Abstract
Purpose
Spinal needle injections are widely applied to alleviate back pain and for anesthesia. Current treatment is performed either blindly with palpation or using fluoroscopy or computed tomography (CT). Both fluoroscopy and CT guidance expose patients to ionizing radiation. Ultrasound (US) guidance for spinal needle procedures is becoming more prevalent as an alternative. It is challenging to use US as the sole imaging modality for intraoperative guidance of spine needle injections due to the acoustic shadows created by the bony structures of the vertebra that limit visibility of the target areas for injection. We propose registration of CT and the US images to augment anatomical visualization for the clinician during spinal interventions guided by US.
Methods
The proposed method involves automatic global and multi-vertebrae registration to find the closest alignment between CT and US data. This is performed by maximizing the similarity between the two modalities using voxel intensity information as well as features extracted from the input volumes. In our method, the lumbar spine is first globally aligned between the CT and US data using intensity-based registration followed by point-based registration. To account for possible curvature change of the spine between the CT and US volumes, a multi-vertebrae registration step is also performed. Springs are used to constrain the movement of the individually transformed vertebrae to ensure the optimal alignment is a pose of the lumbar spine that is physically possible.
Results
Evaluation of the algorithm is performed on 10 clinical patient datasets. The registration approach was able to align CT and US datasets from initial misalignments of up to 25 mm, with a mean TRE of 1.37 mm. These results suggest that the proposed approach has the potential to offer a sufficiently accurate registration between clinical CT and US data.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Spine needle injections are commonly used for analgesia to relieve pain and anesthesia to remove nerve sensation. An example is an injection into the facet joint, which is used to treat chronic lower back pain. Between 25 and 45 % of chronic lower back is attributed to the facet joint [3]. Injections into this region are particularly challenging due to the deep location, proximity to nerves and the narrow joint space. These challenges make it difficult to provide accurate injection into the joint when the procedure is performed without guidance. The current standard to guide the injection is fluoroscopy or computed tomography (CT). However, there are several drawbacks to these image modalities, including the risks posed by ionizing radiation to the patient and the physician.
Another common percutaneous spinal intervention is epidural injection. It is used before surgery as an alternative to general anesthesia, as well as in obstetrics during delivery. Fluoroscopy may only be used in non-obstetric cases because of the ionizing radiation. Palpation can be used as an alternative; however, this can be challenging in obese patients or those with a diseased spine [1]. In these cases, a different guidance modality is required.
Ultrasound (US) guidance for spinal needle insertions is gaining interest, since US is a non-ionizing and more accessible imaging modality compared to fluoroscopy or CT [2]. US has not become the standard of care in spinal injections due to the difficulty in the interpretation of anatomy. There are many ultrasound artifacts limiting the visibility of facet joints or intervertebral spaces. These include attenuation and scattering due to the thick muscles and ligaments around the spine, and acoustic shadows from the bony processes of vertebrae. To enhance the quality of US images, three-dimensional (3D) anatomical information (e.g., from a preoperative CT) can be fused with the intraoperative US images through image registration.
To move toward a clinically acceptable US-guided system for percutaneous spine interventions, the objective of this paper is to register the preoperative diagnostic CT to the intraoperative US using in vivo patient data. Over the past two decades, several research groups have attempted to tackle this challenging problem [6–8, 11, 12, 19, 22, 23]. To the best of our knowledge, these approaches have been validated either on single vertebra registration or on phantom and cadaver experiments. Robust and clinically feasible registration of multiple vertebrae with clinical data has not been reported to date. The preliminary results of our approach on five subjects were presented in [14]. In this paper, we evaluate the approach on 10 subjects, provide a detailed description of the methodology and perform extensive experiments to evaluate the role of the various components of our method on the final CT to US registration accuracy.
Accurate registration of CT and US data will enable intraoperative US needle guidance, and potentially reduce or eliminate the need for intraoperative fluoroscopy or CT which restrict current spinal interventions to specialized facilities. In contrast, US scanners have no radiation risks and are accessible and portable including new pocket-sized scanners. Reliable US guidance for spinal interventions would also improve the accuracy and accessibility of such procedures, and lower the associated costs.
Materials and methods
Data
Data from the lumbar spine of 10 subjects are used to validate the proposed registration algorithm. The registration accuracy is highly dependent on the quality of the acquired US images. Therefore, a protocol is created to set guidelines for US imaging parameters and freehand US data acquisition. The aim of the protocol is to minimize variability in US image quality between subjects, operators and imaging centers. The protocol also ensures that the setup time is minimal to adhere to the current clinical practice. The demographics of the 10 subjects recruited are shown in Table 1.
CT data acquisition
Only subjects with previous CT scans were recruited to avoid any additional radiation exposure. Approval was obtained from Institutional Research Ethics Boards (IREB). Informed consent was acquired from the subjects who participated in the study. Preoperative CT data were collected at St. Paul’s Hospital, Vancouver, BC, Canada, and at Kingston General Hospital, Kingston, ON, Canada. The subjects were in the supine position during CT data acquisition, and the CT images were provided as anonymized DICOM files from the hospitals’ picture archiving and communication systems. Some subjects had the entire lumbar vertebrae visible in their CT, while others had only a portion of the vertebrae visible. Table 2 lists the lumbar vertebrae visible, and hence used for registration, for each subject.
Tracked US data acquisition
The hardware used to acquire the US images and associated tracking data is composed of a SonixTouch US scanner (Analogic, MA, USA) equipped with a Guidance Positioning System (GPS) extension (Ascension DriveBay EM position tracker, VT, USA), a C5-2 GPS curvilinear US transducer (Analogic, MA, USA) and a 800 EM tracking sensor (Ascension, VT, USA) used as the subject coordinate reference. The reference sensor is affixed on the subject’s skin above the T12 vertebra. The GPS extension has an adjustable arm that attaches to the US machine. This allows the position of the EM transmitter to be as close to the reference sensor as possible. The US transducer is tracked by the EM transmitter through an embedded pose sensor. A schematic of the hardware setup for the data acquisition system for tracked US is seen in Fig. 1. Subjects are set in the prone position for US acquisition. To help minimize changes in curvature between the CT acquired in the supine position and the US acquired in the prone position, a medium-sized pillow is placed under the subject’s stomach during the US data acquisition. The imaging and subject setup time is minimal (around five minutes) in order to adhere to the clinical environment and avoid any changes to the current clinical workflow.
Data acquisition system for tracked US. SonixTouch (Ultrasonix, Richmond, BC) ultrasound scanner; GPS extension (Ascension DriveBay electromagnetic tracker); tracked C5-2 curvilinear transducer (Ultrasonix); 3D position sensors affixed to the transducer and subject. This figure is modified from [21]
The sonographer scans the subject and adjusts the US imaging parameters, such as depth, starting with preset suggestions. The preset values were created alongside the sonographer based on US images of volunteers. Only minor manual adjustments to the imaging parameters for each subject are needed which reduces intraoperative imaging time. The sonographer then landmarks the T12 vertebra and sacrum as well as right and left extremes of the L1 transverse processes to determine the US scanning region. This ensures that the entire lumbar region of the spine is scanned. The reference holder is attached approximately 3 cm above the T12 vertebra landmark. For data acquisition, the US transducer moves slowly and smoothly, while keeping complete contact between the subject’s skin and the ultrasound transducer (minimum of twenty seconds per acquisition). By moving slowly, a dense set of 2D US images are acquired during the US scan. This decreases the degree of interpolation needed during the 3D volume reconstruction of the 2D US images and consequently improves the US volume’s image quality for CT to US registration. From the ultrasound scans of volunteers, we also determined a scanning style that produced reliable US signals across different individuals with various spinal curvatures. We refer to this style as the sagittal zigzag scan where the US transducer starts at the left L1 transverse process and moves across to the right L1 transverse process. It then moves down and across in the opposite direction to acquire US image of the entire lumbar spine.
The imaging software comprises of the freely available and open-source PLUS toolkit [10]. PLUS enables the synchronization of the US image data from the US machine and tracking data from the EM tracker. Prior to US data acquisition, we use the fCal application in PLUS with a double N-wire phantom to calibrate the US transducer for depths between 6 and 9 cm. Once an appropriate depth of the US transducer is selected by a trained sonographer to visualize the spine anatomy, PLUS connects to the hardware using the configuration file associated with the depth chosen. Following data acquisition, an US volume is reconstructed using the PLUS toolkit taking into account the physical pixel size. Only the portion of the lumbar spine visible in preoperative CT is included in the final US volume.
Preoperative CT to intraoperative US registration
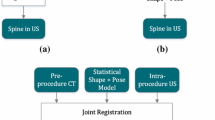
The registration pipeline involves both intensity-based and point-based registration of the bone surfaces to harness the advantages of each method. The aim of the registration workflow is to maximize the similarity using both voxel information and features (points) extracted from the input volumes to find the closest alignment between the CT and US. The general overview of the registration pipeline is illustrated in Fig. 2.
The approach involves three major steps: (i) automatic global intensity-based registration to align the CT and US using voxel intensity information (step 1 in Fig. 2); (ii) automatic global point-based registration using point set correspondences (step 2 in Fig. 2); and (iii) automatic multi-vertebrae point-based registration to account for possible curvature change of each vertebra along the lumbar spine step 3 in Fig. 2). Preprocessing of the US and CT volumes is required prior to registration as enhancement of bone surfaces and elimination of the variability of intensity values in the CT and US volumes will improve the final registration results. Details of the registration are presented below.
Data preprocessing
CT To automatically enhance the CT bone surface, we filter the CT data in the frequency domain using local phase image processing. In the frequency domain, the local amplitude describes the local strength of the image signal and the local phase represents the local structural features, such as discontinuities in the image. Typically, access to these local properties in 2D and 3D is accomplished through the construction of a quadrature pair of oriented band-pass filters. These filters have a \(90^\circ \) phase shift with respect to each other [17]. The Log-Gabor filter is used as the band-pass quadrature filter on the CT slices [7]. As a result of CT local phase filtering, a single feature is extracted, namely the step edge corresponding to a sharp change in the intensity of an image. Step edges occur at soft tissue to bone interfaces. A simple raycasting is done following phase filtering in the posterior to anterior direction of a CT slice such that the first bone pixel encountered for each column is saved as bone and anything below that pixel is saved as background. The raycasting helps to remove the bright intensity values that exist in CT, but do not exist in US, since US signals cannot propagate through the first proximal bone surface. A sagittal CT slice from two different subjects overlaid with the local phase bone surface enhanced from the CT followed by the raycasting is seen in Fig. 3. Finally, to improve the capture range of registration, an inverse Euclidean distance map is calculated on the enhanced bone surface CT image by computing the Euclidean distance between each pixel and the nearest nonzero pixel.
For multi-vertebrae registration (step 3 of our approach), the vertebrae are segmented from the CT data preoperatively. We use the method of Rasoulian et al. [20] for this purpose.
US Local phase filtering of the US volume is performed to automatically enhance the bone surface relative to the soft tissue. The US local phase filtering differs from the CT phase filtering in that multiple edge features (step edge, line, corner, junction) have to be extracted due to the complex shape of the vertebraes appearance in US. Rather than assuming one orientation as is done in the CT local phase filtering, the US local phase filtering allows for the simultaneous estimation of the orientation and local phase information. Details of the description and implementation of this approach are provided in [7]. Subsequently, raycasting is performed from the anterior to posterior direction in order to remove any soft tissue visible above the bone signals. A sagittal US slice of two of the subjects with enhanced bone surface overlaid is shown in Fig. 4.
The bone surface of the US data has to be further segmented prior to the second step of our registration approach. We modified an algorithm originally proposed by Foroughi et al. [5]. In our approach, we use the phase-filtered US data and identify a single pixel bone surface. The bone surface pixels are enhanced by a combination of two main bone features: high acoustic impedance and acoustic shadowing. Pixels with low intensities (shadows under bones) are therefore expected below a pixel of high intensity if the pixel of high intensity is a part of the bone surface. Continuity and smoothness of the bone surface are established by minimizing a cost function using dynamic programming [5]. Our modified algorithm shows fewer false positive bone surface pixels detected, which is critical for accurate point-based registration. An US slice is overlaid with the single pixel US bone surface using the original algorithm and our modified algorithm, respectively, in Fig. 5.
Automatic global intensity-based registration of CT and US
The open-source medical imaging software 3D Slicer is used to automatically register a preprocessed CT to a preprocessed US using an intensity-based approach [15, 16]. The General Registration (BRAINS) module within 3D Slicer version 4.2 that employs the BRAINSFit algorithm is used [9]. Although the original approach was developed for multi-modality registration for the brain, it is used successfully for a variety of multi-modality applications [4, 9]. In the BRAINSFit method, mutual information is used as the similarity metric for registration. Optimal alignment is achieved when the pixel intensity information each input image contains about the other is maximized. The registration is initialized given the transformation of the center of geometry of a preprocessed CT volume to the center of geometry of a preprocessed US volume. This assumes that the center of geometry in both image modalities represents similar structures. A rigid intensity-based registration is performed using the default parameters of the module.
Automatic global point-based registration using point set correspondences
An existing probabilistic point-based registration algorithm, Coherent Point Drift (CPD) [13], is used. One of the benefits for this choice is that it has a closed-form solution. CPD aligns the CT and US point sets using Gaussian mixture models and expectation maximization optimization.
Automatic multi-vertebrae point-based registration
Vertebrae are rigid bodies; however, the intervertebral disks are deformable. CT images are taken in the supine position, whereas the US images are taken in the prone position. To correct for the possible curvature differences between CT and US data, we perform a multi-vertebrae rigid registration. The CPD rigid registration algorithm is modified to define a novel multibody approach, where at each iteration, vertebrae are transformed individually. As a result, it is possible that the vertebrae can be transformed into a pose of the lumbar spine that is not physically possible. To overcome this challenge, points are preoperatively chosen manually between two adjacent vertebrae. Each point is then duplicated where one point belongs to the superior vertebra and the other to the inferior vertebra. These points act as a spring to constrain the pose of the vertebrae during registration. When the registration first begins, the distance between two points that represent a spring is zero. To incorporate the springs in the multi-vertebrae point-based registration, the energy of the springs is added as a regulatory term to the optimization function in the registration pipeline.
To elaborate, in our approach, each vertebra is transformed by an independent rigid transformation, \(T_n\). We assume that point sets of the nth vertebra model are represented by \(X^n\). Since our algorithm does not use any a priori knowledge on the division of ultrasound points to individual vertebrae, the ensemble of target points on the ultrasound is represented by Y. In the absence of any regularization model, the traditional CPD algorithm optimizes the following cost function:
finding the appropriate \(T_n\) for the nth vertebrae that best matches each vertebrae model to the target point set.
The cost function E has the form of \(\sum {p\times \Vert X^n \!-\!T_n(Y)\Vert ^2}\) [13], where p is the posterior probability.
In order to incorporate a biomechanical model, one can add springs between each two adjacent vertebrae. The springs, S, are represented by two ends, between any two adjacent vertebrae. For easier representation, we assume that \(S^n\) are the spring end points belonging to the nth vertebra. Given that springs are only established between adjacent vertebrae, we add R as a regularization term as described below:
where I is the number of springs on each vertebra. Note that the springs that end on the nth vertebrae are transformed by \(T_n\). Combining this regularization term with the registration term will result in:
CPD is an iterative algorithm, which updates the transformation as the certainty of the registration increases. In our proposed method, the cost function is split into terms associated with each vertebra that are optimized separately. In other words, the cost function is optimized with respect to each transformation, \(T_n\), separately and consecutively. Removing terms that are not a function of \(T_n\), the cost function for the nth transformation can be written as:
In order to minimize Eq. 4 using the CPD closed-form solution, we combine the regularization term with the registration term, where the variable p is substituted with the constant \(\alpha \) for all spring points, i. In each iteration of the CPD algorithm, \(T_1,\dots ,T_N\) are updated consecutively. The term \(\alpha \) determines the contribution of the springs. Values of \(\alpha \) between \(2^{-3}\) and \(2^{7}\) were tested; a value of \(2^{5}\) provided the most accurate registration for all clinical datasets and was chosen.
Validation
A gold standard alignment is not possible for the clinical data used in this work as the recruited subjects have pre-existing CT, and fiducial markers that are visible in both CT and US cannot be used. In place of a gold standard, anatomical landmarks on the lamina of each vertebra are identified on the US images. Two operators choose these anatomical landmarks: one orthopedic surgeon and one physician with spine anatomy expertise. The landmarks chosen by the two operators are pooled together. In an F-test performed for one subject, interoperator variability failed to show significance (F \(=\) 0.98).
In the absence of fiducial markers, we assume the CT and US have the optimal alignment following registration. The landmark points chosen in US are assumed to correspond to the same landmarks in the CT and are visually confirmed. To determine the accuracy and precision of the registration method, the CT and the points representing the lamina landmarks are perturbed by a transformation selected randomly from a uniform distribution of 5\(^\circ \) rotation about each axis and 5 mm translation along each axis. The transformation is applied to the entire lumbar spine that is visible in the CT. The initial misalignment is determined by calculating the target registration error (TRE) between the original position of the lamina landmark points and the position of the landmarks after the initial perturbation. To determine the capture range for the registration pipeline, 20 tests are performed with misalignment errors randomly generated within the range 0–25 mm. Registration is then performed, and the final TRE is calculated as the root mean square between the transformed lamina landmark points and their original positions. A test run for one subject is shown as an example in Fig. 6 to demonstrate how this validation works pictorially.
One case of quantitative validation of the CT to US alignment using lamina landmarks placed on the US. Original positions of landmarks are shown in yellow after the alignment of the CT to the US and the transformed positions are shown in blue. a CT aligned to original position of US lamina landmarks; b CT and lamina landmarks transformed by initial perturbation; c CT and lamina landmarks transformed by CT to US transformation found through registration
A qualitative clinical validation is also investigated. Here, a point is added on the posterior dura between two adjacent vertebrae in the US images by both operators. This is where clinicians aim their needle for spinal anesthesia and thus provide a clinically relevant validation. If the points selected are in the correct region, as seen in Fig. 7, after registering the CT to the US, the registration is potentially suitable for spinal interventions.
Results and discussion
As mentioned before, Table 2 lists the lumbar vertebrae included in the registration for each subject. The runtime for each of the main registration components of the pipeline is shown in Table 3. The registration was performed on a Lenovo ThinkCenter, with Intel i5-3570 quad-core CPU and 16 GB of RAM.
As mentioned above, to report quantitative registration accuracies, capture range experiments were performed for all 10 subject datasets. This involved perturbing CT data that are aligned to US by 20 random transformations and re-registering the two volumes. From capture range experiments, an average of 97 % success rate is achieved. Rasoulian et al. [18] defines 2–4 mm as a clinically acceptable accuracy for epidural injections. The space within a facet joint is reported to be between 2 and 4 mm [19]. Success is defined as achieving a mean TRE of 2 mm or less to provide a conservative estimate of clinical acceptability. The mean TRE, maximum point distance and total success rate for each subject are depicted in Table 4. From the results, it is evident that the mean TRE is well below 2 mm.
The qualitative validation of registration involves overlaying contours of the CT on the sagittal, transverse and coronal planes of the US. As an example, the qualitative validation for Subject 7 is illustrated in Fig. 8. A 3D rendering of the CT is also shown in this figure. In Fig. 8, the points on the posterior dura between each two adjacent vertebrae represent the target areas for spinal anesthesia. As mentioned, the posterior dura points are placed on the US images by two operators to clinically validate the CT alignment after registration. From Fig. 8, the CT contours align in all three planes and the points on the posterior dura are all within the target area for spinal anesthesia. Points placed on the posterior dura between adjacent vertebrae may be on different US slices, and therefore, all of the points may not be visible in the coronal slice shown. Qualitative validation provides a clinically relevant result to support the quantitative validation.
The registration pipeline is extensive in that it involves global intensity-based, global point-based and multi-vertebrae point-based registration. We evaluate the effect of each of the three main components of the registration approach (steps 1–3, Fig. 2) on the accuracy of the final alignment of CT and US data. For this purpose, results for the following capture range experiments are outlined:
-
1.
Full registration pipeline.
-
2.
Registration pipeline without the global intensity-based step (i.e., step 1).
-
3.
Registration pipeline without the global point-based step (i.e., step 2).
-
4.
Registration without the multi-vertebrae point-based step (i.e., step 3).
To determine if there is a significant difference in the TRE using the proposed pipeline compared to a modified version, p values are calculated using the Wilcoxon signed-rank test, where \(p<0.01\) is considered significant.
The effect of the intensity-based registration step
CT to US registration results are evaluated without the global intensity-based registration (step 1) and shown in Table 5. The global intensity-based registration involves aligning the bone surfaces in the CT and US that are automatically enhanced using local phase filtering. From the capture range experiments, Subject 1, Subject 4 and Subject 5 failed to show a significant difference in the TRE without the intensity-based registration (\(p = 0.02\), \(p = 0.03\), \(p = 0.39\), respectively). Subjects 2 and 3, and Subjects 6–10 did show a significant difference with an increased TRE (\(p<0.001\), \(p<0.001\), \(p<0.001\), \(p<0.001\), \(p<0.001\), \(p<0.001\), \(p<0.001\), respectively). Removing the intensity information and using solely point information is problematic for these subjects as there is limited US bone visibility in these datasets. Table 5 shows that only Subjects 4 and 5 have a mean TRE less than 2 mm. Subject 1’s mean TRE is negatively affected by three runs of the capture range experiment. These three misalignments have a large rotation about both the z and y axes that the pipeline could not correct without intensity information. It is therefore more robust to include the global intensity-based registration as the first step to provide a coarse initial registration between the CT and US.
The effect of the global point-based registration step
Following the global intensity-based registration to attain a coarse estimate of the alignment between the CT and US, global point-based registration is performed in our proposed approach. Next, we consider whether the global point-based registration step is necessary. From the results of capture range experiments (Table 6), although Subject 6 did show a significant difference in the accuracy (\(p < 0.001\)), the mean TRE for that subject is still below the clinical threshold of 2 mm. Subject 4 was the only subject that failed to show a significant difference in the accuracy (\(p = 0.052\)). Table 6 shows that the mean TRE for most subjects are too high for clinical use in percutaneous spinal interventions. Subject 4 had 6/20 runs that were unsuccessful and negatively affected the mean TRE. In these cases, the intensity-based registration incorrectly aligned the CT vertebral bodies to the lamina of the US. The global point-based registration corrects instances where the global intensity-based step does not provide a close initial alignment between the CT and US. Since the final registration step is constrained by the springs, movement occurs between adjacent vertebrae. A close global alignment is required as there is limited global movement of vertebrae in the multi-vertebrae registration.
The effect of multi-vertebrae point-based registration step
In the last step of registration, springs are used to constrain the multi-vertebrae point-based registration. As vertebrae are the rigid bodies transformed individually at each iteration of the registration, without the springs, registration can result in a pose of the lumbar spine that is not physically possible. This includes two vertebrae intersecting (collision). An example from one run of Subject 10 is shown in Fig. 9. We considered the effect of removing multi-vertebrae point-based registration from the proposed registration pipeline. From the capture range experiments (Table 7), all subjects had a significant loss in accuracy when multi-vertebrae registration is not performed in the pipeline (\(p = 0.001\) for Subject 5 and \(p < 0.001\) for all other subjects). Only Subject 4 has a clinically acceptable mean TRE value.
Registration result after one run for Subject 10 showing a spine pose that is not physically possible. The left panel shows a superior to inferior view of the lumbar spine; the right panel shows a posterior to anterior view of the lumbar spine. Here, L1 and L2 are colliding and that L2, L3, L4, L5 have moved away from each other more than what is physically possible
Conclusion and future work
We presented a novel registration pipeline for the lumbar spine that accurately aligns preoperative CT with intraoperative US, and validated it using 10 clinical datasets. By aligning the CT with the US, anatomical information that is not visible in US is provided to the clinician to guide spine needle interventions without having to expose the physician or patient to more radiation intraoperatively. To the best of our knowledge, this is the first work where multiple vertebrae are registered between CT and US using clinical data.
The proposed registration pipeline shows great promise for guiding percutaneous spine needle procedures, but further improvements are needed for its clinical use. The location of the springs that constrain multi-vertebrae registration is chosen manually preoperatively; however, improvements can be made to automate spring selection for a clinical workflow. In addition, in the multi-vertebrae point-based registration step, the weight \(\alpha \) is constant for all spring points; this could be adaptive depending on where the spring points are between the vertebrae. Finally, the registration pipeline is extensive so it would be useful if steps were only included if the registration accuracy was not clinically acceptable with a simpler version of the pipeline. The registration pipeline needs to be executed on a larger number of patient datasets to further demonstrate its robustness.
References
Balki M, Lee Y, Halpern S, Carvalho J (2009) Ultrasound imaging of the lumbar spine in the transverse plane: the correlation between estimated and actual depth to the epidural space in obese parturients. Anesth Analg 108(6):1876–1881
Conroy P, Luyet C, McCartney C, McHardy P (2013) Real-time ultrasound-guided spinal anaesthesia: a prospective observational study of a new approach. Anesthesiol Res Pract 2013:525 818–525 824
Falco F, Manchikanti L, Datta S, Sehgal N, Geffert S, Onyewu O, Zhu J, Coubarous S, Hameed M, Ward S, Sharma M, Hameed H, Singh V, Boswell M (2012) An update of the effectiveness of therapeutic lumbar facet joint interventions. Pain Physician 15:E909–E953
Fedorov A, Tuncali K, Fennessy FM, Tokuda J, Hata N, Wells WM, Kikinis R, Tempany CM, Tempany CM (2012) Image registration for targeted MRI-guided transperineal prostate biopsy. J Magn Reson Imaging 36(4):987–9926
Foroughi P, Boctor E, Swartz M, Taylor R, Fichtinger G (2007) P6d–2 ultrasound bone segmentation using dynamic programming. In: Ultrasonics symposium New York, NY, USA, pp 2523–2526
Gill S, Abolmaesumi P, Fichtinger G, Boisvert J, Pichora D, Borshneck D, Mousavi P (2012) Biomechanically constrained groupwise ultrasound to CT registration of the lumbar spine. Med Image Anal 16(3):662–674
Hacihaliloglu I, Rasoulian A, Rohling R, Abolmaesumi P (2013) Statistical shape model to 3D ultrasound registration for spine interventions using enhanced local phase features. In: Medical image computing and computer-assisted intervention-MICCAI, pp 361–368
Herring J, Dawant B, Maurer C, Muratore D, Galloway R Jr, Fitzpatrick J (1998) Surface-based registration of CT images to physical space for image-guided surgery of the spine: a sensitivity study. IEEE Trans Med Imaging 17(5):743–752
Johnson H, Harris G, Williams K (2007) Brainsfit: mutual information registrations of whole-brain 3D images, using the insight toolkit. Insight J
Lasso A, Heffter T, Pinter C, Ungi T, Fichtinger G (2012) Implementation of the plus open-source toolkit for translational research of ultrasound-guided intervention systems. In: MICCAI workshop on systems and architectures for computer assisted interventions, Nice, France, pp 1–12
Moore J, Clarke C, Bainbridge D, Wedlake C, Wiles A, Pace D, Peters T (2009) Image guidance for spinal facet injections using tracked ultrasound. In: Yang GZ, Hawkes D, Rueckert D, Noble A, Taylor C (eds) MICCAI 2009. LNCS 5761, pp 516–523
Muratore DM, Russ JH, Dawant BM, Galloway R Jr (2002) Three-dimensional image registration of phantom vertebrae for image-guided surgery: a preliminary study. Comput Aided Surg 7(6):342–352
Myronenko A, Song X (2010) Point set registration: coherent point drift. IEEE Trans Pattern Anal Mach Intell 32(12):2262–2275
Nagpal S, Abolmaesumi P, Rasoulian A, Ungi T, Hacihaliloglu I, Osborn J, Borschneck D, Lessoway V, Rohling RN, Mousavi P (2014) CT to US registration of the lumbar spine: a clinical feasibility study. In: Stoyanov D, Collins L, Sakuma I, Abolmaesumi P, Jannin P (eds) IPCAI 2014. LNCS 8498, pp 108–117
Pieper S, Halle M, Kikinis R (2004) 3D Slicer. In: IEEE symposium on biomedical imaging: nano to macro 2004, pp 632–635
Pieper S, Lorensen B, Schroeder W, Kikinis R (2006) The NA-MIC kit: Itk, vtk, pipelines, grids and 3D slicer as an open platform for the medical image computing community. In: IEEE symposium on biomedical imaging: nano to macro 2006, pp 698–701
Rajpoot K, Grau V, Noble JA (2009) Local-phase based 3D boundary detection using monogenic signal and its application to real-time 3D echocardiography images. In: IEEE symposium on biomedical imaging: from nano to macro 2009, pp 783–786
Rasoulian A, Rohling R, Abolmaesumi P (2012) Probabilistic registration of an unbiased statistical shape model to ultrasound images of the spine. In: proceedings of SPIE medical imaging, Orlando, Florida, USA, pp 8316:83161P-1-83161P-6
Rasoulian A, Abolmaesumi P, Mousavi P (2012) Feature-based multibody rigid registration of CT and ultrasound images of lumbar spine. Med Phys 39(6):3154–3166
Rasoulian A, Rohling R, Abolmaesumi P (2013) Lumbar spine segmentation using a statistical multi-vertebrae anatomical shape+pose model. IEEE Trans Med Imaging 32(10):1890–1900
Ungi T, Abolmaesumi P, Jalal R, Welch M, Ayukawa I, Nagpal S, Lasso A, Jaeger M, Borschneck DP, Fichtinger G, Mousavi P (2012) Spinal needle navigation by tracked ultrasound snapshots. IEEE Trans Biomed Eng 59(10):2766–2772
Winter S, Brendel B, Pechlivanis I, Schmieder K, Igel C (2008) Registration of CT and intraoperative 3D ultrasound images of the spine using evolutionary and gradient-based methods. IEEE Trans Evolut Comput 12(3):284–296
Yan CXB, Goulet B, Pelletier J, Chen SJ-S, Tampieri D, Collins DL (2011) Towards accurate, robust and practical ultrasound-CT registration of vertebrae for image-guided spine surgery. Int J Comput Assist Radiol Surg 6(4):523–537
Acknowledgments
This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC), Canadian Institutes of Health Research (CIHR) and the NSERC Collaborative Research and Training Experience (CREATE) program. We would like to thank Kingston General Hospital Imaging Department, and the Human Mobility Research Centre, specifically Fiona Howells, Leone Ploeg, Karen Pearson, Barbara Delaney, and Catherine Logan Munro for their assistance with patient recruitment, and data acquisition.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conict of interest.
Ethical standard
The study was approved by the institutional Research Ethics Board.
Informed consent
All participants provided informed consent to be part of the study.
Rights and permissions
About this article
Cite this article
Nagpal, S., Abolmaesumi, P., Rasoulian, A. et al. A multi-vertebrae CT to US registration of the lumbar spine in clinical data. Int J CARS 10, 1371–1381 (2015). https://doi.org/10.1007/s11548-015-1247-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-015-1247-5