L. Fan, 13 Feb. 2012, (BJFC-S788; living culture, CFCC89982).
Notes: Cytospora chrysosperma is the most commonly recorded Cytospora species, with a wide host range (Deng 1963; Tai 1979; Wei 1979; Chen 2002; Zhuang 2005; Adams et al. 2006; Fan et al. 2014b). Fan et al. (2014b) treated BJFC-CGHs-10 (living culture CFCC89600) as a reference specimen, as it has not yet been epitypified. In the current study, isolate CFCC89982 was shown to be C. chrysosperma based on phylogenetic analyses and morphological characters. This is the first record of C. chrysosperma on U. pumila in China.
Cytospora carbonacea Fr., Syst. mycol. (Lundae) 2(2): 544 (1823). (Fig. 5)
Morphology of Cytospora carbonacea from Ulmus pumila (BJFC-S630). a, b Habit of ascomata on twig. c Transverse sections through ascomata. d, e Habit of conidiomata on twig. f Transverse sections through conidiomata. g Longitudinal sections through ascomata. h Longitudinal sections through conidiomata. i Asci. j Ascospores. k Conidiophores. l Conidia. m Colonies on PDA at 3 days (left) and 30 days (right). Scale bars: A, D = 1 mm; B, E = 500 μm; C, F–H = 200 μm; I–L = 5 μm
Host/Distribution: Pathogen on twigs and branches of U. pumila. Known from U. americana and U. campestris in Germany, and U. minor in Iran (Fries 1823; Fotouhifar et al. 2010)
Ascostromata immersed in the bark, erumpent, not crowded, circular to ovoid, (840–)900–1180(−1230) μm (av. = 980 μm, n = 20) diam. Disc pale brown to black, circular to ovoid, (240–)260–330(−380) μm (av. = 320 μm, n = 20) diam, with 4–8 ostioles arranged circinately in a disc, brown to black, (76–)80–100.5(−106) μm (av. = 95.5 μm, n = 20) diam; 4–10 perithecia arranged circinately in black entostromata, flask-shaped to spherical, (300–)320–390(−410) μm (av. = 350 μm, n = 20) diam. Asci free, clavate to elongate obovoid, (49.5–)50–59.5(−60) × 4–4.5(−5) μm (av. = 54.5 × 4.5 μm, n = 20), 8-spored. Ascospores elongate-allantoid, thin-walled, hyaline, aseptate, (14–)14.5–17.5(−18) × (4–)4.5–5 μm (av. = 16 × 4.5 μm, n = 50). Conidiomata immersed in bark, erumpent in a large area, discoid, with large multiple locules. Disc grey to black, nearly flat, circular to ovoid, (310–)350–380(−420) μm (av. = 360 μm, n = 20) diam, with multiple ostioles per disc, with columnar stroma crossed over the pycnidia. Ostiole medium grey, prominent, (46.5–)50–57(−62) (av. = 54.5 μm, n = 20) diam. Locules complex multi-loculed, subdivided frequently by invaginations with common walls, (940–)1100–1450(−1510) μm (av. = 1270 μm, n = 20) diam. Conidiophores hyaline, unbranched or occasionally branched at the base, (11–)12–20(−21.5) μm (av. = 15 μm, n = 20). Conidia hyaline, eguttulate, elongate-allantoid, aseptate, (8.5–)9–13(−13.5) × (1.5–)2–3 μm (av. = 11 × 2 μm, n = 50).
Cultures: Cultures initially white, slow-growing, becoming pale to dark brown after 7–10 days. The colony was flat, felt-like, with a uniform texture, compact. Conidiomata formed randomly on the agar surface.
Materials examined: CHINA, Shaanxi Province: Yulin City, 38°15′02.66″N, 109°44′31.79″E, elev. 1080 m.a.s.l., on twigs and branches of U. pumila, coll. X.L. Fan, 26 May 2012, (BJFC-S1071; living culture, CFCC50055); 38°15′01.73″N, 109°44′30.27″E, elev. 1071 m.a.s.l., on twigs and branches of U. pumila, coll. X.L. Fan, 26 May 2012, (BJFC-S1072; living culture, CFCC50056); CHINA, Qinghai Province: Haidong city, Pingan county, 36°28′50.48″N, 102°10′03.29″E, elev. 2208 m.a.s.l., on twigs and branches of U. pumila, coll. X.L. Fan, 15 August 2012, (BJFC-S630; living culture, CFCC89947); Haidong city, Huzhu county, Weiyuan, 36°50′54.64″N, 101°57′41.89″E, elev. 2601 m.a.s.l., on twigs and branches of U. pumila, coll. X.L. Fan, 15 August 2012, (BJFC-S631; living culture, CFCC50059); CHINA, Heilongjiang Province: Qiqihar City, Longsha Park, 47°35′06.68″N, 123°95′05.97″E, elev. 231 m.a.s.l., on twigs and branches of U. pumila, coll. X.L. Fan, 13 July 2011, (BJFC-S339; living culture, CFCC50058).
Note: Cytospora carbonacea has previously been recorded from Ulmus americana, U. campestris and U. minor in Germany and Iran (Fotouhifar et al. 2010). The first report of this species in China was from Syzygium aromaticum (Zhang et al. 2014a, b). The present study is the first report of C. carbonacea from U. pumila also providing detailed descriptions of both sexual and asexual morphs. In the present study, isolates CFCC50055, CFCC50056, CFCC50058, CFCC50059 and CFCC89947 were shown to be C. carbonacea based on phylogenetic analyses and morphological characters. Fresh collections of C. carbonacea are needed from Ulmus sp. in USA (Fries 1823) for epitypification purposes to fix the application of the name.
Cytospora ribis Ehrenb., Sylv. mycol. berol. (Berlin): 28 (1818). (Fig. 6)
Morphology of Cytospora ribis from Ulmus pumila (BJFC-S671). a, b Habit of conidiomata on a twig. c Transverse sections through conidiomata. d Longitudinal sections through conidiomata. e Conidiophores. f Conidia. g Colonies on PDA at 3 days (left) and 30 days (right). Scale bars: A = 1 mm; B = 500 μm; C, D = 200 μm; E, F = 5 μm
Host/Distribution: Pathogen on twigs and branches of U. pumila. Known from Elaeagnus angustifolia, Lepidium latifolium, Platanus orientalis, Thuja orientalis in Iran, Platanus orientalis, Ribes mandshuricum in Poland, and Ribes rubrum in the Netherlands (Saccardo 1884; Mulenko et al. 2008; Fotouhifar et al. 2010).
Conidiomata immersed in bark, erumpent in a large area, discoid, with large multiple locules. Disc grey to black, nearly flat, circular to ovoid, (350–)380–410(−470) μm (av. = 400 μm, n = 20) diam, with one to four ostioles per disc. Ostiole medium grey, prominent, (120–)130–160(−180) μm (av. = 150 μm, n = 20) diam. Locules complex multi-loculed, subdivided frequently by invaginations with common walls, (1440–)1470–1800(−1890) μm (av. = 1650 μm, n = 20) diam. Conidiophores hyaline, unbranched or occasionally branched at the bases, (17–)17.5–18(−18.5) μm (av. = 17.5 μm, n = 20), occasionally conidiophores 38.5–39.5(−40) μm (av. = 39 μm, n = 20). Conidia hyaline, eguttulate, elongate-allantoid, aseptate, (3.0–)3.5–4.5(−5) × 1–1.5 μm (av. = 4× 1 μm, n = 50).
Cultures: Colonies remaining white, growing rapidly, covering the dish after 7–10 days. Colonies flat, felt-like, with a regular edge, texture uniform; conidiomata sparse, irregularly distributed over agar surface.
Materials examined: CHINA, Qinghai Province: Xining City, 36°38′32.51″N, 101°44′42.89″E, elev. 2419 m.a.s.l., on twigs and branches of U. pumila, coll. X.L. Fan, 16 Aug. 2012, (BJFC-S671; living culture, CFCC50026); ibid. (living culture, CFCC50027).
Notes: Cytospora ribis has been reported from Iran, Poland and the Netherlands (Mulenko et al. 2008; Fotouhifar et al. 2010). However, these records lacked detailed descriptions and illustrations (Mulenko et al. 2008; Fotouhifar et al. 2010). Isolates CFCC50026, CFCC50027 were shown to be C. ribis based on phylogenetic analysis and morphological characters. This finding represents a new host record for China. Cytospora ribis has not been epitypified, and fresh collections are needed from Ribes rubrum L. in Germany for epitypification purposes.
Discussion
The current study identified four species (C. carbonacea, C. chrysosperma, C. pruinopsis sp. nov. and C. ribis) associated with canker disease on Ulmus pumila in northern China. Cytospora carbonacea and C. chrysosperma have been reported previously from Ulmus spp., but without detailed morphological observations (Conway and Morrison 1983; Fotouhifar et al. 2010; Zhang et al. 2014a, b). In addition, two other species were isolated from Ulmus pumila, namely C. pruinopsis sp. nov. and C. ribis. In the phylogenetic analysis, C. ribis formed a distinct clade with high support values (MP-BS/ML-BS/BPP = 98/99/100) (Fig. 2). Cytospora pruinopsis sp. nov. has a single conidiomatal locule, with one ostiole per disc, and is thus easily distinguishable from the other three species. Furthermore, based on phylogenetic analyses, these four species also proved to be distinct (Fig. 2).
Although species of Cytospora have been commonly reported from Ulmus spp. in several countries, these records have largely been lacking in detailed morphological and molecular data. The species can be differentiated, however, based on the characteristics of their conidiomata and conidial dimensions. Six species (C. ambiens, C. carbonacea, C. chrysosperma, C. leucostoma, C. pulchella and C. sacculus) associated with Cytospora canker disease of Ulmus spp. can be distinguished. Cytospora carbonacea has multiple conidiomatal ostioles, and can thus easily be differentiated from the other species in this study. C. chrysosperma has multi-loculate conidiomata, subdivided by invaginations into irregular chambers sharing common walls, which distinguishes it from C. sacculus. Cytospora pulchella has larger conidia than C. ambiens and C. sacculus (6–8 × 1.5–2 vs. 4.5–4.9 × 1–1.2 μm and 3.6–5.2 × 0.9–1.2 μm). Cytospora leucostoma can be separated based on the presence of a conceptacle compared with other Cytospora spp. from Ulmus.
The new species C. pruinopsis is similar to C. pruinosa (Fr.) Sacc. in that it has a single locule with a central ostiole. It differs from C. pruinosa, however, in its wing-like ectostroma around the ostiole and smaller conidia (3 × 0.8 vs. 5–6 × 1.2 μm in C. pruinosa) (Adams et al. 2006). Based on phylogenetic analyses, C. carbonacea is most closely related to C. elaeagni, which has been recorded from Elaeagnus angustifolia in China, Germany and North America (Saccardo 1889; Chen 2002; Zhuang 2005). Cytospora pruinopsis can be distinguished from C. elaeagni based on its ostiolar morphology (multiple ostioles vs. a single ostiole), larger mean conidial size (11 × 2 vs. 7.7 × 2.6 μm) and smaller mean conidiophore length (14.9 vs. 22.5 μm).
Cytospora ribis was newly reported from Ulmus pumila in this study, which also represents a new host association for this species. C. carbonacea has previously been reported from U. americana L. and U. campestris L. in Germany and U. minor Mill. in Iran (Fotouhifar et al. 2010). In the present study C. carbonacea was isolated from U. pumila, suggesting that Ulmus spp. may be its primary host. C. chrysosperma is the type species of Cytospora, and occurs globally on a wide range of hosts (Adams et al. 2005; Fan et al. 2014a). Previous records show five plant genera listed as hosts of this species in China—chiefly, Salix and Populus, but also Castanea, Morus, and less frequently, Ulmus (Deng 1963; Tai 1979; Wei 1979; Chen 2002; Zhuang 2005). The present study clarified the presence of four Cytospora species isolated from U. pumila in northern China, of which one species proved to be new to science. To fully elucidate the Cytospora spp. that occur on woody hosts, a more exhaustive sampling of other hosts from other regions of the world will be needed to help clarify the host range and distribution of these important canker pathogens.
References
Adams GC, Surve-Iyer RS, Iezzoni AF (2002) Ribosomal DNA sequence divergence and group I introns within the Leucostoma species L. cinctum, L. persoonii, and L. parapersoonii sp. nov., ascomycetes that cause Cytospora canker of fruit trees. Mycologia 94:947–967
Adams GC, Wingfield MJ, Common R, Roux J (2005) Phylogenetic relationships and morphology of Cytospora species and related teleomorphs (Ascomycota, Diaporthales, Valsaceae) from Eucalyptus. Stud Mycol 52:1–144
Adams GC, Roux J, Wingfield MJ (2006) Cytospora species (Ascomycota, Diaporthales, Valsaceae), introduced and native pathogens of trees in South Africa. Australas Plant Pathol 35:521–548
Carbone I, Kohn L (1999) A method for designing primer sets for speciation studies in filamentous Ascomycetes. Mycologia 91:553–556
Chen MM (2002) Forest fungi phytogeography, forest fungi phytogeography of China, North America, and Siberia and international quarantine of tree pathogens. Sacramento, California, USA
Conway KE, Morrison LS (1983) Diseases and decay fungi in windbreaks in Oklahoma. Plant Dis 67:289–291
Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G (2004) MycoBank: an online initiative to launch mycology into the 21st century. Stud Mycol 50:19–22
Crous PW, Hawksworth DL, Wingfield MJ (2015) Identifying and naming plant-pathogenic fungi: past, present, and future. Annu Rev Psychol 53:12.1–12.21
Deng SQ (1963) Fungi of China. Beijing, China. (in Chinese)
Desjardins P, Hansen JB, Allen M (2009) Microvolume protein concentration determination using the NanoDrop 2000c spectrophotometer. J Vis Exp 33:1–3
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Dudka IO, Heluta VP, Tykhonenko YY, Andrianova TV, Hayova VP, Prydiuk MP, Dzhagan VV, Isikov VP (2004) Fungi of the Crimean peninsula. Institute of Botany, National Academy of Sciences of Ukraine, Ukraine
Ehrenberg CG (1818) Sylvae Mycologicae Berolinenses. Formis Theophili Bruschcke, Berlin (In Latin)
Fan XL, Liang YM, Ma R, Tian CM (2014a) Morphological and phylogenetic studies of Cytospora (Valsaceae, Diaporthales) isolates from Chinese scholar tree, with description of a new species. Mycoscience 55:252–259
Fan XL, Tian CM, Yang Q, Liang YM, You CJ, Zhang YB (2014b) Cytospora from Salix in northern China. Mycotaxon 129:303–315
Fan XL, Hyde KD, Liu M, Liang YM, Tian CM (2015a) Cytospora species associated with walnut canker disease in China, with description of a new species C. gigalocus. Fungal Biol 119:310–319
Fan XL, Hyde KD, Yang Q, Liang YM, Ma R, Tian CM (2015b) Cytospora species associated with canker disease of three anti-desertification plants in northwestern China. Phytotaxa 197:227–244
Fotouhifar KB, Hedjaroude GA, Leuchtmann A (2010) ITS rDNA phylogeny of Iranian strains of Cytospora and associated teleomorphs. Mycologia 102:1369–1382
Fries EM (1823) Systema mycologicum vol 2. Greifswald, Germany. (in Latin)
Gadgil PD (2005) Fungi on trees and shrubs in New Zealand. Fungi of New Zealand Volume 4. Hong Kong, China
Gilman JC, Tiffany LH, Lewis RM (1957) Iowa Ascomycetes II. Diaporthaceae: Valsaceae. Iowa State Coll J Sci 31:623–647
Glass NL, Donaldson GC (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol 61:1323–1330
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321
Hillis DM, Bull JJ (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol 42:182–192
Katoh K, Toh H (2010) Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26:1899–1900
McNeill J, Barrie FR, Buck WR, et al (2012) International code of nomenclature for algae, fungi, and plants (Melbourne Code). Regnum Veg 154
Mulenko W, Majewski T, Ruszkiewicz-Michalska M (2008) A preliminary checklist of micromycetes in Poland. W. Szafer Institute of Botany, Polish Academy of Sciences
O’Donnell K (1993) Fusarium and its near relatives. In: Reynolds DR, Taylor JW (eds) The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systenatics. CAB International, Wallingford, pp 225–233
Posada D, Crandall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818
Rambaut A, Drummond A (2010) FigTree v.1.3.1. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh
Rannala B, Yang Z (1996) Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J Mol Evol 43:304–311
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Rossman AY, Adams GC, Cannon PF, Castlebury LA, Crous PW, Gryzenhout M, Jaklitsch WM, Mejia LC, Stoykov D, Udayanga D, Voglmayr H, Walker DM (2015) Recommendations of generic names in Diaporthales competing for protection or use. IMA Fungus 6:145–154
Saccardo PA (1884) Sylloge Fungorum vol 3. Typis Seminarii, Italy (in Latin)
Saccardo PA (1889) Sylloge Fungorum vol 14. Typis Seminarii, Italy (in Latin)
Spaulding P (1961) Foreign diseases of forest trees of the world. US Dep Agric Agric Handb 197:1–361
Spielman LJ (1983) Taxonomy and biology of Valsa species on hardwoods in North America, with special reference to species on maples. Cornell University, New York
Spielman LJ (1985) A monograph of Valsa on hardwoods in North America. Can J Bot 63:1355–1378
Swofford DL (2003) PAUP*: Phylogenetic analysis using parsimony (* and other methods). Version 4.0b10. Sunderland, England, UK
Tai FL (1979) Sylloge Fungorum Sinicorum. Beijing, China. (in Chinese)
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Wang YL, Lu Q, Decock C, Li YX, Zhang XY (2015) Cytospora species from Populus and Salix in China with C. davidiana sp. nov. Fungal Biol 119:420–432
Wei JC (1979) Identification of fungus handbook. House, Shanghai, China. (in Chinese)
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Snisky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. San Diego, USA, pp 315–322
Wingfield MJ, De Beer ZW, Slippers B, Wingfield BD, Groenewald JZ, Lombard L, Crous PW (2012) One fungus, one name promotes progressive plant pathology. Mol Plant Pathol 13:604–613
Zhang QT, Lu Q, He M, Decock C, Zhang XY (2014a) Cytospora palm sp. nov. (Diaporthales, Ascomycota), a canker agent on Cotinus coggygria (Anacardiaceae) in Northern China. Cryptogam Mycol 35:211–220
Zhang YB, You CJ, Fan XL, Tian CM (2014b) Taxonomy and phylogeny of Cytospora in Northeast China. Mycosystema 33:806–818 (in Chinese)
Zhuang WY (2005) Fungi of northwestern China. Mycotaxon Ltd, Ithaca
Acknowledgments
This study is financed by National Natural Science Foundation of China (Project No.: 31170603). We are grateful to Chungen Piao and Minwei Guo (China Forestry Culture Collection Center [CFCC], Chinese Academy of Forestry, Beijing) for support of strain preservation during this study. Thanks are also due to Yongping An (Forest Diseases and Insect Pests Prevention Station of Guyuan, Ningxia, China) and Huaizhang Yao (Environment Protection Bureau of Yulin Economic Development Area, Shaanxi, China) for assistance with specimen collection during this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Section Editor: Roland Kirschner
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table 1
(DOC 105 kb)
Rights and permissions
About this article
Cite this article
Yang, Q., Fan, XL., Crous, P.W. et al. Cytospora from Ulmus pumila in Northern China. Mycol Progress 14, 74 (2015). https://doi.org/10.1007/s11557-015-1096-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11557-015-1096-1