Abstract
A new poroid wood-inhabiting basidiomycete, Perenniporia nanlingensis, collected in Guangdong Province, southern China, is described and illustrated on the basis of three collections. Both the morphology and phylogenetic analysis of rDNA ITS sequences support this new species. Macroscopically, the new species is characterized by an annual growth habit, resupinate basidiocarps with pinkish buff to cinnamon-buff pore surface when dry. Microscopically, it has a trimitic hyphal system, slightly dextrinoid and cyanophilous skeletal and binding hyphae, and its basidiospores are ellipsoid, truncate, strongly dextrinoid and cyanophilous, 9.0–9.8 × 5.0–5.9 μm.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Perenniporia Murrill is a large, cosmopolitan genus. According to the modern definition it is characterized by ellipsoid to distinctly truncate basidiospores, which usually are thick-walled, cyanophilous and variably dextrinoid; its hyphal structure is di- to trimitic with clamp connections on generative hyphae and its vegetative hyphae are cyanophilous and variably dextrinoid (Decock and Stalpers 2006). About ninety species have been described or transferred to the genus, and most of them occur in the tropics or subtropics (Cui et al. 2007; Dai et al. 2002; Decock 2001; Decock and Figueroa 2000; Decock and Ryvarden 1999, 2000, 2003; Decock et al. 2000, 2001; Gilbertson and Ryvarden 1987; Hattori and Lee 1999; Núñez and Ryvarden 2001; Ryvarden and Gilbertson 1994; Xiong et al. 2008).
Knowledge of Perenniporia in China was summarized by Dai et al. (2002). To date, 29 species of Perenniporia have been recorded from China (Cui et al. 2006, 2007, 2008, 2010; Dai and Penttilä 2006; Dai et al. 2003, 2004, 2007; Wang et al. 2009; Xiong et al. 2008). During an investigation of wood-inhabiting fungi in southern China, an additional undescribed species of Perenniporia was found. To confirm the affinity of the new taxon and infer its evolutionary relationships among similar species of Perenniporia, phylogenetic analyses were performed based on ITS sequences.
Materials and methods
Morphological studies
The studied specimens are deposited at the herbarium of the Institute of Microbiology, Beijing Forestry University (BJFC). The microscopic routine used in the study follows Dai (2010). Sections were studied at magnification up to ×1000 using a Nikon Eclipse E 80i microscope and phase contrast illumination. Drawings were made with the aid of a drawing tube. Microscopic features, measurements and drawings were made from slide preparations stained with Cotton Blue and Melzer’s reagent. Spores were measured from sections cut from the tubes. In presenting the variation in the size of the spores, 5% of measurements were excluded from each end of the range, and are given in parentheses. In the text the following abbreviations are used: KOH = 5% potassium hydroxide, CB = Cotton Blue, CB + = cyanophilous, L = mean spore length (arithmetic average of all spores), W = mean spore width (arithmetic average of all spores), Q = variation in the L/W ratios between the specimens studied, n = number of spores measured from given number of specimens. Special color terms are from Anonymous (1969) and Petersen (1996).
Molecular procedures and phylogenetic analyses
The fungal taxa used in this study are listed in Table 1. Phire Plant Direct PCR Kit (Finnzymes) procedure was used to extract total genomic DNA from the fruitbody and for the polymerase chain reaction (PCR), and PCR amplification was confirmed on 1% agarose electrophoresis gels stained with ethidium bromide (Stöger et al. 2006). DNA sequencing was performed at Beijing Genomics Institute. All newly generated sequences have been submitted to GenBank and are listed in Table 1.
Sequences were aligned with additional sequences from GenBank (Table 1) using BioEdit (Hall 1999) and ClustalX (Thomson et al. 1997). Alignment was manually adjusted to allow maximum alignment and minimize gaps. In the study, nuclear ribosomal RNA genes were used to determine the phylogenetic position of the new species. The internal transcribed spacer (ITS) regions were amplified with the primers ITS4 and ITS5 (Gardes and Bruns 1993; White et al. 1990).
Maximum parsimony and MrBayesian analysis were applied to the ITS dataset. All characters were weighted and gaps were treated as missing data. Maximum parsimony analysis (PAUP* version 4.0b10) was used (Swofford and PAUP* 2002). Trees were inferred using the heuristic search option with TBR branch swapping and 1,000 random sequence additions. Max-trees were set to 5,000, branches of zero length were collapsed and all parsimonious trees were saved. Clade stability was assessed using a bootstrap (BT) analysis with 1,000 replicates (Felsenstein 1985). Descriptive tree statistics tree length (TL), consistency index (CI), retention index (RI), rescaled consistency index (RC), and homoplasy index (HI) were calculated for all trees generated under different optimality criteria. Bayesian analysis with MrBayes3.1.2 (Ronquist and Huelsenbeck 2003) implementing the Markov Chain Monto Carlo (MCMC) technique and parameters predetermined with MrMODELTEST2.3 (Posada and Crandall 1998; Nylander 2004) was performed and the parameters in MrBayes were set as follows: lset nst = 6, and rates = gamma. Four simultaneous Markov chains were run with 1,500,000 generations, starting from random trees, and keeping one tree every 1,000th generation.
Results
Taxonomy
Perenniporia nanlingensis B.K. Cui & C.L. Zhao, sp. nov. (Fig. 1)
MycoBank no.: MB 561625
Carpophorum annuum, resupinatum. Facies pororum cremea bubalina vel roseo-bubalina; pori rotundi, 6–7 per mm. Systema hypharum trimiticum, hyphae generatoriae fibulatae, hyphae skeletales subiculi 2.6–5.2 μm in diam. Sporae hyalinae, ellipsoideae, truncatae, dextrinoideae, CB+, 9.0–9.8 × 5.0–5.9 μm.
Type. — CHINA. Guangdong Province, Ruyang County, Nanling Nature Reserve, on dead angiosperm tree, 16.IX.2009 Cui 7589 (holotype in BJFC); Cui 7541 & 7620 (paratype in BJFC).
Etymology. — nanlingensis (Lat.): referring to the mountain name in southern China.
Fruitbody. — Basidiocarps annual, resupinate, adnate, not easily separated from substrate, corky, without odour or taste when fresh, becoming hard corky upon drying, up to 35 cm long, 10 cm wide, 5.5 mm thick at center. Pore surface cream-buff to yellowish buff when fresh, pinkish buff to cinnamon-buff upon drying; pores round, 6–7 per mm; dissepiments thick, entire. Sterile margin narrow, cream-buff, up to 1 mm wide. Subiculum cream to buff, thin, up to 0.5 mm thick. Tubes concolorous with pore surface, hard corky, up to 5 mm long.
Hyphal structure. — Hyphal system trimitic; generative hyphae with clamp connections; skeletal and binding hyphae weakly detrinoid, CB+, tissues unchanged in KOH.
Subiculum. — Generative hyphae infrequent, hyaline, thin-walled, usually unbranched, 2.5–3.8 μm in diam; skeletal hyphae dominant, hyaline, thick-walled with a wide to narrow lumen, branched, interwoven, 2.6–5.2 μm in diam.; binding hyphae hyaline, thick-walled, frequently branched, flexuous, interwoven, 1.5–2.5 μm in diam.
Tubes. — Generative hyphae infrequent, hyaline, thin-walled, unbranched, 2.8–3.5 μm in diam.; skeletal hyphae dominant, hyaline, thick-walled with a wide to narrow lumen, branched, interwoven, 2.9–5 μm; binding hyphae hyaline, thick-walled, frequently branched, interwoven, 1.1–2.7 μm in diam. Cystidia absent, but fusoid cystidioles present, hyaline, thin-walled, 10–13 × 4.2–6 μm; basidia barrel-shaped, with four sterigmata and a basal clamp connection, 10.2–11.1 × 8.2–9.3 μm; basidioles dominant, in shape similar to basidia, but slightly smaller.
Spores. — Basidiospores ellipsoid, distinctly truncate, hyaline, thick-walled, smooth, strongly dextrinoid, CB+, (8.8–)9.0–9.8(–10.0) × (4.9–)5.0–5.9(–6.0) μm, L = 9.32 μm, W = 5.39 μm, Q = 1.7–1.76 (n = 90/3).
Molecular phylogeny
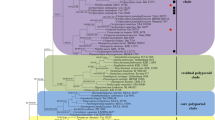
The ITS dataset included sequences from 50 fungal specimens representing 26 taxa. The dataset had an aligned length of 577 base pairs with 303 parsimony informative positions. Parsimony analysis yielded one parsimonious tree (TL = 362, CI = 0.503, RI = 0.765, RC = 0.385, HI = 0.497) that is shown in Fig. 2. Bayesian analysis resulted in average standard deviation of split frequencies = 0.006778. In phylogenetic reconstruction, the sequences of Polyporus arcularius (Batsch) Fr. obtained from GenBank (AF516524) was used as outgroup. The ITS strict consensus tree (Fig. 2) generated by Bayesian analysis and Maxium Parsimony showed sequences of Perenniporia nanlingensis were grouped together with other species of Perenniporia as a monophyletic cluster with strong support.
Discussion
The phylogenetic analysis showed that the three samples of Perenniporia nanlingensis formed a distinct lineage and showed phylogenetic distance from other taxa. Both morphology and rDNA data confirmed that the three samples represent a new species in Perenniporia.
Perenniporia nanlingensis is characterized by an annual growth habit, resupinate basidiocarps with pinkish buff to cinnamon-buff pore surface, a trimitic hyphal system with slightly dextrinoid and cyanophilous skeletal and binding hyphae, and its basidiospores are ellipsoid, truncate, strongly dextrinoid and cyanophilous, 9.0–9.8 × 5.0–5.9 μm.
Perenniporia roseoisabellina (Pat. & Gaillard) Ryvarden and P. nanlingensis share similar truncate basidiospores, but the former differs from the latter in larger pores (2–3 per mm) and nondextrinoid hyphae (Hattori and Lee 1999). Perenniporia subaurantiaca (Rodway & Cleland) P.K. Buchanan & Ryvarden resembles P. nanlingensis in both having similar sized pores and basidiospores, but the former has a pale brown pore surface and non-truncate basidiospores (Hattori and Lee 1999).
Perenniporia nanlingensis may be confused with P. subacida (Peck) Donk by sharing resupinate basidiocarps and a buff to cinnamon-buff pore surface (Núñez and Ryvarden 2001), but the latter species is distinguished from P. nanlingensis by having perennial basidiocarps and smaller basidiospores (4.3–5.4 × 3.2–4.1 μm, Dai et al. 2002).
Perenniporia straminea (Bres.) Ryvarden is similar to P. nanlingensis, both having an annual growth habit, resupinate basidiocarps, similar pores (6–7 per mm), and truncate basidiospores. However, P. straminea is distinguished from P. nanlingensis by having distinctly smaller basidiospores (3.2–4 × 2.4–3 μm, Cui et al. 2010).
Phylogenetically, Perenniporia japonica (Yasuda) T. Hatt. & Ryvarden is so far the most closely related species to P. nanlingensis. The former species, however, differs from P. nanlingensis by having rhizomorphs and smaller basidiospores, 4–5.2 × 3–3.9 μm (Dai et al. 2002).
References
Anonymous (1969) Flora of British fungi. Colour identification chart. Her Majesty´s Stationery Office, London
Cui BK, Dai YC, Decock C (2006) Two species of Perenniporia (Basidiomycota, Aphyllophorales) new to China. Fungal Sci 21:23–28
Cui BK, Dai YC, Decock C (2007) A new species of Perenniporia (Basidiomycota, Aphyllophorales) from eastern China. Mycotaxon 99:175–180
Cui BK, Yuan HS, Dai YC (2008) Wood-rotting fungi in eastern China 1. Polypores from Wuyi Mountains, Fujian Province. Sydowia 60:25–40
Cui BK, Zhao CL, Li HJ, He SH (2010) Polypores from Chebaling Nature Reserve, Guangdong Province. Mycosystema 29:834–840
Dai YC (2010) Hymenochaetaceae (Basidiomycota) in China. Fungal Divers 45:131–343
Dai YC, Cui BK, Huang MY (2007) Polypores from eastern Inner Mongolia, northeastern China. Nova Hedwig 84:513–520
Dai YC, Härkonen M, Niemelä T (2003) Wood-inhabiting fungi in southern China 1. Polypores from Hunan Province. Ann Bot Fenn 40:381–393
Dai YC, Niemelä T, Kinnunen J (2002) The polypore genera Abundisporus and Perenniporia (Basidiomycota) in China, with notes on Haploporus. Ann Bot Fenn 39:169–182
Dai YC, Penttilä R (2006) Polypore diversity of Fenglin Nature Reserve, northeastern China. Ann Bot Fenn 43:81–96
Dai YC, Wei YL, Wang Z (2004) Wood-inhabiting fungi in southern China 2. Polypores from Sichuan Province. Ann Bot Fenn 41:319–329
Decock C (2001) Studies in Perenniporia. Some Southeast Asian taxa revisited. Mycologia 93:774–759
Decock C, Buchanan P, Ryvarden L (2000) Revision of some Australasian taxa of Perenniporia (Basidiomycota, Aphyllophorales). Aust Syst Bot 13:823–844
Decock C, Figueroa H (2000) Studies in Perenniporia, Navisporus ortizii, a synonym of Perenniporia matius, and a note on Navisporus and Perenniporia in Cuba. Cryptogam Mycol 21:153–162
Decock C, Figueroa H, Ryvarden L (2001) Studies in Perenniporia. Perenniporia contraria and its presumed taxonomic synonym Fomes subannosus. Mycologia 93:196–204
Decock C, Ryvarden L (1999) Studies in neotropical polypores. Some coloured resupinate Perenniporia species. Mycol Res 103:1138–1144
Decock C, Ryvarden L (2000) Studies in neotropical polypores 6. New resupinate Perenniporia species with small pores and small basidiospores. Mycologia 92:354–360
Decock C, Ryvarden L (2003) Perenniporiella gen. nov. segregated from Perenniporia, including a key to neotropical Perenniporia species with pileate basidiomes. Mycol Res 107:93–103
Decock C, Stalpers J (2006) Studies in Perenniporia: Polyporus unitus, Boletus medulla-panis, the nomenclature of Perenniporia, Poria and Physisporus, and a note on European Perenniporia with a resupinate basidiome. Taxon 53:759–778
Felsenstein J (1985) Confidence intervals on phylogenetics: an approach using bootstrap. Evolution 39:783–791
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118
Gilbertson RL, Ryvarden L (1987) North American polypores 2. Megasporoporia – Wrightoporia. Fungiflora, Oslo
Hall TA (1999) Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hattori T, Lee S (1999) Two new species of Perenniporia described from a lowland rainforest of Malaysia. Mycologia 91:525–531
Núñez M, Ryvarden L (2001) East Asian polypores 2. Polyporaceae s. lato. Synop Fungorum 14:165–522
Nylander JAA (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University
Petersen JH (1996) Farvekort. The Danish Mycological Society’s colour-chart. Foreningen til Svampekundskabens Fremme, Greve
Posada D, Crandall KA (1998) Modeltest: Testing the model of DNA substitution. Bioinformatics 14:817–818
Ronquist F, Huelsenbeck JP (2003) MRBAYES 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Ryvarden L, Gilbertson RL (1994) European polypores 2. Synop Fungorum 7:394–743
Stöger A, Schaffer J, Ruppitsch W (2006) A rapid and sensitive method for direct detection of Erwinia amylovora in symptomatic and asymptomatic plant tissues by polymerase chain reaction. J Phytopathol 154:469–473
Swofford DL, PAUP* (2002) Phylogenetic analysis using parsimony (*and other methods). Version 4.0b10. Sinauer Associates, Sunderland, Massachusetts
Thomson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Wang B, Dai YC, Cui BK, Du P, Li HJ (2009) Wood-rotting fungi in eastern China 4. Polypores from Dagang Mountains, Jiangxi Province. Cryptogam Mycol 30:233–241
White TJ, Burns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols, a guide to methods and applications. Academic, San Diego
Xiong HX, Dai YC, Cui BK (2008) Perenniporia minor (Basidiomycota, Aphyllophorales), a new polypore from China. Mycotaxon 105:59–64
Acknowledgements
Special thanks are due to Drs. Ping Du, Haijiao Li and Wei Wang (BJFC, China) for assistance on field trips and to Dr. Xiaoyong Liu (HMAS, China) for help in molecular analysis. We express our gratitude to Prof. Yucheng Dai (IFP, China) for revising the Latin description and improving the manuscript. The research is financed by the National Natural Science Foundation of China (Project Nos. 30900006, 31093440 and 30910103907).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhao, CL., Cui, BK. A new species of Perenniporia (Polyporales, Basidiomycota) described from southern China based on morphological and molecular characters. Mycol Progress 11, 555–560 (2012). https://doi.org/10.1007/s11557-011-0770-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-011-0770-1