Abstract
The neglected false truffle species Hydnotrya bailii Soehner (Ascomycetes, Discinaceae) is re-described and separated from its sister taxon Hydnotrya tulasnei by morphological and phylogenetic analyses based on internal transcribed spacer rDNA sequences. The most distinct morphological and ecological characters are small globose, rather than kidney-like, ascomata as known from the sister taxon H. tulasnei, strictly monoseriate ascospores and montane habitats. Phylogenetic analyses resulted in two clearly separated clusters that revealed the ectomycorrhizal specificity of H. bailii to Picea abies and that H. tulasnei is preferably associated to Fagus sylvatica. We also show that H. bailii was already present in mycorrhizal samples but until now could not be correctly assigned. Our analyses also indicate cryptic diversity within H. cerebriformis and other, morphologically not yet characterized, Hydnotrya groups. An emended determination key for all Hydnotrya species known from Central Europe is provided.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The evolution of mycorrhizal mutualism had a profound impact on terrestrial life. Fossil records indicate that mycorrhizae facilitated the colonization of the land by plants, and today over 90% of the plant species form mycorrhizal associations (Smith and Read 2008). Ectomycorrhizal fungi display a great variety in their phenotypic appearance (Hibbett et al. 1997). Mycologists very early noted the morphological similarity between gasteroid hypogeous and epigeous representative fruiting bodies (Hesse 1890; Zeller and Dodge 1924). The shift between these forms is known as gastromycetation (Albee-Scott 2007; Binder and Bresinsky 2002; Thiers 1984). Within the Ascomycetes, this evolutionary process is solely known from the Morchellaceae-Discinaceae-Helvellaceae-Tuberaceae clade (Laesso and Hansen 2007).
To date, three genera are known in the family Discinaceae: Discina, Gyromitra and Hydnotrya. Inconsistent species concepts of the hypogeous ectomycorrhizal genus Hydnotrya have lead to systematic rearrangements in the past (Montecchi and Sarasini 2000; Moser 1963; Soehner 1959). Identification of Hydnotrya species has undergone intensive questioning and controversial descriptions in the last century (Hesse 1890; Hollos 1911; Montecchi and Sarasini 2000; Moser 1963; Soehner 1959; Szemere 1965; Trappe 1975; Zhang 1991). For instance, the type species Hydnotrya tulasnei (Berk.) Berk. & Broome (1846) synonymises H. carnea (Corda) Zobel (1854) and H. intermedia Buchholtz (1904). The genus is distributed across the Northern hemisphere (Europe, North America and Asia). At present, five European Hydnotrya species are accepted, among which H. cerebriformis and H. michaelis are restricted to mountainous coniferous forests. This indicates that certain Hydnotrya species are specific ectomycorrhizal partners of coniferous trees.
In accordance with this hypothesis, the German mycologist Ert Soehner presented a valid Latin description of a novel Hydnotrya species, H. bailii, in his “Tuberaceen-Studien V” (Soehner 1959). The new species was found in association with Picea abies in mountainous forests. Soehner noted that H. carnea and H. intermedia (synonyms of H. tulasnei) can be distinguished from H. bailii by lacking strictly monoseriate ascospores. Unfortunately, Soehner's legacy was neglected by later taxonomists (Montecchi and Sarasini 2000; Szemere 1965). Because the valid species diagnosis of H. bailii had been overlooked, discriminating characters had not been recognized again. Accordingly, each more recently treated spruce-associated specimen with similarity to H. tulasnei was assigned to this species. In contrast, H. tulasnei is not associated with spruce but with Fagus sylvatica, Pinus spp. and Corylus avellana.
Here, we reconsider the taxonomical status of the neglected species Hydnotrya bailii, based on morphological and molecular characteristics. We re-evaluate its host specificity and provide an overview of the currently described and cryptic Hydnotrya species diversity, as well as a morphological key of all Central European species.
Material and methods
Morphological studies
The taxonomic descriptions are based on dried material collected by the authors and original material collected by Ert Soehner deposited at the Botanische Staatssammlung München (Table 1). Microscopic characteristics of eighteen specimens were observed from 10–20 µm thick microtome cross-sections of dried specimens mounted in 5% KOH (w/v) and additionally in cotton blue in lactic acid. Tissue measurements were made with 40x and 100x oil immersion lenses and repeated 20 times.
DNA isolation, PCR and sequencing
Two fruiting body collections of Hydnotrya bailii, one collection of Hydnotrya cerebriformis, one collection of Hydnotrya tulasnei and 14 ectomycorrhizal root tips of Hydnotrya tulasnei were sequenced (Table 1). Total genomic DNA was extracted from 50 mg ascomata using the Masterpure® Fungal Genomic DNA Kit following the manufacturer’s protocol. The ITS rDNA region was amplified with PCR primers ITS1 and ITS4 (White et al. 1990). The PCR reactions were run on a Biorad thermal cycler with the following settings: initial denaturation for 2 min at 95°C followed by 35 cycles of: 30 s denaturation at 95°C, annealing at 60°C for 30 s, extension for 1 min at 72°C and final extension at 72°C for 10 min. Alternatively, fruiting body tissue and mycorrhized root tips were homogenized with a glass micromortar and a micropestle. DNA isolation, PCR and sequencing were performed as previously described (Münzenberger et al. 2009) with the primers ITS1F (Gardes and Bruns 1993) and ITS4 (White et al. 1990).
Phylogenetic analysis
Sequences obtained as described above were complemented using a nomenclature-based search for Discinaceae ITS sequences in Genbank (http://ncbi.nlm.nih.gov/), as well as a search against NCBI's nucleotide collection using Blast (Altschul et al. 1990) and the Genbank sequence EU784276 as query. We further considered all Blast hits with a score larger than the score of the best hit placed within the sister genus, Gyromitra. To ensure sufficient sequence overlap between all accessions, sequences comprising only (or almost only) the ITS2 part were removed from the combined dataset (which comprised more ITS1-only than ITS2-only sequences).
The sequences thus obtained were aligned with the fast but accurate POA software (version 2; Lee et al. 2002) in progressive alignment mode. In order to cope with alignment ambiguity caused by the need to include potentially distant outgroup sequences for rooting, we followed a two-step approach. To be able to use the Gyromitra sequences as outgroup, the first alignment comprised all selected Discinaceae accessions, but was cleaned from ambiguously aligned regions using Gblocks (Castresana 2000). After phylogenetic trees had been inferred from the first alignment as described below, a second alignment was constructed with POA in the same way, but was restricted to those ingroup sequences that appeared sufficiently close to H. bailii and H. tulasnei in the first phylogenetic trees. Accordingly, the second alignment could be used throughout its entire length in phylogenetic analysis, and rooting of the resulting trees could be done according to the results from the first analysis.
Phylogenetic analysis under the maximum-likelihood (ML) criterion (Felsenstein 1981) was done with RAxML version 7.0.4, using its novel rapid bootstrap option with subsequent search for the best tree under the GTRMIX approach (Stamatakis et al. 2008). GTRMIX uses the fast but accurate GTRCAT model approximation during heuristic search but the full GTR+GAMMA model for the final likelihood computation (Stamatakis 2006). Bootstrapping under the maximum-parsimony (MP) criterion (Fitch 1971) was done with PAUP* version 4.0b10 (Swofford 2002), treating gaps as missing data, collapsing branches of zero minimum length, and using 10 rounds of random sequence addition followed by TBR branch swapping per bootstrap replicate. In both ML and MP bootstrapping, 1000 replicates were conducted. Trees were inferred from the two alignments in exactly the same manner. To quantify the separation between H. tulasnei and H. bailii observed in the inferred trees, we calculated pairwise uncorrected distances with PAUP* (treating gaps as missing data) and determined the maximum within-cluster and minimum between-cluster distances (the “barcoding gap”) for these two clades using OPTSIL (Göker et al. 2009) in input cluster quality mode.
Sequence alignments and phylogenetic trees are included in the online supplementary material available at http://dx.doi.org/10.1007/s11557-009-0625-1.
Results
Comparative morphological study
The original specimens collected by Soehner and Bail (Table 1) and deposited as H. bailii at the Botanische Staatssammlung München were, regarding their micro- and macromorphology, nearly identical to our recent collections. The most striking morphological differences to the sister taxon H. tulasnei are the small globose fruiting body (Figs. 1 and 2) and the monoseriate assembly of the slightly smaller ascospores (Figs. 3 and 4). Most specimens of H. tulasnei compared to have larger fruiting bodies, and their ascospores do not strictly follow monoseriate but often biseriate assembly. These morphological characteristics were also present in the original material collected by Soehner and determined as H. intermedia, which is now regarded as a synonym of H. tulasnei.
A dichotomous key was developed and tested by using specimens from H. cerebriformis, H. michealis, H. tulasnei and H. bailii. Specimens of the two other European species H. confusa and H. cubispora known only from the British islands were not included.
Phylogenetic hypothesis
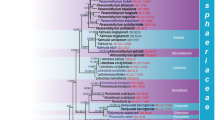
The first alignment comprised 56 sequences and 2237 characters, most of its length being caused by few sequences containing insertions within the ITS1 and others comprising parts of the large subunit rDNA. Gblocks reduced this alignment to 395 unambiguously aligned characters. The resulting best ML tree had a log likelihood of −2547.675 and is shown in Fig. 5 together with ML and MP bootstrap values. While the separation of ingroup (Hydnotrya) and outgroup (Gyromitra) sequences was moderately to well supported (99/70%), strong support (96–100%) was achieved for a basal split of Hydnotrya in a clade comprising Genbank sequences from H. michaelis and one from “H. variiformis” and another clade comprising the remaining specimens (H. tulasnei, H. bailii, H. cerebriformis, “H. variiformis”, H. cubispora and various environmental and undetermined samples). Because the H. michaelis clade was almost as distant to the clade containing the remaining Hydnotrya as the outgroup, the second alignment was restricted to this large Hydnotrya subclade, and rooting of the second tree was done according to this subclade's basal split into H. tulasnei, H. bailii and some environmental samples on the one hand and H. cerebriformis, “H. variiformis”, H. cubispora and other undetermined samples on the other hand.
Phylogenetic tree inferred under the maximum-likelihood (ML) criterion from 395 unambiguously aligned characters and rooted with the sister genus Gyromitra (Discinaceae). Numbers on the branches represent support values from 1,000 replicates under the ML (left) and maximum-parsimony (right) criterion. The branches are scaled in terms of the expected number of substitutions per site. Sequences from Genbank are indicated by their accession number in parentheses after their “organism” modifier as found in the Genbank flatfiles. Labels of sequences newly obtained in the course of this study include the assigned taxon name and the isolation number. Note the considerable distance between H. michaelis/“H. cf. variiformis” and the other Hydnotrya collections. Triangles indicate collections from Europe, squares indicate collections from North America and dots indicate collections from Asia
The best ML tree inferred from the taxonomically restricted but full-length second alignment had a log likelihood of −4381.652 and is shown in Fig. 6 together with ML and MP bootstrap values. The topology is nearly identical to the corresponding subtree of the tree shown in Fig. 5, but, as expected, the support values are significantly higher for some groupings. H. bailii and H. tulasnei are revealed as well-separated and well-supported (100%) sister groups of each other (90/68%). Both include undetermined Genbank samples; the H. bailii cluster also includes one Genbank sequence (AM261522) identified as H. tulasnei. Pairwise uncorrected distances within the H. tulasnei cluster were at most 1.03%; within the H. bailii cluster, at most 0.27%; between the two clusters, a minimum of 4.33%. Further sequences from undetermined samples form a grade at the base of the H. bailii/H. tulasnei clade, most of them being also quite distant to each other and forming highly supported (95–100%) subclades. Within the outgroup, our three European collections of H. cerebriformis appear as even more distant from their North American counterparts (represented by GenBank accessions) than H. bailii from H. tulasnei, also hinting at cryptic diversity within Hydnotrya.
Phylogenetic tree inferred under the maximum-likelihood (ML) criterion from a full ITS alignment including closely related Hydnotrya sequences selected and rooted according to the tree in Fig. 5. Numbers on the branches represent support values from 1,000 replicates under the ML (left) and maximum-parsimony (right) criterion. The branches are scaled in terms of the expected number of substitutions per site. Sequences from Genbank are indicated by their accession number in parentheses after their “organism” modifier as found in the GenBank flatfiles. Labels of sequences newly obtained in the course of this study include the assigned taxon name and the isolation number. Triangles indicate collections from Europe, squares indicate collections from North America and dots indicate collections from Asia
Taxonomic description
Because Soehner’s description was neglected by more recent studies, we here repeat the Latin diagnosis given in 1959, and provide the first English diagnosis.
Hydnotrya bailii Soehner 1959
Latin diagnosis
Ascomata hypogaea, primum subglobosa, subtomentosa, cum 1–3 cavernis, postea tuberose excavate, carnosa, 1–2(−2,5) cm in diam. metientia. Gleba canaliculis et vinculis gyroso-labyrintheis composite, cubicula hymenio ascisque subhymenialibus crebris et paraphysibus vestita. Parietes 1 mm crassi foris subalbidis. Asci cylindracei apice rotundati, 250–300 × 35–40 µm longi et lati, 8-spori, paraphyses ascos superantes, Sporae exacte uniseriatae, sphaericae, verrucosae, (27.5-)30–34(−37.5) µm cum sculptura in diam metientes.
Holotype
Zackenfall, Sudeten mountain range (1880, leg. Bail: Rabenhorst, Hb. Mycol. ed. 2. no. 321-M: Holotypus, Botanische Staatsammlung München).
Etymology and diagnosis
The name of the species refers to the collector of the holotype, Bail. Young ascomata ochre, later magenta reddish, when ageing dark brown to brown-black, irregularly wrinkled and elongated, bulbous, uneven, with deep furrows often with multiple lobes, with one or many irregular vent-like disruptive openings of elliptic to crater-like shape or many smaller ones leading to the inner space, openings usually found at the apex, seldom at basal site, surface waxy velvety, lacking a basal hole. Peridia 150–200 µm thick.
Gleba ochre to magenta reddish to dark brown; depending on ascomata age, strongly convoluted cavities, with white velvety coating: Paraphyses small, cylindrical (250–300 µm). Asci cylindrical, 250–300 × 30–40 µm, eight-spored, numerous asci in subhymenium. Ascospores strictly monoseriate, (27.5-) 30–34 (−37.5) µm (with ornamentation), globose, ripened spores brown-reddish, blistered warty. Ascomata 1–2(−2.5) cm in size with pleasant aromatic smell.
Ecology
The holotype is known from montane spruce (Picea abies) forests. The present collections, which confirm the host specificity postulated by Soehner (1959), were made near Schierke (Harz; Saxony-Anhalt; Germany) and near Hinterzarten (Black Forest; Baden-Württemberg, Germany).
Specimens studied for comparisons
Holotype Hydnotrya bailii (Soehner) (leg. Bail 1880), Hydnotrya bailii (Soehner) 2064 Simmerberg (Herb. Soehner), Hydnotrya bailii Zuckermantel (1905). All specimens are deposited at the Botanische Staatsammlung München (corresponding curator Dr. D. Triebel).
Discussion
Based on morphological data and ITS sequence analysis we here confirm the neglected taxon Hydnotrya bailii (Soehner 1959) as a distinct species. H. bailii is morphologically differentiated from Hydnotrya tulasnei by its smaller and globose fruiting body, strictly monoseriate ascospores, specificity to spruce and to a montane-boreal habitat. We do not consider the lack of an ITS sequence from the holotype or other material determined by Soehner as Hydnotrya bailii (which would clarify the issue of sequence identity with the present material) as an obstacle for recognizing the two species as distinct. Morphological comparison confirmed the identity of our recently collected specimens and those investigated by Soehner (1959) regarding the characteristics listed by Soehner as typical for H. bailii.
Hydnotrya species such as Hydnotrya cerebriformis Harkn. (1889), Hydnotrya michaelis (E. Fisch) Trappe (1975), Hydnotrya cubispora (E.A. Bessey & B.E. Thomps.) Gilkey (1939) (Lack 2003) and Hydnotrya confusa Spooner (1992) can be easily distinguished by their ascospore ornamentation. In contrast, H. tulasnei and H. bailii are more difficult to distinguish due to their similar tissue anatomy. This might explain the complete absence of H. bailii in the current literature and databases (e.g. Mycobank, Index Fungorum). Difficult access to the original publication (Soehner 1959) and the small number of mycologists in Europe collecting hypogeous fungi may also account for this problem.
There are strong indications that Hydnotrya tulasnei is ectomycorrhizal with preference for broad leafed trees. Tedersoo et al. (2006) analyzed beech ectomycorrhizae whose ITS sequences were identical to sporocarp sequences of Hydnotrya tulasnei found in a Danish beech forest (AJ969616 and AJ969620 from mycorrhized root tips, AJ969621 from a sporocarp). Another sporocarp sequence (EU784276) of original material collected by Hawker (1954) was morphologically determined as Hydnotrya tulasnei (Brock et al. 2009). These four sequences fall into our Hydnotrya cluster (Figs. 5 and 6). The sequences of the H. tulasnei cluster are prevalently from broad-leafed trees but never from spruce. Sequences of specimens reported to be spruce-associated Hydnotrya species, do not fall into our H. tulasnei cluster. Vohnik et al. (2007) identified a spruce-associated sporocarp from a mountainous area of Southern Bohemia in the Czech Republic (AM261522) as H. tulasnei based on 99% sequence identity with Genbank entry AJ534700. This sequence stems from a mycorrhized root tip collected in an Estonian mixed forest also containing spruce (Tedersoo et al. 2003). Both sequences fall unambiguously into our H. bailii cluster. We have to conclude that (Vohnik et al. 2007) and (Tedersoo et al. 2003) collected specimens of H. bailii. This supports our observation that H. bailii is separated from H. tulasnei not only by morphological characters and ITS-sequence but also by preference for coniferous trees and a montane to boreal distribution.
Regarding other Hydnotrya species, our results reveal that H. cerebriformis requires a taxonomic revision, as our European collections are clearly differentiated from the North American ones (represented by the Genbank sequences in Figs. 5 and 6) in our phylogenetic reconstructions using the ITS locus. The branches separating the two clades are even longer than those separating H. bailii from H. tulasnei (Figs. 5 and 6). Cryptic Hydnotrya species from distinct and biogeographically diverse sampling sites across Europe (Peintner et al. 2007, EF040877 unpublished: EU816614) North America (Dickie et al. 2009, EU880227 unpublished: AY684066, DQ420632, DQ 437704) and Japan (Ishida et al. 2007, AB218071, Ogura-Tsujita and Yukawa 2008, AB428790) and forming several clades within a grade next to the H. tulasnei/H. bailii cluster also indicate that the species diversity of Hydnotrya is far greater than previously assumed. Morphologically well-characterized species such as Hydnotrya confusa that were not yet sequenced may account for some of the observed “cryptic” diversity. Most likely some Hydnotrya species still remain undiscovered. We therefore recommend intensifying the search for fruiting bodies at these sampling sites to clarify the presence of such unknown taxa most likely morphologically similar to H. tulasnei. Furthermore, the investigation of collections from other areas is required, other continents as well as other regions of Europe such as the Mediterranean, the Balkans and the Carpathian areas. For now, the 50th anniversary of Hydnotrya bailii should be dedicated to the resurrection of this valid species whose discriminating characters were not recognized during the past 50 years.
Dichotomous key to the central European Hydnotrya species
-
1. Ascospores in general ellipsoid, 24–35 × 13–26 µm, (quotient: 1.3–1.8) brown, irregularly warty, hyaline when younger. Ptycothecium strongly folded inwards, brown to brownish-yellow, sometimes carmine, hollow, with reddish gleba and white hymenial surface. So far only reported from montane coniferous habitats................................................................Hydnotrya michaelis
-
1′. Ascospores in general globose to cubic..................................................................................2
-
2. Ascospores brown to yellow-brown with dense spiny ornamentation, 28–35 µm in diameter*. Ptycothecium dark brown-carmine coloured, strongly folded inwards. Gleba uncoloured, with white hymenial surface. Associated with montane conifers.........................Hydnotrya cerebriformis
-
2′. Ascospores, cubic, 20–45 µm in diameter*........................................Hydnotrya cubispora
-
2″. Ascospores ornamented with large warts and coarse harsh surface, mono and biseriate, 21–38 µm in diameter*.................................................................................................................................3
-
3. Ascospores biseriate, rarely monoseriate, 25–38 µm in diameter*. Ptycothecium uneven, reddish-ochre with brown notes, kidney-like, lobated, surface not much folded inwards, 1–6 cm in width. Gleba rose-whitish, later dark red with white hymenial surface. Broadest host range of all Hydnotrya species, but in general associated with Fagus, Corylus and Pinus in lowlands and montane habitats..........................................................................................Hydnotrya tulasnei
-
3′. Ascospores strictly monoseriate (when young rarely biseriate), 21–36 µm in diameter*. Ptycothecium uneven, reddish-ochre-brown, lobated, infolded on the surface, in general rather globose than wide, 1–1.5 cm, gleba rose-whitish, later dark red with white hymenial surface. Strictly associated with Picea abies in montane habitats..........................................Hydnotrya bailii
*Including ornamentation
References
Albee-Scott SR (2007) Does secotioid inertia drive the evolution of false-truffles? Mycol Res 111:1030–1039. doi:10.1016/j.mycres.2007.08.008
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Binder M, Bresinsky A (2002) Derivation of a polymorphic lineage of Gasteromycetes from boletoid ancestors. Mycologia 94:85–98
Brock M, Döring H, Bidartondo MI (2009) How to know unknown fungi: the role of a herbarium. New Phytol 181:719–724. doi:10.1111/j.1469-8137.2008.02703.x
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552
Dickie IA, Dentinger BTM, Avis PG, McLaughlin DJ, Reich PB (2009) Ectomycorrhizal fungal communities of oak savanna are distinct from forest communities. Mycologia. doi:10.3852/08-178
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Fitch WM (1971) Towards defining the course of evolution: minimal change for a specified tree topology. Syst Zool 20:406–416
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118. doi:10.1111/j.1365-294X.1993.tb00005.x
Göker M, García-Blázquez G, Voglmayr H, Tellería MT, Martín MP (2009) Molecular taxonomy of phytopathogenic fungi: a case study in Peronospora. PLoS One 4:e6319. doi:10.1371/journal.pone.0006319
Hawker L (1954) British hypogeous fungi. Phil Trans Roy Soc London 237:429–546
Hesse R (1890) Die Hypogaeen Deutschlands Band II. Verlag L Hoffstetter, Halle
Hibbett DS, Pine EM, Langer E, Langer G, Donoghue MJ (1997) Evolution of gilled mushrooms and puffballs inferred from ribosomal DNA sequences. PNAS 94:12002–12006
Hollos L (1911) Magyarország földalatti gombái, Szarvasgombaféléi (Fungi Hypogaei Hungariae). Budapest, Kiadja
Ishida TA, Nara K, Hogetsu T (2007) Host effects on ectomycorrhizal fungal communities: insight from eight host species in mixed conifer-broad leaf forests. New Phytol 174:430–440. doi:10.1111/j.1469-8137.2007.02016.x
Laesso T, Hansen K (2007) Truffle trouble: what happened to the Tuberales. Mycol Res 111:1075–1099. doi:10.1016/j.mycres.2007.08.004
Lack B (2003) Hydnotrya cubispora in Wales. Field Mycol 4:4
Lee C, Grasso C, Sharlow MF (2002) Multiple sequence alignment using partial order graphs. Bioinformatics 18:452–464
Montecchi A, Sarasini M (2000) Fungi ipogei d’Europa. AMB, Trento
Moser M (1963) Ascomyceten (Schlauchpilze) Band IIa. Gustav Fischer Verlag, Stuttgart
Münzenberger B, Bubner B, Wöllecke J, Sieber TN, Bauer R, Fladung M, Hüttl RF (2009) The ectomycorrhizal morphotype Pinirhiza sclerotia is formed by Acephala macrosclerotiorum sp. nov., a close relative of Phialocephala fortinii. Mycorrhiza, doi:10.1007/s00572-009-0239-0
Ogura-Tsujita Y, Yukawa T (2008) Epipactis helleborine shows strong mycorrhizal preference towards ectomycorrhizal fungi with contrasting geographic distributions in Japan. Mycorrhiza 18:331–338. doi:10.1007/s00572-008-0187-0
Peintner U, Iotti M, Klotz P, Bonuso E, Zambonelli A (2007) Soil fungal communities in a Castanea sativa (chestnut) forest producing large quantities of Boletus edulis sensu lato (porcini): where is the mycelium of porcini? Environ Microbiol 9:880–889. doi:10.1111/j.1462-2920.2006.01208.x
Smith SE, Read DJ (2008) Mycorrhizal symbiosis, Academic press
Soehner E (1959) Tuberaceen Studien V, Mitteilungen der Botanischen Staatssammlung München
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690
Stamatakis A, Hoover P, Rougemont J (2008) A rapid bootstrap algorithm for the RAxML web servers. Syst Biol 75:758–771. doi:10.1080/10635150802429642
Swofford DL (2002) PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0 b10. Sinauer Associates, Sunderland
Szemere L (1965) Die unterirdischen Pilze des Karpatenbeckens. Akadémiai Kiadó, Budapest
Thiers HD (1984) The secotioid syndrome. Mycologia 76:1–8
Tedersoo L, Kõljalg U, Hallenberg N, Larsson KH (2003) Fine scale distribution of ectomycorrhizal fungi and roots across substrate layers including coarse woody debris in a mixed forest. New Phytol 159:153–165. doi:10.1046/j.0028-646x.2003.00792.x
Tedersoo L, Hansen K, Perry BA, Kjøller R (2006) Molecular and morphological diversity of pezizalean ectomycorrhiza. New Phytol 170:581–596. doi:10.1111/j.1469-8137.2006.01678.x
Trappe JM (1975) Generic synonyms in the Tuberales. Mycotaxon 2:109–122
Vohnik M, Fendrych M, Kolarik M, Gryndler M, Hrselova H, Albrechtova J, Vosatka M (2007) The ascomycete Meliniomyces variabilis isolated from a sporocarp of Hydnotrya tulasnei (Pezizales) intracellularly colonises roots of ecto- and ericoid mycorrhizal host plants. Czech Mycol 59:215–226
White TJ, Bruns TD, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR Protocols: a guide to methods and applications. Academic Press, New York, pp 315–322
Zeller SM, Dodge CW (1924) Leucogaster and Leucophlebs in North America. Ann Mo Bot Gard 11:389–410
Zhang BC (1991) Morphology, cytology and taxonomy of Hydnotrya cerebriformis (Pezizales). Mycotaxon 42:155
Acknowledgements
We thank M. Roth, Müncheberg for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
These files are unfortunately not in the Publisher's archive anymore:
-
Supplementary 1 (NEX 73.3 kb)
-
Supplementary 2 (NEX 138 kb)
Rights and permissions
About this article
Cite this article
Stielow, B., Bubner, B., Hensel, G. et al. The neglected hypogeous fungus Hydnotrya bailii Soehner (1959) is a widespread sister taxon of Hydnotrya tulasnei (Berk.) Berk. & Broome (1846). Mycol Progress 9, 195–203 (2010). https://doi.org/10.1007/s11557-009-0625-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-009-0625-1