Abstract
Purpose
While 3D patient-specific digital models are currently available, thanks to advanced medical acquisition devices, there is still a long way to go before these models can be used in clinical practice. The goal of this paper is to demonstrate how 3D patient-specific models of anatomical parts can be analysed and documented accurately with morphological information extracted automatically from the data. Part-based semantic annotation of 3D anatomical models is discussed as a basic approach for sharing and reusing knowledge among clinicians for next-generation CAD-assisted diagnosis and treatments.
Methods
We have developed (1) basic services for the analysis of 3D anatomical models and (2) a methodology for the enrichment of such models with relevant descriptions and attributes, which reflect the parameters of interest for medical investigations. The proposed semantic annotation is ontology-driven and includes both descriptive and quantitative labelling. Most importantly, the developed methodology permits to identify and annotate also parts-of-relevance of anatomical entities.
Results
The computational tools for the automatic computation of qualitative and quantitative parameters have been integrated in a prototype system, the SemAnatomy3D framework, which demonstrates the functionalities needed to support effective annotation of 3D patient-specific models. From the first evaluation, SemAnatomy3D appears as an effective tool for clinical data analysis and opens new ways to support clinical diagnosis.
Conclusions
The SemAnatomy3D framework integrates several functionalities for 3D part-based annotation. The idea has been presented and discussed for the case study of rheumatoid arthritis of carpal bones; however, the framework can be extended to support similar annotations in different clinical applications.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Nowadays, a wide range of advanced techniques is available, which can create accurate and detailed 3D anatomical models from digital imaging (MRI, CT, MicroCT, etc.). Three-dimensional patient-specific models (3D-PSM) are expected to be extremely useful in many applications such as diagnosis, biomechanical simulation, computer-assisted surgery, prosthesis fitting and even legal medicine. Nevertheless, there is still a long way to go before these models can be used in clinical practice.
We believe that one reason for the slow uptake of 3D-PSM is the lack of integration between digital data and medical knowledge. In other words, the semantic annotation of 3D-PSM is not a mature technique yet: tools are lacking, which, on the one side, support the analysis of 3D-PSM for the extraction of relevant parameters and, on the other side, support the storing of these parameters in a structured manner together with the 3D-PSM itself. In fact, the common practice of medical doctors is still to describe the clinical findings in a separate text report or in a data collection form, which are mostly unstructured. This makes aggregate and automatic analysis of 3D-PSM from multiple sources difficult and heavily relying on manual intervention.
The goal of this paper is to demonstrate how patient-specific 3D models of anatomical parts can be processed and documented with morphological information extracted automatically from the data. Part-based semantic annotation of patients’ 3D models of anatomy is discussed as a basic approach enabling, sharing and reusing knowledge among clinicians for the next-generation CAD-assisted diagnosis and treatments. To this end, we have developed (1) a methodology for the enrichment of 3D anatomical models with relevant descriptions and attributes, which reflect the parameters of interest for medical investigations; (2) basic services for the morphological analysis of 3D anatomical models; and (3) a data model and format which supports the storage and sharing of annotated 3D-PSM in medical repositories.
The proposed semantic annotation includes both a descriptive and a quantitative annotation: the first relies on anatomical concepts and relationships formally defined in reference ontologies and ad hoc ones; the latter refers to attributes corresponding to numerical measures of morphological parameters. Most importantly, the methodology developed allows us to identify and annotate also the parts-of-relevance (PoRs) in 3D surface reconstructions of anatomical entities. The computational tools for 3D shape analysis support the automatic quantitative annotation. The set of computational tools developed has been integrated in a prototype system, the SemAnatomy3D framework, which demonstrates the functionalities described.
Semantic annotation is well known in radiology and biomedicine: the annotation of clinical data generally involves the analysis of image features (e.g. texture, regions, contours) for the identification of regions of interest (ROI) in 2D/3D medical images and the annotation of the ROI with clinical information, such as measured diagnostic parameters and/or clinical findings.
Over the years, it has been shown how annotations based on ontologies may have important benefits over the ones stored in an unstructured manner (e.g. clinical notes, diagnosis reports). Ontology-driven annotations allow machines to analyse and access such information, opening the way to large-scale data and information mining in the medical domain. Several ontologies have been proposed in the biomedical domain (e.g. FMA [1], RadLex [2]), expressing a wide range of concepts and providing a constructed model of biomedical domain [3].
Among the most interesting tools available in the state of the art aiming at semantic annotation of medical data, we can mention iPad [4], which extends the functionality of the image viewing platform OsiriX [5] to add semantic tags from the RadLex ontology [2] to 2D medical scans through a simple user interface and stores the annotation in the Annotation and Image Markup schema (AIM) [6]. However, the process is mostly manual and can only support the annotation of 2D Dicom images. The Medico system [7] applies an automatic detection of anatomical structures within CT scans of the human torso and maps them to the concepts that are derived from FMA/ICD10 [8]/RadLex. This approach is applicable only to CT datasets of the human torso (i.e. 3D volumes) and verified only within a small set of sample images.
We remark the importance of extending the ontology-driven annotation to 3D-PSM and the difference between image annotation and 3D annotations. Annotating regions of an image and of a 3D surface model is conceptually the same process, but identifying the regions of interest in a 3D model has a higher complexity and the selection methods applicable to images are not straightforwardly extendable to 3D [9].
In the biomedical community, only few existing initiatives couple 3D anatomical models with their semantic formalizations (e.g. anatomical label, functionality, anatomical features) to support specific medical applications [10]. An example is theBodyParts3D [11] platform, which integrates canonical 3D anatomical models with the FMA structured knowledge for training purposes and does not include patient-specific anatomical information to support clinical investigation. Going one step further, My Corporis Fabrica [12] extends FMA with patient-specific 3D geometrical data and biomechanical parameters. The goal is to derive a patient-specific 3D representation from a formal description of anatomy to support the simulation of anatomical joint functions. Primal picture [13] is a commercial platform that presents an initiative to link structured knowledge not only to the whole canonical 3D models but also to its relevant subparts mainly for educational purpose. Finally, 3DSlicer, a medical image visualization tool [14], attempts to annotate the 3D patient-specific organs models segmented from images by a hierarchical structure of pre-defined anatomical labels.
In available systems, however, there is no support to analyse and annotate a 3D-PSM and its PoRs with anatomical characterizations and parameters, properly formalized and reflecting specific domain knowledge: SemAnatomy3D aims to bridge this gap. This work is rooted in the extensive analysis of the research challenges pertaining to the development of semantic 3D media [15], where the medical field was recognized as an exemplary one: the rich annotation of digital 3D data can support the automation of large-scale clinical studies and provide an additional instrument of investigation for doctors and scientists, stimulating new medical reasoning and correlations [16].
Finally, we want to remark the importance and novelty of our contribution in terms of data model and format for the storage of annotated 3D-PSM: the definition of a suitable data model for sharing part-based annotation of 3D-PSM has not been yet proposed as a standard solution in the state of the art. Our contribution sets the basis for the development of such a standard. This is an important technical contribution for a full exploitation of annotation, and possibly even for the development of markup language for 3D-PSM.
The paper is organized as follows. In “User requirements for SemAnatomy3D design” section, the user requirements that served as a basis for the SemAnatomy3D framework are reported and “SemAnatomy3D framework” section describes the components and the functionalities of the proposed tool. In “Evaluation and conclusive remarks” section, we discuss how the current version of SemAnatomy3D is able to answer to the case study on Rheumatoid Arthritis, which we select as a convenient usage scenario. Finally, conclusive remarks are given indicating the future research directions.
User requirements for SemAnatomy3D design
The development of SemAnatomy3D started within the framework of different international and national research projects characterized by the attempt to integrate expertise in geometric modelling and analysis with knowledge formalization and medical applications (MultiScaleHuman [17], MEDIARE [18], Politecmed [19] consortia). This context constituted a rich and heterogeneous background for the elicitation and analysis of physicians’ requirements, spanning from clinicians, radiologists, orthopaedists and rheumatologists to tissue engineers and hand surgeons. It was also complemented by the developers of medical imaging equipment and computer-aided diagnosis (CAD) software and by external research groups [20, 21].
The requirement elicitation phase has been addressed through the preparation and distribution of questionnaires for collecting the basic requirements, opinions, perspectives and desiderata from 20 experts of the community, regarding the integration between the formalization of the medical knowledge focusing on musculoskeletal diseases and digital data.
The first investigation showed a clear demand for the development of CAD-like systems encapsulating tools for a semantically rich and interoperable clinical annotation system able to:
-
identify and measure clinical parameters based on the geometric/morphological characterization of the shape of organs, anatomical elements or their parts;
-
devise formal methods to assess the similarity among shapes to support the retrieval of similar clinical cases in order to speed up the diagnosis process and support comparative analysis among known cases;
-
gather information about specific patients to ease the evaluation of their follow-up in order to highlight temporal trends of pathology markers, possibly depending on current therapy;
-
perform statistical analysis over a significantly large population of patients to trigger the possible detection of new correlation patterns and speed up the screening of large populations for abnormal cases.
In the second phase of the requirement analysis, we conducted face-to-face meetings with the experts to achieve a deeper understanding about the clinical data analysis pipeline and to identify the interactions needed with the system. In particular, we used the diagnosis of Rheumatoid Arthritis (RA) of the wrist joint as an illustrative scenario to discuss how part-based semantic annotation of 3D medical data may be designed to support clinical investigation. RA, a chronic inflammatory disorder, affects the lining of small joints, causing a painful swelling that can eventually result in bone erosion and joint deformity. Then, some important diagnostic analyses are carried out monitoring morphological features of the bones and the wrist joint itself, and this constitutes a valuable use case to verify the potential of SemAnatomy3D.
In this particular scenario, our goal has been to identify: all the anatomical landmarks for carpal bones to formulate a reliable conceptualization; the measures or descriptions doctors associated with anatomical landmarks; how the characterization of anatomical landmarks influences diagnosis; and the grouping factors that are important for devising a statistical analysis of patient’s carpal bones.
In the next sections, we will describe the SemAnatomy3D framework and how it is able to address the clinical investigation of RA.
SemAnatomy3D framework
The use case scenarios described in “User requirements for SemAnatomy3D design” section indicate a need for an expressive annotation of 3D-PSM. To fulfil this requirement, we defined an annotation pipeline and developed a prototype graphical tool called the SemAnatomy3D, which consists of the following main components,
-
1.
3D visualization and interaction—it supports loading and visualizing a single or a set of 3D surface models and permits direct interactions with the 3D models, e.g. zoom, rotate, selection of subparts in the 3D model. We used Java Swing to support cross-platform style of interactive user interface and Visualization Toolkit (VTK) [22] to build the 3D interaction and visualization widgets.
-
2.
Ontology loader module—it allows loading the conceptualization stored as .owl files from the local file system, and it implements a keyword-based browsing of the concepts. For the loading and navigation of the ontology, we used the Jena2 ontology API [23].
-
3.
Annotation services—the module supports a manual and descriptive annotations of 3D-PSM, using a template-based method. Further, a set of geometric and shape analysis tools have been incorporated for quantitative characterization. We utilized VTK data structures for implementing geometric and shape analysis methods.
-
4.
SemAnatomy3D knowledge base—the core of the knowledge base is the SemAnatomy3D data model and its related format for storing annotated 3D models, the .sem3D file. The annotation produced during the SemAnatomy3D workflow together with the 3D subpart identifiers produced, is automatically stored in the SemAnatomy3D knowledge base. For updating the knowledge base, we utilized the SPARQL functionalities embedded in Jena2 ontology API [24].
The SemAnatomy3D workflow for the carpal bones annotation is shown in Fig. 1. In the following subsections, we will expound the SemAnatomy3D platform by presenting the formalized context of our case study, by discussing part-based annotation functionalities and by detailing how the part-based 3D annotations are stored in the SemAnatomy3D knowledge base.
The formalization of the context
From the survey of biomedical ontologies, we realized that it is difficult to enforce the use of a single view/perspective on the underlying knowledge domain, each of which reflects a specific interest of the experts involved in the clinical analysis. Moreover, most of the reference biomedical ontologies provide a standard terminology without defining quantitative attributes, properties and relations among the concepts.
To support the carpal bone case study, we have studied and identified reference ontologies (or their parts) to be reused and integrated in our knowledge space. Nevertheless, the reference ontologies extensively model the whole human anatomy and, therefore, are very complex to manage. In addition, we are focused only on the knowledge related to the carpal bones and their relevant anatomical landmarks.
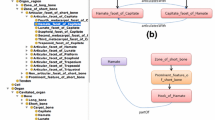
An interest is growing in the emerging area of modular ontologies where the emphasis is on either extracting and managing modules of ontologies relevant to a particular application scenario or developing them independently and integrating into a larger ontology [25, 26]. However, we prefer to define an ad hoc Carpal bone Ontology to realize a conceptualization at the granularity required, and appropriate for its practical usage. More precisely, we extracted the FMA’s anatomy formalization related only to the carpal bones (Figs. 2, 3) and extended it with part-hood and articulation relations between facets and the properties of anatomical concepts (e.g. bone volume, bone surface area).
The Carpal bone Ontology is defined around of two main anatomical concepts that are required by our case study: FMA:Carpal_bone (Fig. 2) that models 8 individual carpal bones, FMA:zone_of_short_bone (Fig. 3) that models the articulation facets as well as prominent anatomical features. The entire formalization related to the scaphoid bone is shown in Fig. 4, where the carpal bone ontology namespace is referred as “CO:”. We defined a similar formalization for each carpal bone and group them under a top concept Carpal_region.
Further, we associated relational properties between the anatomical concepts. For example, Fig. 5 shows the CO:hasAriculationFacet and CO:Articulates With relations between FMA:Hamate and FMA:Capitate, and Fig. 6 depicts the CO:partOf relation between FMA:Hamate and its subpart FMA:Hook_ of_Hamate. To this end, the parameters that can be computed from the 3D bone models have been formalized in the Carpal bone Ontology mainly as the properties of the Carpal_region, FMA:Carpal_bone and FMA:zone_of_short_bone. The pathological markers have not been included in the Carpal bone Ontology.
This formalism represents the anatomical information in a form that able to support reasoning, inference and assertion. Particularly, when such knowledge is associated directly with the patient 3D data, it will allow for a dynamic navigation not only of the knowledge but also of the linked 3D geometry, with possibilities to extend the reasoning to the geometric aspects. We believe that it will be highly potential for the automation of large-scale clinical studies.
Descriptive annotation
The descriptive annotation of 3D-PSM aims to describe the data and information contained in the models by means of the concepts/terms defined in the reference ontologies. To this end, SemAnatomy3D includes functionalities to associate descriptive information with 3D subparts either via interactive mode or via controlled mode. Interactive mode is more flexible but manual, while controlled mode is automatic yet gives less adaptability. Both modalities are important: the manual annotation mechanism may be used to associate even completely free-text annotations with 3D parts, and may be also used to fine-tune the controlled annotations, if needed.
It is important to underline that in our case study, different PoRs in the carpal bones can have varying topological dimension, such as surface patch—articular and non-articular facets of the bone, prominent features (e.g. scaphoid tubercle, hook of hamate, ligament insertion sites); edges—boundaries between anatomical landmark regions, contours indicating abnormalities/disease-affected regions (e.g. eroded regions; vertices—feature points of the bone, such the tip of a protruded facet, extreme pressure point) (Fig. 7).
Fully interactive annotation
As said before, the interactive mode is flexible, and the annotator can define and select any type of PoRs within a 3D surface from the interactive tool palette (Fig. 8), where interaction tools such as smart-cut, draw, paint and delete strokes, picking of points, are offered. In this process, there are methods to assist the annotation: for instance, the user can simply drag the mouse and select the articulation facets and the prominent features of scaphoid over a patient-specific model by using the smart-cut tool, and the system automatically computes the entire cut in the 3D surface including the region that is not visible from the viewpoint, and the selected portion of the model is coloured accordingly. Further, the system allows fine-tuning of the PoRs boundary by using simple interaction tools, such as paint and delete strokes (mouse click). After having identified the PoR, the user can annotate it with the conceptual tags selected from the ontology loaded in the system, the carpal bone ontology in our case study. SemAnatomy3D allows on the fly loading of any ontology (in .owl) and the navigation of large reference ontologies using iterative keyword search mechanism to locate quickly the conceptual tags needed to complete the annotation, supported also by the auto-completion functionality of the terms.
Controlled annotation
As an alternative approach, we have developed an automatic template-based method that associates automatically conceptual tags of the Carpal bone Ontology with parts of 3D-PSM. In this process, a 3D template model for each carpal bone is used, which is generated according to the methodology presented in [27]. The templates, built from a statistical analysis of real patient data, can be considered as the average healthy shape of the sample population which capture the healthy shape variability while preserving important anatomical landmark features. Given the template, the basic idea is to transfer the annotation of the template automatically onto the 3D-PSM, by co-registering the annotated 3D template with the 3D-PSM, and transferring to the latter the positions of anatomical landmarks.
For instance, to find the articulation regions and prominent feature of the scaphoid in a 3D-PSM, we register our parametric scaphoid template against the targeted 3D-PSM using a non-rigid transformation and propagate the annotation onto the target model. To this end, we apply a non-rigid variation of the Iterative Closest Point algorithm initialized with a coarse alignment using centroid to find the matching between template and target model. Using the nearest neighbour search method, the annotation is automatically propagated from the vertices of the annotated template to the closest vertices of the target mesh. After propagation, the system detects the boundary of each annotated surface fragment and allows modifications of the PoRs boundary using simple interaction tools described in the previous section. This automatic method only supports annotation with the controlled terms that have been pre-associated with the parametric 3D template model. Following a similar approach [27], templates for other anatomical parts could be developed to automatize the annotations of large sets of 3D-PSM.
Quantitative annotation
In our case study, the annotation entails not only the description of 3D-PSM and its PoRs at the conceptual level, but also the association of numeric values that reflect the characterization of the patient’s anatomy at the geometric and structural level. Thanks to the 3D representation of the patient anatomy, it is possible to compute a wide range of parameters, which provide a rich morphological characterization. We formalized the significant parameters as attributes in the Carpal bone Ontology, and within SemAnatomy3D framework we support automatic measurements of these attributes from the 3D models, or their subparts, by means of popular geometric and shape analysis methods proposed in the literature. We name this process as quantitative annotation of 3D-PSM.
In Table 1, we present the attributes that we consider as significant to provide a rich characterization of the 3D bone model. Note that some of these measurements (marked in italic) are already considered important in the clinical analysis of carpal bone, e.g. bone volume (BV), bone surface area (BS). We added a few supplementary parameters that have not been correlated with any clinical measurement but can present a specific perspective of the bone morphology, e.g. curvature map, compactness. Finding correlation between various 3D characterizations and pathological markers is clearly an interesting research direction to explore, but it is not within the scope of this paper. We believe there is a great potential in exploiting the results of geometry processing in the medical area, and the implementation of these descriptors was important to show to the physicians what could be done. Finally, we are using these geometric descriptors, and several others, to build another approach to the automatic annotation of anatomical landmarks based on machine learning [18].
To date, we developed two types of computational tools to address the proposed quantitative annotation of 3D-PSM (Fig. 9)
-
1.
quantitative measurements used to quantify the parameters in Table 1, plus another set of morphological operators such as the geodesic distance between anatomical landmarks, which could be used to further analyse the shape of 3D bones.
-
2.
(dis)similarity measurements we implemented a method to compute the difference between two bones. There are two ways to use this measure: (1) relying on the template, we can measure the deviation of the 3D-PSM from the normality, useful for instance to indicate a pathological situation. Defining the normality in anatomy is a non-trivial task because of the huge variability in shape of anatomical structures, even within the healthy population.
Therefore, to measure the deviation from normality, we consider a healthy average model of a sample population built from the homogeneous classes of 3D bone reconstruction [27] and co-register it (rigidly) with the target data. The measurement signifies the vertex-wise distance between the healthy template and target data. It offers the possibility not only to measure an overall average/max distance with the normality but also to classify various subparts of the 3D models according to the scalar value of the distance; it also permits the automatic identification of abnormal regions.
The same measure is also used to monitor the differences among the same bone instance acquired in two different acquisitions, thus supporting the documentation of the follow-up analysis. In this case, instead of using a generic healthy template, we can directly use the baseline model to evaluate the changes in follow-up data. In Fig. 10, we present our preliminary result where the “Erosion Map” of the model with t2 timestamp has been computed by registering the baseline model with t1 timestamp, and the red colour represents the regions where erosion escalated compared to the baseline model.
Coding the 3D part-based annotation
The result of each annotation session is saved as a set of instances in SemAnatomy3D knowledge base. Our goal is to encode the whole and part-based 3D annotation so that we are able to set up a concrete bridge between semantic information and patient’s geometry for supporting storage, reuse, sharing and part-based retrieval of 3D-PSM.
In this context, the main objectives are: (1) storing the reference to the annotated 3D geometry without any duplication of the whole data and (2) directly linking the references with the original 3D model to allow quick retrieval. Within the SemAnatomy3D framework, we realize this concept by two primary ingredients: sem3D file format and Sem3D annotation data model.
In the current practice, there is no such standard format available which fulfils the requirement to store 3D part-based annotation by reference. Thus, we develop our own standard—sem3D, to store the geometric data related to the identifiers of the annotated 3D geometry. The format sem3D follows an index-based approach, that is, it uses simply the reference to the geometric primitives of the original mesh model to avoid any duplication of potential large data files. The sem3D format can code 3D fragment identifiers of varying topological dimensional: for instance, a 3D surface fragment is stored as the list of the indices of the cells (triangle) of the complete mesh model which belong to the fragment. In summary, sem3D works as a pointer to the geometry, which does not store redundant information. With this approach, a sem3D file storing a surface fragment containing 717 cells and 379 points has \(<1\)KB. However, the index-based alone is not sufficient to establish a proper link with conceptual information and has to be coupled with a formal annotation data model.
Unfortunately, the common data models [28, 29] are mostly designed for text files and image annotation. Thus, they are not applicable for annotating 3D-PSM and their varying dimensional subparts. The Sem3D annotation data model is an extended version of the Open Annotation data model (OA) developed by W3C Open Annotation Community Group, to fulfil the main requirements of SemAnatomy3D annotation framework: storing the annotation of varying topological dimensional 3D fragments and supporting whole and part-based annotation with descriptive and quantitative attributes.
In the Sem3D annotation data model, we realize the link between the 3D fragment identifier stored as .sem3D file and the 3D model using the path defined by the relations between sem3D:3DModel and sem3D:3DFragment. To describe this idea, we present a snapshot of the SemAnatomy3D knowledge base in Fig. 11, which is related to saving of a 3D-PSM of the scaphoid bone and its 3D surface fragment annotated as FMA:scaphoid_tubercle. With this scheme, we store only the index of the annotated geometry as .sem3D file; however, the semantic relations interconnect the 3D-PSM and the annotated region in such a manner that allows quick referencing of geometry and topology for the retrieval and rendering. Therefore, the Sem3D annotation data model facilitates interoperability, querying, reasoning and discovery of 3D-PSM as a whole and its subparts.
Evaluation and conclusive remarks
The user requirement analysis indicated quite sharply that physicians are ready to envisage the digital patient concept, equipped with the software tools needed to analyse and manage the digital model of patients, up to their usage complex simulation processes. SemAnatomy3D realizes what we envisage as an important step towards this goal, by the integration of geometry processing and knowledge formalization methods. It is obviously not a product which can be used now in clinical practice, and rather, it is a demonstrator of what could be achieved exploiting the availability of accurate 3D reconstructions of organs and anatomy directly from patients’ data.
A necessary step to move forward along this direction is represented by a thorough evaluation of our proposal in the medical, and possibly clinical, context. Given the complexity of the functionalities offered by the SemAnatomy3D prototype, also its validation by experts is a relatively complex task and it should touch all the facets of the platform. At the current stage of development of SemAnatomy3D, the evaluation cycle is focusing on the following: degree of satisfaction in terms of availability of relevant analysis functionalities; expressivity of the formalization adopted; and accuracy of the anatomical characterization and computed parameters/attributes.
The results of this first cycle will give us important suggestions and indication for the further refinements and improvements of the functionalities offered. The evaluation started already, with a pool of ten experts with interest in the case study, in particular radiologists, hand surgeons, anatomists and rheumatologists. The evaluation setting is the following: experts are first asked to attend a short presentation of the platform, including motivations for its development and brief overview of the functionalities; the data set used in the case study is illustrated, and examples of 3D reconstructions are shown. After a demonstration of an annotation session, experts are asked to perform themselves an annotation. During the process, conducted with the assistance of our team, we ask questions and promote discussions aimed at gathering the users feedback on the high-level requirements listed in “User requirements for SemAnatomy3D design” section.
In the following, as a conclusion of the paper, we summarize the results of the evaluation, which is still ongoing, and highlight future development. We want to remark that while images are surely commonplace in their clinical practice, the usage of 3D reconstructions by physicians is still relatively rare and used in practice mostly by researchers or surgeons for intervention planning and monitoring. Therefore, as a first important feedback, we mention that the implementation of the SemAnatomy3D platform per se was perceived as very useful. The prototype had a prominent role in our discussions with experts, allowing us to make concrete examples of what could be achieved and to stimulate reasoning on new perspectives for scientific investigation in the medical field.
Is SemAnatomy3D able to address the users’ requirements?
To drive the discussion, we have used as guidelines the four high-level requirements (see “User requirements for SemAnatomy3D design” section) and analysed with the experts if the SemAnatomy3D and its annotation results are suitable to support these activities.
Requirement 1
Identify and measure clinical parameters based on the geometric/morphological characterization of the shape of organs, anatomical elements or their parts.
The most appreciated aspect was the possibility to annotate and analyse not only the single bones of the carpus but also the whole district. When consulting a surface fragment of a 3D-PSM annotated as an articulation facet, a clinician is likely to be willing to consult adjacent facets of the fragment with which it articulates. This use case was particularly significant for erosion analysis in rheumatoid arthritis, where the co-analysis of all involved articulation facets may give an effective visual evaluation of the pathological conditions. Useful queries mentioned by experts ranged from simple ones, such as “Show the inter-bone distance map of patient XX, and highlight all the “FMA:Articulation_facets_of_short_bone” with average erosion value larger than 2.5?” to rather complex, yet intriguing, ones pointing to the possibility to simulate the functionality of the carpus. To answer the first query and to visualize its results are quite easy, thanks to the conceptualization adopted, which models all the joint quantitative parameters. Thus, we first retrieve all 3D models in the carpus of the patient XX, visualize its attribute “Inter-bone joint space metrics” and then select the articulation facets whose “Erosion score” attribute is higher than the prescribed one. Biomechanical simulation is out of the scope of SemAnatomy3D. However, we remark that the possibility to manipulate rich 3D characterizations of the carpus really stimulated the experts and demonstrated the potential of 3D representations of anatomy for more complex studies, such as biomechanical simulations.
Requirement 2
Devise formal methods to assess the similarity among shapes to support the retrieval of similar clinical cases in order to speed up the diagnosis process and support comparative analysis among known cases.
The answers were positive for all our experts: the fine-grained annotation of 3D-PSM is considered a very informative characterization, which allows documenting each significant anatomical feature. Consequently, users appreciated the fact that search and retrieval of annotated 3D-PSM could exploit a fused search, which integrates text-based search and content-based retrieval together. This hybrid retrieval technique, made it possible only thanks to the part-based annotation model, can support advanced queries, such as “Retrieve all records of clinical cases where the “RAD:scaphoid_tubercle” had a shape similar to the “RAD:scaphoid_tubercle” of patient XYZ, and where the “RAD:scaphoid_tubercle” was 30 – 40% eroded and detected as affected by degenerative joint disease”. The query above was perceived as highly innovative and with potential to set up novel comparative analysis of 3D-PSM.
Requirement 3
Gather information about specific patients to ease the evaluation of their follow-up in order to highlight temporal trends of pathology markers, possibly depending on current therapy.
The conceptualization related to the anatomical landmarks and quantitative parameters was judged exhaustive to document appropriately the anatomical and pathological conditions of patients, at least from the perspective of the bone conditions. Indicators not related to shape properties, such as pain, were mentioned as missing. Such kinds of indicators will be considered in future development of the platform and would localize, for instance, the region in the 3D-PSM where the difficulties in movements or pain are concentrated.
Concerning the follow-up, the functionality offered to evaluate the “distance to normality” was appreciated as a tool to evaluate quantitatively also the differences between two different stages of pathology evolution in the same patient. The discussion on this aspect triggered further suggestions to set up a library of tools to quantify automatically the evolution of parameters relevant for the follow-up (e.g. the “Inter-bone joint space metrics” and the “Erosion score”). The improvement here will involve the conceptualization of the follow-up concept itself, with the definition of a proper set of attributes.
Requirement 4
Perform statistical analysis over a significantly large population of patients to trigger the possible detection of new correlation patterns and speed up the screening of large populations for abnormal cases.
We discussed this question with the experts after their experience of the platform, and after having shown them what the knowledge base stores in terms of instance and documented 3D-PSM. Showing them examples of complex queries, such as the one described in Requirement 2, it was easier for the experts to understand how the queries could be run on a set of repositories exposing their data sets annotated with the same ontological scheme and stored with the same data model. Although the physicians had not expertise in semantic and knowledge technologies, they realized that the semantic annotation pushed forward by SemAnatomy3D could be highly innovative and with potential to set up and run large-scale analysis of 3D-PSM.
Is the formalization of the carpus satisfactory?
The focused and fine-grained domain formalization was judged pragmatic and useful, especially thanks to the quantitative parameters included at the conceptual level. Experts appreciated the fact that the advantage of using formal constructs was clearly demonstrated and gave suggestions on how to extend further the conceptualization by means of extending the formalization to the whole wrist joint, adding the layer involving ligaments and associating a measure of accuracy with the quantitative parameters.
Is the controlled annotation meaningful?
A large part of experts’ interest in SemAnatomy3D was captured by the template-guided annotation of 3D-PSM. The functionality indeed has been recognized as important to automatize the semantic annotation of 3D-PSM.
The template-based annotation has been presented in [27], and its validation is surely fundamental. For the sake of the initial validation, we conducted a first qualitative evaluation by checking the differences between the annotations conducted by the experts, and discussing with them the results of the controlled annotation.
For the manual annotation, the experts were assisted also by the visualization of the original MRI images, which originated the 3D-PSM. For each anatomical landmark, the physicians were asked to draw the location and boundaries of the anatomical landmarks and their annotated 3D model was stored in the SemAnatomy3D knowledge base. While the collection of the ground truth is still ongoing, we would like to report a first feedback we got from the first experts, one radiologist and one anatomist, with whom we run the manual annotation. The inter-subject variability and the inherent fuzziness of the landmark demarcation lines made them comment that accuracy in terms of alignment of the boundaries is not an issue. Correctness of the location of landmarks is more relevant to them. Once completed the collection of the ground truth, we will run a more systematical analysis of the differences of the manual annotations to the template-driven ones, using some of the metrics proposed in [30].
Are the quantitative parameters meaningful and accurate?
The quantitative attributes associated with our anatomical taxonomy were presented to the experts and perceived by them as belonging to two broad classes: a first set represents parameters that are already known to be relevant in clinical practice and useful to document anatomical and/or clinical investigations (e.g. bone volume, erosion score); a second class of parameters does not have clear clinical/anatomical significance (e.g. Gaussian curvature map) but are perceived as useful for gathering new insights into the quantification of important clinical markers. Belonging to the first class, the erosion score is evaluated visually in the current clinical practice, based on the ratio between the volume of the erosion and the hypothetically healthy bone, analysing all the slices in the image stack. The same score is implemented in SemAnatomy3D literally, as the volume difference between the 3D-PSM and the template. The availability of this tool to measure directly the erosion score has been perceived as very useful, but its accuracy is still to be assessed. To this end, we are collecting a data set of pathological 3D-PSM with erosion scores computed according to the standard OMERACT RAMRIS [31] criteria, so that we can map and compare the differences. We remark that manual evaluation of bone erosions is considered a tedious, time-consuming and not a fully repeatable task. Considering the big amount of patients suffering from RA, this functionality has a large potential impact and we are planning to perform a critical evaluation of the results.
Concerning the geometric parameters that do not have an immediate clinical relevance, the experts had the feeling they could be useful but without a precise opinion on them. We believe that the evaluation of these parameters is not to be asked to physicians: we are currently using an extended set of attributes to be fed to a machine learning system in order to see whether they correlate well either with the location of anatomical landmarks or with pathological markers. A complete discussion of these results will be subject of further studies and publications.
References
Rosse C, Jose LV, Jr Mejino (2007) The foundational model of anatomy ontology. Anat Ontol Bioinform Princip Pract 6:59–117
Mejino JLV, Rubin DL, Brinkley JF (2008) FMA-RadLex: an application ontology of radiological anatomy derived from the foundational model of anatomy reference ontology. In: Proceedings of AMIA symposium
Lia Z, Yanga MC, Ramani K (2009) A methodology for engineering ontology acquisition and validation. Artif Intell Eng Des Anal Manuf 23(1):37–51
Rubin DL, Rodriguez C, Shah P, Beaulieu C (2008) iPad: semantic annotation and markup of radiological images. In: AMIA 2008 symposium proceedings, pp 626-630
OsiriX Dicom viewer. [Online]. http://www.osirix-viewer.com/
Mongkolwat P, Kleper V, Talbot S, Rubin D (2014) The National Cancer Informatics Program (NCIP) Annotation and Image Markup (AIM) Foundation Model. J Digit Imaging 27(6):692–701
Seiferta S, Kelma M, Moeller M, Mukherjee S, Cavallarod K (2010) Semantic annotation of medical images. Med Imaging 7628:2010
World Health Organization and others (1992) The ICD-10 classification of mental and behavioural disorders: clinical descriptions and diagnostic guidelines. World Health Organization, Geneva
Spagnuolo M, Falcidieno B (2009) 3D media and the semantic web. IEEE Intell Syst 24:90–96
Banerjee I, Catalano CE, Robbiano F, Spagnuolo M (2014) Accessing and representing knowledge in the medical field: visual and lexical modalities. In: Magnenat-Thalmann N, Ratib O, Choi HF (eds) 3D multiscale physiological human. Springer, London, pp 297-316
Mitsuhashi N, Fujieda K, Tamura T, Kawamoto S, Takagi T, Okubo K (2009) BodyParts3D: 3D structure database for anatomical concepts. Nucleic Acids Res 37:D782–D785
Palombi O, Bousquet G, Jospin D, Reveret L, Faure F (2009) My corporis fabrica: a Unified ontological, geometrical and mechanical view of human anatomy. In: 2nd workshop on 3D physiological human, 3DPH2009, vol 5903. Zermatt, Switzerland, pp 207–219
Primal pictures. [Online]. https://www.primalpictures.com
Fedorova A, Beichelb R, Kalpathy-Cramerc J, Finetd J, Fillion-Robind J, Pujola S, Bauerb C, Jenningsc D, Fennessya F, Sonkab M, Buattib J, Aylwardd S, Millere JV, Pieperf S, Kikinisa R (2012) 3D Slicer as an image computing platform for the quantitative imaging network. Magn Reson Imaging 30(9):1323–1341
Chiara Eva Catalano, Mortara Michela, Spagnuolo Michela, Falcidieno Bianca (2011) Semantics and 3D media: current issues and perspectives. Comput Gr 35:869–877
Catalano CE, Robbiano F, Parascandolo P, Cesario L, Vosilla L, Barbieri F, Spagnuolo M, Viano G, Cimmino MA (2013) Exploiting 3D part-based analysis, description and indexing to support medical applications. Med Content-Based Retr Clin Decis Support 7723:21–32
MutliscaleHuman FP7 partners. [Online]. http://multiscalehuman.miralab.ch/partners
MEDIARE project. [Online]. http://www.research.softeco.it/mediare.aspx
Politecmed consortium. [Online]. REF-http://www.camelotbio.com/politecmed
IMGINE. [Online]. https://team.inria.fr/imagine/
Gaslini Children’s Hospital. [Online]. http://www.gaslini.org/servizi/notizie/notizie_homepage.aspx
VTK wiki. [Online]. http://www.vtk.org/Wiki/VTK
(2011–2013) Apache Jena framework. [Online]. http://jena.apache.org/index.html
ARQ-A SPARQL Processor for Jena. [Online]. http://jena.apache.org/documentation/query/
Agibetov A, Catalano CE, Patane’ G, Spagnuolo M (2015) Ontology modularization for multi-scale biomedical data management and visualization. In: CARS workshop on multiscale digital patient (MDP)
Agibetov A, Vaquero RMM, Friese K, Patanè G, Spagnuolo M, Wolter F (2014) Integrated visualization and analysis of a multi-scale biomedical knowledge space. In: Proceedings of the EuroVis workshop on visual analytics, pp 25–29
Banerjee I, Laga H, Patané G, Kurtek S, Srivastava A, Spagnuolo M (2015) Generation of 3D canonical anatomical models: an experience on carpal bones. In: New trends in image analysis and processing—ICIAP 2015, Genova, In press
Kahana J, Koivunen MR, Prud’Hommeauxb E, Swick RR (2002) Annotea: an open RDF infrastructure for shared Web annotations. Comput Netw 39(5):589–608
Hunter J, Cole T, Sanderson R, Van de Sompel H (2010) The open annotation collaboration: a data model to support sharing and interoperability of scholarly annotations. In: Digital humanities, pp. 175–178
Chen X, Golovinskiy A, Funkhouser T (2009) A benchmark for 3D mesh segmentation. ACM Trans Gr 28(3):73
Østergaard M, PeterfConaghan PC, McQueen F, Bird P, Ejbjerg B, Shnier R, O’Connor P, Klarlund M, Emery P, Genant H, Lassere M, Edmonds J (2003) OMERACT rheumatoid arthritis magnetic resonance imaging studies. Core set of MRI acquisitions, joint pathology definitions, and the OMERACT RA-MRI scoring system. J Rheumatol 30(6):1385–1386
Acknowledgments
This study was funded by the FP7 Marie Curie Initial Training Network “MultiScaleHuman”: Multi-scale Biological Modalities for Physiological Human Articulation (2011–2015), contract MRTN-CT-2011-289897. This work is also partially funded by the Project FAS—MEDIARE “Nuove metodologie di Imaging Diagnostico per patologie reumatiche”.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Imon Banerjee, Chiara Eva Catalano, Giuseppe Patané, Michela Spagnuolo declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Informed consent
This article does not contain any identifiable patient’s information.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 (mp4 66102 KB)
Rights and permissions
About this article
Cite this article
Banerjee, I., Catalano, C.E., Patané, G. et al. Semantic annotation of 3D anatomical models to support diagnosis and follow-up analysis of musculoskeletal pathologies. Int J CARS 11, 707–720 (2016). https://doi.org/10.1007/s11548-015-1327-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-015-1327-6