Abstract
Capers (Capparis spinosa) are affected by white blister rust attributed to Albugo capparidis or, applying a broad species concept, to Albugo candida. Within the past 3 years, a great diversity within Albugo parasitic to the Brassicaceae has been observed. This has led to the description of two new specialized species within the parasites to Brassicaceae and the confirmation that Albugo lepidii is distinct from Albugo candida. In addition, it has been realized that Albugo candida has a broad host spectrum within the Brassicaceae, extending to the closely related Cleomaceae. Through molecular phylogenetic analysis of cox2 sequences and morphological comparison, it is demonstrated that the host range of A. candida extends to the Capparaceae. These findings are both relevant for practical plant pathology and raise questions regarding the mechanisms involved in the exceptional broad host range of Albugo candida, compared to other Albugo species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Capers are widely grown in the Mediterranean and elsewhere for their edible flower buds. The caper bush (Capparis spinosa L.) is attacked by relatively few pathogens with one of the most prevalent being white blister rust caused by members of the Albuginaceae. This family of plant parasites is highly distinctive from the second obligate parasitic group of oomycetes, the Peronosporaceae (Riethmüller et al. 2002; Hudspeth et al. 2003; Thines and Spring 2005; Thines et al. 2008). The Albuginaceae contain four distinct lineages (Albugo s.str., parasitic to Brassicales; Albugo s.l., parasitic to Convolvulaceae; Pustula, parasitic to Asterales s.l.; and Wilsoniana, parasitic to Caryophyllales). Within the Albuginales, about 40 species responsible for white blister rust disease of economically important agricultural crops and common weeds have been described (Biga 1955; Choi and Priest 1995). The most widely recognized species, Albugo candida (Pers.) Roussel, had been thought to be the exclusive white rust pathogen of the Brassicaceae, infecting as many as 63 genera and 241 species (Mukerji 1975; Saharan and Verma 1992). Only recently was it realized that a high degree of genetic diversity is present within Albugo on Brassicaceae (Choi et al. 2006; Voglmayr and Riethmüller 2006) and that several of the observed lineages might constitute distinct species. Following the recent lectotypification of A. candida, the taxonomic status of which had previously been unclear (Choi et al. 2007), two specialized Albugo species parasitic to Brassicaceae have been described within Albugo (Choi et al. 2007, 2008). It was also demonstrated that A. candida s.str. has a broad host range extending over more than a dozen genera of the Brassicaceae and into the Cleomaceae, as the type of A. chardonii W. Weston (Chardón and Toro 1930; Vanev et al. 1993) was found to be nested within A. candida (Choi et al. 2007). Albugo candida had also been reported from Capparaceae, (Choi and Priest 1995), but since no clear-cut morphological difference was given between A. candida and A. capparidis (de Bary) Kuntze, the second species of Albugo reported from Capparaceae, the identification of A. candida remained unsecured. Given that Albugo parasitic to Brassicales includes several distinct species (Choi et al. 2007, 2008) on the one hand, and the broad host spectrum for A. candida on the other, it was the aim of this study to clarify if the host spectrum of A. candida extends to Capparaceae, or if the lack of morphological characters for distinguishing A. candida from A. capparidis stated in prior studies (e.g., de Bary 1863; Berlese and DeToni 1888) are the reason for the reports of A. candida on Capparaceae.
Materials and methods
DNA was extracted for analysis of the cox2 locus from two specimens of A. chardonii, (including the type; Plant Pathology Herbarium of Cornell University—CUP), and six specimens of Albugo capparidis (US National Fungus Collection—BPI; and the Jardin Botanique National de Belgique—BR).
Specimens examined: Albugo chardonii: Colombia, Dept. Cundinamarca, wet meadow above Salto de Tequendama, on Cleome anomala H.B. & K., 6 July 1929, Carlos Eugenio Chardón (CUP-CO-000668, Typus); USA, NY, Ontario Co., Geneva, on Cleome hassleriana Chodat cv. ‘Rose Queen’, 28 May 2002, Karen Snover (CUP-065770). Albugo capparidis: Ethiopia, Agaro, Kaffa Province, 15 km N. W. of Jimma, edge of woodland along low road to, alt. 1780 m. 7°43′N 36°46′E, on Capparis sp., 5 January 1962, F.G. Meyer (BPI 185279); Italy, Rome, on living leaves of Capparis sp., May 1904 (BPI 184346; other herbaria: D. Saccardo Mycotheca Italica 1461); Italy, Environs de Roma, Mont Palatino, on Capparis rupestris Sibth. & Sm. (now, a synonym of C. spinosa), September 1887, E. Bommer and M. Rousseau (BR 075128-50); Italy, Bovezzano, on Capparis rupestris (now, a synonym of C. spinosa), Sept 1887, E. Bommer and M. Rousseau (BR 075128-51); Italy, Verona, on leaves of Capparis spinosa, Sept 1878, E. Bommer and M. Rousseau (BR 075128-52); USA, Hawaii, Kaalualu and Waiohinu, halfway between, on living leaves and stems of Capparis sp., 7 September 1929, Otto Degener (BPI 184347; other herbaria: Plants of Hawaii Herbarium Otto Degener 3761).
Morphological studies
Herbarium specimens were first moistened with 70% alcohol, and then the white blister rust organs were transferred with glass needles to a droplet of 60% lactic acid pipetted onto a microscope slide. These preparations were slowly warmed up, covered with coverslips and examined in brightfield- and DIC-light microscopy, using an Olympus BX51 microscope (Olympus, Tokyo, Japan) for measurements, and a Zeiss AX10 microscope (Carl Zeiss, Göttingen, Germany) mainly for photographs.
DNA extraction, amplification and sequencing
DNA was extracted from sporangiophores and sporangia formed on the lower surface of the infected leaves. DNA extraction was performed according to the methodology described in Lee and Taylor (1990). For cox2 amplification, the primers designed by Hudspeth et al. (2000) were employed. The PCR products were purified using a QIAquick gel extraction kit (Qiagen, Hilden, Germany) and sequenced on an automatic sequencer (ABI Prism TM 377 DNA Sequencer), using the BigDye™ (Applied Biosystems, Foster City, CA, USA) cycle sequencing kit, version 3.1.
Phylogenetic analysis
Thirty-four sequences of partial cox2 mtDNA, including A. capparidis, A. chardonii, and A. candida, were analyzed in this study. Information for the sequences is shown in Table 1. Herbaria abbreviations follow Holmgren and Holmgren (1998). The newly obtained cox2 sequences were edited using the DNASTAR computer package (Lasergene, Madison, WI), version 5.05. Alignment of the sequences was performed using CLUSTAL X (Thompson et al. 1997), which is possible without ambiguity because of the presence of only few indels that are restricted to the outgroup. Phylogenetic trees were obtained from the data using maximum likelihood (ML) and maximum parsimony (MP) methods. For ML inference, RAxML (Stamatakis 2006) version 7.0.3 was used with all parameters set to default values, using the GTRMIX variant. A MP heuristic search was performed with 1,000 random sequence additions and branch swapping by tree bisection-reconnection (TBR), using PAUP* version 4b10 (Swofford 2002). For both analyses, the relative robustness of the individual branches was estimated by bootstrapping (BS) using 1,000 replicates. Bootstrapping was conducted based on heuristic searches by 100 rounds of random sequence addition and subsequent TBR branch swapping. The alignments and tree obtained were deposited in Treebase (accession number S2358).
Results
Morphological analysis
Eight specimens of Albugo chardonii on Cleome anomala (1) and C. hasslerana (1) and white blister rusts labelled as A. capparidis on Capparis spinosa (3) and C. sp. (3) were morphologically compared with A. candida from various brassicaceous hosts. In Cleome anomala infected by A. chardonii, the leaf surface had sparse yellow to straw-colored lesions, with sori mostly on the lower surface, but occasionally on the upper surface. The sori were formed subepidermal, color primarily white, but occasionally light-yellow, shape elongated and irregular, often confluent, covering mostly large areas of the lower and rarely upper side of the leaf (Fig. 1a). The grouped sporangiophores were hyaline, cylindrical or clavate, 25–43 μm long, 10–15 μm wide (n = 131), thick-walled, especially towards the base (up to 5.5 μm; Fig. 1b, c). The sporangia were arranged in basipetal chains, hyaline, globose to subglobose, (13.8–)15.5–20.9(−23.5) μm diam. (n = 108), the primary sporangia were similar to the secondary sporangia, although the former exhibit a slightly thicker wall (1.2–2 μm) than the latter (0.5–0.8 μm) (Fig. 1d, e, respectively). Resting organs were not seen. Similar symptoms were observed on Capparis materials. The sori were formed on both upper and lower surface of infected leaves, and were subepidermal, colour white to yellowish, shape elongated and irregular, often confluent (up to 10 mm) and sometimes rounded, with irregularly torn epidermis. The morphological characteristics of sporangiophores and sporangia are indeed identical to those of A. chardonii, excluding minor difference in sporangial size; (13.3–)14.5–18.4(–22.5) μm diam. (n = 115) in the primary sporangia, which were similar to the secondary sporangia, although the former exhibit a slightly thicker wall (1.2–1.8 μm) than the latter (0.5–1.0 μm). Resting organs were not seen. The morphological characteristics of A. chardonii and the specimens labelled as A. capparidis are identical to those of A. candida s.str. from Arabis, Brassica, Capsella, Lunaria, Raphanus, Sisymbrium, and Thlaspi. Therefore, two species could not be differentiated from A. candida by their morphological comparison.
Albugo chardonii (a–e) on Cleome anomala and A. capparidis (f–h) on Capparis spinosa. a Sori on infected leaves; b, c, f sporangiophore; d, g primary sporangia; e, h secondary sporangia. Scale bars 15 µm for b, c, f; 20 µm for d, e, g, h. Source: CUP CO-000668 (typus) for A. chardonii; BR 75128-50 for A. capparidis
Molecular analysis
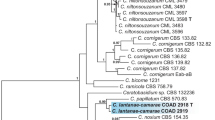
The phylogenetic relationship among the Albugo species was inferred from the ML and MP analyses of the aligned sequences of the partial cox2 mtDNA. A ML tree is presented in Fig. 2. Out of 572 total characters, 133 were parsimony-informative, and parsimony analysis resulted in 117 most parsimonious trees of 370 steps with a consistency index (Kluge and Farris 1969) of 0.6513 and a retention index (Farris 1989) of 0.8740. Since no statistically significant differences were found between the tree topologies of the ML and MP analyses, only the ML tree is shown in Fig. 2. The phylogenetic inferences revealed that the sequences from Capparis spinosa, Capparis sp., Cleome anomala, and C. hasslerana were embedded within A. candida materials from Arabidopsis, Arabis, Aubrietia, Berteroa, Biscutella, Brassica, Capsella, Diplotaxis, Eruca, Erysimum, Heliophila, Iberis, Lunaria, Raphanus, Sisymbrium, and Thlaspi, with maximum support in both analyses. The close relationship of A. chardonii, the specimens labeled as A. capparidis and A. candida was also supported by high sequence similarity of 99.8–100% (none or 1 out of 572 characters different). The level of sequence identity between these three species exceeds that found among A. candida materials from various brassicaceous hosts, indicating that they are indeed to be considered conspecific.
Phylogenetic tree of Albuginaceae species inferred by ML analysis using the partial cox2 mtDNA. ML and MP BS values above 50% are given above/below the branches, respectively. The number of nucleotide changes between taxa is represented by branch length and the scale bar equals the number of nucleotide substitutions per site. Albugo specimens from Cleome and Capparis are in bold
Discussion
For the white blister rust parasitic on Cleome anomala, the epithet “chardoni” was first introduced as a dedication to Carlos Eugenio Chardón. This epithet is generally used in most taxonomy and plant pathology related works. However, ICBN Article 60.11 (McNeill et al. 2006) requires that when a male personal name ends with a consonant, the epithet should be formed by adding -ii- (stem augmentation plus the genitive inflection) in conformity with Recommendation 60C.1. If known not to be so, the termination is to be corrected orthographically. Therefore, the termination was corrected here as “chardonii” throughout.
Until now, Albugo strains infecting Cleome species were regarded as both A. candida and A. chardonii, because of unclear morphological difference. The only previously reported morphological difference between A. chardonii and A. candida is the thickness of the sporangial wall. A. candida has been reported to have a uniform wall thickness of the sporangia, while those of A. chardonii have wall thickening across the base and sides (Chardón and Toro 1930; Choi and Priest 1995). In the present observation, however, the wall thickness of the sporangia showed no obvious morphological difference of the two species. The wall thickness in the sporangia of A. chardonii was mostly uniform, while only a few sporangia, particularly secondary ones, had irregular thickened walls, the basal and lateral portions being slightly thicker than the apical part. In a previous study (Constantinescu and Thines 2006) and the present one, however, this morphological feature was also commonly observed in most materials of A. candida. Phylogenetic analyses (Choi et al. 2007; this study) have shown A. chardonii grouped together with A. candida s.str. from various plants of the Brassicaceae with a high level of sequence homology. This hints at the possibility that the two species are indeed conspecific. Interestingly, the morphological and molecular identity among them is in agreement with previous inoculation experiments; Safeeulla (1952) and Jörstad (1964) reported that A. candida can attack plants in the Cleomaceae, and recently Khunti et al. (2000) showed that an isolate from Brassica juncea could successfully infect Cleome viscosa. Therefore, we conclude that A. chardonii is to be regarded a synonym of A. candida.
Similar to the situation in Cleome, Albugo from Capparis was also classified either as A. capparidis or as A. candida. A. capparidis was formally introduced by de Bary (1863), based on a variety of A. candida previously described by Rabenhorst (1844). But de Bary notes in his description that the species was “in speciminibus C. Candida [A. candida] omnino similia”. Also Pirotta (1884, as cited in Fischer 1892) and Berlese and DeToni (1888) could not find differences between Albugo accessions from Brassicaceae and from Capparaceae. In the monographic work of Biga (1955), the species is recognised on the basis of host preference, but both sporangia and oospore features are identical to A. candida var. macrospora. The ornamentation of the oospore wall has proven to be possibly the most important character to distinguish between closely related species in the Albuginaceae (Voglmayr and Riethmüller 2006; Choi et al. 2007, 2008). Interestingly, Choi and Priest (1995), who unfortunately do not mention any specimens they might have scrutinized, cite Biga (1955) and Saccardo (1888) and state the oospore ornamentation was “tuberculate”, a feature that could indicate that this species might be unrelated to A. candida, depending on the interpretation of the character state “tuberculate”. As Voglmayr and Riethmüller (2006) have shown that a specimen from Cleome was placed distinct from A. candida among the specialized species, two lineages can obviously parasitize the genus Cleome, A. candida and an additional possibly undescribed species. Our findings presented here demonstrate that A. candida may also parasitize Capparis spinosa and its natural host range therefore extends to the family Capparaceae. Whether the situation in Capparaceae is similar to the situation found in Cleomaceae, where two distinct Albugo species may occur as parasites, has to be revealed by future studies. In addition, it will be necessary to determine and investigate the specimen on which Rabenhorst (1844) had based the variety later granted species rank by de Bary (1863), to clarify if A. capparidis, similar to A. chardonii, needs to be relegated into synonymy with A. candida.
References
Berlese AN, DeToni JB (1888) Phycomyceteae. In: Saccardo PA (ed) Sylloge Fungorum, 7(1). P.A. Saccardo, Pavia, Italy, pp 181–322
Biga MLB (1955) Review of the species of the genus Albugo based on the morphology of the conidia. Sydowia 9:339–358
Chardón CE, Toro RA (1930) Mycological explorations of Colombia. J Dept Ag Porto R 14:222–224
Choi D, Priest MJ (1995) A key to the genus Albugo. Mycotaxon 53:261–272
Choi YJ, Hong SB, Shin HD (2006) Genetic diversity within the Albugo candida complex (Peronosporales, Oomycota) inferred from phylogenetic analysis of ITS rDNA and COX2 mtDNA sequences. Mol Phylogenet Evol 40:400–409. doi:10.1016/j.ympev.2006.03.023
Choi YJ, Shin HD, Hong SB, Thines M (2007) Morphological and molecular discrimination among Albugo candida materials infecting Capsella bursa-pastoris world-wide. Fungal Divers 27:11–34
Choi YJ, Shin HD, Ploch S, Thines M (2008) Evidence for uncharted biodiversity in the Albugo candida complex, with the description of a new species. Mycol Res 112:1327–1334
Constantinescu O, Thines M (2006) Dimorphism of sporangia in Albuginaceae (Chromista, Peronosporomycetes). Sydowia 58:178–190
de Bary A (1863) Recherches sur le développement de quelques champignons parasites. Ann Sci Nat (Bot) Sér 4 20:5–148
Farris JS (1989) The retention index and the rescaled consistency index. Cladistics 5:417–419. doi:10.1111/j.1096-0031.1989.tb00573.x
Fischer A (1892) Phycomycetes. In: Fischer A, Hauck F, Limpricht G, Luerssen C, Richter P, Winter G (eds) Dr. L. Ranenhorst’s Kryptogamen-Flora von Deutschland, Österreich und der Schweiz vol. 1, 2nd edn. von Eduard Kummer, Leipzig, Germany
Holmgren PK, Holmgren NH (1998). Index herbariorum: a global directory of public herbaria and associated staff. New York Botanical Garden. http://sciweb.nybg.org/science2/indexHerbariorum.asp. Continuously updated
Hudspeth DSS, Nadler SA, Hudspeth MES (2000) A COX2 molecular phylogeny of the Peronosporomycetes. Mycologia 92:674–684. doi:10.2307/3761425
Hudspeth DSS, Stenger D, Hudspeth MES (2003) A COX2 phylogenetic hypothesis of the downy mildews and white rusts. Fungal Divers 13:47–57
Jörstad I (1964) The Phycomycetous genera Albugo, Bremia, Plasmopara, and Pseudoperonospora in Norway, with an appendix containing unpublished finds of Peronospora. Nytt Mag Bot 11:47–82
Khunti JP, Khandar RR, Bhoraniya MF (2000) Studies on host range of Albugo cruciferarum the incitant of white rust of mustard. Agric Sci Dig 20:219–221
Kluge AG, Farris JS (1969) Quantitative phyletics and the evolution of anurans. Syst Zool 30:1–32. doi:10.2307/2412407
Lee SB, Taylor JW (1990) Isolation of DNA from fungal mycelia and single spores. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic, New York, pp 282–287
McNeill JF, Barrie F, Burdet HM, Demoulin V, Hawksworth DL, Marhold K, Nicolson DH, Prado J, Silva PC, Skog JE, Wiersema J, Turland NJ (eds) (2006) International code of botanical nomenclature (Vienna Code). Regnum vegetabile 146. Koeltz, Königstein
Mukerji KG (1975) Albugo candida. IMI Descriptions of Pathogenic Fungi and Bacteria 46, No. 460
Rabenhorst GL (1844) Deutschlands Kryptogamen-Flora. Erster Band, Pilze, Leipzig, Germany
Riethmüller A, Voglmayr H, Göker M, Weiß M, Oberwinkler F (2002) Phylogenetic relationships of the downy mildews (Peronosporales) and related groups based on nuclear large subunit ribosomal DNA sequences. Mycologia 94:834–849. doi:10.2307/3761698
Saccardo PA (1888) Cystopus capparidis. In: Saccardo PA (ed) Sylloge Fungorum, 7. P.A. Saccardo, Pavia, Italy, pp 236–237
Safeeulla KM (1952) Morphological and cytological studies of Albugo species on Ipomoea hederacea. Curr Sci 18:287–288
Saharan GS, Verma PR (1992) White rusts: a review of economically important species. International Development Research Centre, Ottawa, Ont
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi:10.1093/bioinformatics/btl446
Swofford DL (2002) PAUP: phylogenetic analysis using parsimony (*and other methods).Version 4.0b10. Sinauer, Sunderland, Mass
Thines M, Spring O (2005) A revision of Albugo (Chromista, Peronosporomycetes). Mycotaxon 92:443–458
Thines M, Göker M, Telle S, Ryley M, Mathur K, Narayana YD, Spring O, Thakur RP (2008) Phylogenetic relationships of graminicolous downy mildews based on cox2 sequence data. Mycol Res 112:345–351. doi:10.1016/j.mycres.2007.10.010
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882. doi:10.1093/nar/25.24.4876
Vanev SG, Dimitrova EG, Ilieva EI (1993) Gybite v Bylgariya. 2 tom. Razred Peronosporales. (Fungi Bulgaricae, vol. 2. Ordo Peronosporales). Bulgarian Academy of Sciences, Sofia. (In Bulgarian)
Voglmayr H, Riethmüller A (2006) Phylogenetic relationships of Albugo species (white blister rusts) based on LSU rDNA sequence and oospore data. Mycol Res 110:75–85. doi:10.1016/j.mycres.2005.09.013
Acknowledgments
The authors are grateful to the curator of the Plant Pathology Herbarium of the Cornell University, New York, USA (CUP) for providing the Albugo specimens investigated in this study. This work was financially supported by a research grant from the Korea Research Foundation (KRF-2003-015-C00611). Financial support by the German Science Foundation (DFG) and the Landesstiftung Baden-Württemberg (Elite Program for Postdocs) for M.T. is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Choi, YJ., Shin, HD. & Thines, M. The host range of Albugo candida extends from Brassicaceae through Cleomaceae to Capparaceae. Mycol Progress 8, 329–335 (2009). https://doi.org/10.1007/s11557-009-0604-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-009-0604-6