Abstract
Regional myocardial function assessment is essential for diagnosis and evaluation of heart disease. The purpose of this study was to enhance the spatial resolution of a speckle tracking echocardiography approach and enable layer-specific analysis of the myocardium. Following validation with software-implemented and mechanical phantoms versus imposed values, short-axis cines were obtained from 50 rats. The cines were post-processed by a speckle tracking commercial program, and the myocardial velocities were processed by a three-dimensional wavelet de-noising program, instead of the built-in smoothing process of the commercial program. Software-implemented phantom measurements yielded rotation errors of 7.5%, 2.9%, and 3.4%, for inner, middle, and outer layers, respectively. Analysis of a shrinking/expanding mechanical phantom yielded strain errors of 3%, 5%, and 7% for the three layers. Bland–Altman analysis showed agreement between the commercial and enhanced programs. Thus, layer-specific analysis is feasible while using echocardiography even on small animals such as rats.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
The mammalian heart performs a complex movement during its contraction due to its geometric fibrous structure. The myocardial fibers are wound in a helical orientation [9], and the fiber orientation changes smoothly from a right-handed helix in the endocardium to a left-handed helix in the epicardium [9, 39, 40]. The contraction and relaxation of this structure of fibers produces a wringing motion, i.e., torsion, with the base and apex rotating in opposite directions. This unique structure explains the inhomogeneity of strain and torsion measurements across the transmural orientation described by tagged-magnetic resonance imaging (MRI) studies [3, 29, 32, 42]. The contribution of the transmural inhomogeneities to left ventricular (LV) function was studied by mathematical models, which demonstrated that larger torsion of the inner layers, relatively to the outer layers, equalizes the sarcomere shortening, mechanical loading [2, 8, 19], and energy demand [4] between the endocardial and epicardial layers.
Different pathologies induce changes in torsion [17, 29, 33, 42]. These changes can occur in a specific region or layer, therefore interrupting the balance between the myocardial layers. Hence, efforts have been made to enhance the spatial resolution of wall motion estimation by tagged-MRI [3, 7, 11, 12, 14, 17, 25, 27, 29, 32, 33, 38, 42], tissue Doppler imaging (TDI) [13, 41], and speckle tracking echocardiography (STE) [18] in order to enable evaluation of the transmural distribution of the LV contraction–relaxation function. Yet, both tagged-MRI and TDI suffer from significant limitations. Cardiac tagged-MRI suffers from limited spatial and temporal resolutions as well as limited availability, and is still an expensive modality. TDI allows valid measurements only for motion along the orientation of the ultrasound beam. The STE method, on the other hand, is angle-independent, and can provide functional measurements in all LV segments. The STE method was validated against different modalities [1, 10, 18, 20, 26, 34, 37], and its sensitivity along the circumferential direction was assessed versus histology on a rat model of acute myocardial infarction [35]. The STE method provides real-time assessment of LV function [26, 37], thus it has the potential of becoming a widespread clinical tool.
Pathologies induced in rats are commonly used as models of human heart disease. The ability to monitor non-invasively the development of myocardial remodeling as a response to induced pathologies or drugs (e.g. doxorubicin [31]) may be challenging but rewarding. The rat heart is small and its heart rate is fast, thus its STE analysis requires relatively fine image quality and high frame rate (more than 180 frames per second for an anesthetized rat). This may be the reason that up till now only a few studies utilized the STE method to evaluate cardiac function in rats, e.g. myocardial infarction [35], doxorubicin cardio-toxicity [31], and diabetic cardiomyopathy [24]. The STE analysis is based on tracking of speckles from frame to frame in a region of myocardial tissue imaged by echocardiography, and averaging the tissue tracking results over the width of the myocardial wall, thus provides strain measurements that are also averaged across the wall. These strain measurements provide sufficient information for many clinical studies. However, there are vital clinical questions that require analysis at a higher spatial resolution, e.g., detection of endocardial ischemia, measurement of endocardial/epicardial rotation, shear etc. The purpose of this study was to evaluate an enhanced revision of the STE commercial program that allows layer-specific analysis, versus the original version that averages the strain across the whole myocardium of a rat. The enhanced revision is termed the layer-specific STE (LS-STE) program. The LS-STE program processes the myocardial velocities, using a wavelet de-noising technique instead of the built-in smoothing process of the STE commercial program. A smoothing process is required due to the noise that exists in the myocardial velocities signals, where most of the noise is caused by inaccurate tracking of the tracking algorithm. In the present study, the accuracy of the LS-STE method was assessed under controlled conditions, using specially designed phantoms. Validation of the accuracy of measuring the tissue rotation in three layers was done using data from different phantoms: (1) a simulation of tissue rotation in software; (2) a simulation of tissue rotation and shrinkage/expansion in software, (3) a rotating mechanical phantom. Validation of the accuracy of measuring the strain in three layers was done using data from different phantoms: (1) a simulation of tissue rotation and shrinkage/expansion in software, (2) a shrinking/expanding mechanical phantom. Following validation by phantoms, the transmural distribution of LV rotation and circumferential strain were measured in standard short axis cross-sections of normal rats by the LS-STE program and by manually applying the STE commercial program to the inner and outer layers of the myocardium as described by Hui et al. [21]. The two methods were compared as another validation of the LS-STE program. Analyzing the transmural distribution of these parameters in human studies will allow early detection of different hypertrophy-causing pathologies, and subtle, non-transmural pathologies. For example, in early stages of aortic-stenosis [12] and in cardio-toxicity due to doxorubicin treatment the sub-endocardium is initially injured [5], and thus, there is an advantage in analyzing the performance of the myocardial layers, while specifically studying the performance of the endocardium.
2 Materials and methods
A STE commercial program called ‘2D-strain’ (EchoPACTM Dimension ‘08, GE Healthcare Inc., Norway) was modified by calculating the functional measurements at three myocardial layers instead of for the whole myocardial wall, thus increasing the transmural resolution. The EchoPAC processes standard echocardiographic two-dimensional (2D) gray-scale cine loops, imposes a grid of points termed “tracking points” (TP) on each frame of the imaged myocardial cross-section, and evaluates the velocities of the TP from the tracked movement of the strong reflectors within the assigned region [36]. The velocity values, calculated at the TP in 2D Cartesian coordinates, are transformed to object-related coordinates; namely, the circumferential and radial orientations of the LV short-axis cross-section. Consequently, a sampled velocity function is obtained with the following dimensions: (1) circumferential orientation sampled at the TP, producing ~40–50 samples. (2) Radial orientation with four samples across the myocardial wall. (3) Time dimension of one cardiac cycle, with frame rate of 225 [frames/s].
2.1 De-noising using wavelet functions
The measured noisy myocardial velocities, before they undergo the smoothing process of the EchoPAC, are used as the raw data for the algorithm presented here. The algorithm was implemented by extending to 3D some 2D routines (dwt, idwt, appcoef, detcoef, dyadup, wavedec, waverec), which are included in the Matlab wavelet toolbox (MathWorks Inc. USA).
The 3D wavelet shrinkage method, used to de-noise the sampled velocity function, consists of three steps:
-
1.
Replacing data points of low tracking quality by a weighted average of their neighboring data points: the speckle tracking algorithm calculates the location of each TP at the next time frame by minimizing the correlation between a kernel and a target block. The correlation coefficient between the kernel and the target block, for each TP, serves as a “tracking quality” measure for each TP, calculated by:
$$ \rho \left( {k,l} \right) = {\frac{{\sum\nolimits_{m = 1}^{M} {\sum\nolimits_{n = 1}^{N} {\left( {b_{m,n}^{j} - \bar{b}^{j} } \right)\left( {b_{m + k,n + l}^{j + 1} - \bar{b}^{j + 1} } \right)} } }}{{\sqrt {\sum\nolimits_{m = 1}^{M} {\sum\nolimits_{n = 1}^{N} {\left( {b_{m,n}^{j} - \bar{b}^{j} } \right)^{2} \sum\nolimits_{m = 1}^{M} {\sum\nolimits_{n = 1}^{N} {\left( {b_{m + k,n + l}^{j + 1} - \bar{b}^{j + 1} } \right)^{2} } } } } } }}} $$(1)b j m,n is the intensity of the (m, n) pixel in the kernel area, b jm+k,n+l is the intensity of the (m + k, n + l) pixel in the target area, \( \bar{b}^{j} \) is the mean intensity and ρ(k, l) is the tracking quality, given by a number between 0 and 1. When the tracking algorithm loses tracking, the data point may obtain large, unrealistic values. To minimize the effect of such poor quality data points (tracking quality below 0.3), they are replaced by a weighted average of their neighboring data points, in both spatial and temporal orientations.
-
2.
Interpolation along the radial orientation: in order to exploit during the de-noising process the full three dimensionality of the data, i.e., also the radial orientation that comprises of only four data points, the data is interpolated by (1:10) along the radial orientation, before being transformed to the wavelet domain. The interpolation affects the de-noising of the results along the direction of the interpolation, thus less Detail coefficients will be zeroed.
-
3.
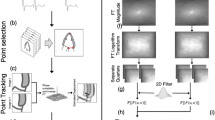
3D wavelet de-noising: the wavelet shrinkage procedure, which utilizes the fast wavelet transform [30], is applied to the interpolated data, using the Daubechies mother wavelet with four vanishing moments. A one-stage discrete wavelet transform of a signal employs high-pass and low-pass filters to retrieve the Approximation and Details coefficients. The wavelet multi-resolution representation is obtained by iterating the decomposition process, with successive Approximations being decomposed in turn, so that one signal is broken down into seven lower resolution components. Since the velocity function is three-dimensional (two spatial dimensions and the temporal dimension), the de-noising process uses simultaneously three 1D filter banks, one for each of the three dimensions. If the data is of size N1 by N2 by N3, applying the 1D analysis filter bank to the third dimension gives eight sub-band data sets, each of size N1/2 by N2/2 by N3/2. Each of the eight details (at each resolution level) contains one of the eight combinations of high and low frequency components of each dimension. One step of the iterative discrete wavelet transform process is sketched in Fig. 1a.
Two-channel sub-band coder with three levels of decomposition. a One level of decomposition creates seven different details (LLH-HHH) and one approximation (LLL) which is decomposed to another seven different details (b). It is described in b which details were thresholded and which were set to zero (Di—details from iteration i)
Following the wavelet analysis, Detail coefficients are thresholded. The Detail coefficients at each level j are treated using soft thresholding. More specifically, for levels j = 1:(J − k) the coefficients are removed completely, and for the last k levels a threshold is determined according to Eq. 2 as seen in Fig. 1b [15, 16],
where N is the length of each Detail at this level, σ is the noise variance estimated using MAD/ 0.6745. MAD stands for the median absolute value of the coefficients of the first level, i.e., of the highest resolution. To obtain the smoothed signal, the inverse of 3D DWT is computed using the reconstruction filters, in an iterative process similar to that of the 2D case. The whole process running time is less than 2 s on a 2.4 GHz dual-core CPU.
The rotation of each myocardial layer is calculated from the locations and velocities of each TP, as described by Notomi et al. [34], and the circumferential strain is calculated as described by Rappaport et al. [36]. The results are presented for three myocardial layers, and not four, as the two mid-wall layers results are averaged. The rotation and circumferential strain are evaluated relatively to end diastolic position of the LV. End diastolic time is defined as the time right before the R wave of the ECG signal.
2.2 Validation process
2.2.1 Software-implemented phantoms
2.2.1.1 Rotating phantom
The phantom was designed and developed using the Matlab software. An US image of a circular phantom was made to rotate artificially, so that the angular velocity changed linearly with the radius, consequently creating shear among the layers
where \( \omega \left( r \right) \) is the angular velocity as function of radius r, and R out and R in are the outer and inner radii, respectively. The circumferential and radial strains were zero. Five cines were created with different ω(R out) values of 0.01–0.03 [rad/fr].
2.2.1.2 Rotating and shrinking/expanding phantom
The phantom was designed and developed using the Matlab software and the Field II simulation program, which produces US images similar to the way US imaging system produces them [22, 23]. The speed of sound was assumed to be 1540 m/s, the central frequency was 2 MHz, kerf was 0.0025 mm, element height was 7 mm, the focus was at 80 mm, the number of elements was 64, and the sector angle = 90°. The number of scatterers was 132100, including ~1200 strong scatterers and ~130900 weak scatterers. The artificial cine simulates apical short axis cross-section of the LV with non-uniform contraction and relaxation. The myocardial ring shrinks/expands and rotates inhomogeneously across the radius as function of time. The cine contains 58 frames. The rotation as function of time is given by
where θ(n, r) is the tissue rotation as function of frame number and radius r, N is the total number of frames (N = 58).
The rotation changes linearly with the radius
where θ(n, r) is the tissue rotation as function of radius r and frames n, and R out(0), R in(0) are the outer and inner radii at the first frame, respectively.
The circumferential strain changes as function of radius and frame number, while the scaling factor of the inner radius is defined as:
where μ(R in(0), n) is the scaling factor of the inner radius as function of time, R in(n) is the inner radius as function of time, n is the frame number, and N is the total number of frames (N = 58).
The ring contracts while preserving its area, which results of the following formula for the circumferential strain
where S c(r, n) is the tissue circumferential strain, r is the radius, n is the frame number, and R in(0) is the inner radius at the first frame.
The peak rotation and peak circumferential strain, obtained by the LS-STE program, were compared to the imposed values of the simulation programs. Error was defined as % from the real value.
2.2.2 Mechanical phantoms
The purpose of creating mechanical phantoms was to create a model of moving tissue in which the reflectors are well seen during the whole movement.
2.2.2.1 Cylindrical rotating phantoms
The phantoms were designed and constructed with different shears generated along the radial orientation of the phantom. The phantoms were constructed from flexible polyurethane–Epusil U-105 mixed with curing agent UC-139 (Polymer Gvulot Ltd. Israel). Silicon Carbid powder-GRIT 1000 (BUEHLER Ltd. USA) was mixed with the uncured material to create small reflectors within the phantoms. During the curing process, the material was degassed (−0.7 bar versus atmosphere pressure), so that air bubbles would not disrupt the US imaging. The material was poured into a mold to create a hollowed cylinder, serrated from the inside and outside, creating very clear reflectors in the inside and outside of the cylinder. The phantoms were created in two sizes: (1) outer diameter of 10 cm and inner diameter of 6 cm modeling a human heart. (2) Outer diameter of 10 mm and inner diameter of 6 mm modeling a rat’s heart. The phantoms underwent a partial curing process during 48 h to create a consolidated flexible structure. A certain section of the periphery of the phantoms was rotated by a Plexiglas disc that was connected to a computer controlled electrical motor, while the inside was held static by a Plexiglas serrated cylinder, which was partially inserted into the central part of the phantom. The ultrasound scan was performed at a location adjacent to the Plexiglas disc, thus the rotation of the scanned area was somewhat smaller than the imposed one.
2.2.2.2 Shrinking/expanding phantom
The phantom was designed and constructed in order to validate the strain measurements. The phantom was constructed from Agar-agar (Pronadisa, Madrid, Spain) and Silicon Carbide powder–GRIT 1000 (BUEHLER Ltd., IL, USA) mixed with degassed water. The phantom was pressed and released against a Perspex wall by a piston, where its motion was imposed by a computer controlled electrical motor. The phantom was scanned through an “acoustic window” in the Perspex wall. The imposed strain values were 10%, 15%, and 20%.
The large cylindrical rotating phantom and the shrinking/expanding phantom were scanned by a VividTM 3 system (GE Healthcare Inc. Norway) using 3S phased array probe, and a cardiac application. The frame rate was 40 frames per second. The small cylindrical rotating phantom was scanned by a VividTM i system (GE Healthcare Inc. Norway) and 10S phased array probe. The frame rate was 225 frames per second. The acquired cines were analyzed by the EchoPAC to produce the non-filtered myocardial velocities within the region in the image termed “myocardial tissue”. The noisy velocities were analyzed by the new method, to produce the LS-STE velocities and the peak rotation and peak circumferential strain values. For the rotating phantoms: a narrow ROI was chosen, encompassing either the inner, middle, or outer layers, thus limiting the measurement to be the average value for a narrow ROI as described by Hui et al. [21]. For the small rotating phantom it was possible to apply only two narrow ROIs, so the results of the EchoPAC were compared only to the inner and outer layers. The peak rotation value for a narrow ROI was calculated by the EchoPAC and was compared to the results of the LS-STE method with a wide ROI, which contained the three radial layers.
2.3 Animal studies
2.3.1 Experimental animals
Animal experiments were approved and conducted according to the institutional animal ethical committee guidelines (ethics number: IL-101-10-2007). In this study, 50 male Sprague-Dawley rats of 3 months old were examined, weighing 289 ± 21 g (Mean ± SD).
2.3.2 Echocardiography protocol
Each rat was anesthetized by an intraperitoneal (IP) injection of ketamine-xylazine mixture and its chest was shaved. The rat was placed in a left lateral decubitus position and scanned via a commercially available echo-scanner—VividTM i ultrasound cardiovascular system (GE Healthcare Inc. Israel) using a 10S phased array pediatric transducer and a cardiac application with high temporal and spatial resolutions. The transmission frequency was 10 MHz, the depth was 2.5 cm, and the frame rate was 225 frames per second. The standard 2D echocardiography included scanning at three short-axis levels: basal (mitral valve), equatorial (papillary muscle tips), and the apical cross-sections. The acquired cines were analyzed by the EchoPAC by choosing 3 ROIs: a wide ROI, which included the whole myocardium, and two narrow ROIs, one for the inner layers and one for the outer layers of the myocardium. The wide ROI was analyzed by the LS-STE program to produce myocardial rotation and circumferential strain values at three myocardial layers. The two narrow ROIs were analyzed by the EchoPAC to produce myocardial rotation and circumferential strain values for the inner half and the outer half of the myocardium as was done by Hui et al. [21]. The results of the LS-STE program (three layers) were compared to the results of the EchoPAC (two layers) by: (1) averaging the LS-STE results of the inner and middle layers and comparing it to the inner ROI, analyzed by the EchoPAC. (2) Averaging the LS-STE results of the outer and middle layers and comparing it to the outer ROI, analyzed by the EchoPAC. An example of de-noised velocities is seen in Fig. 2. The velocities are displayed on the short axis cross-section.
2.3.3 Statistical analysis
Comparison between the LS-STE program and the STE commercial program was performed using the Bland–Altman analysis [6]. The mean value and limits of agreement (average ± 1.96 * SD) are presented for each plot. The precision of the mean and limits of agreement are given by a confidence interval. For the different combinations of the levels and layers, the two parameters (rotation and circumferential strain) were compared by two-way analysis of variance (ANOVA) using Sigma Stat 3.11 (Systat Software, Inc., San-Jose, CA, USA). Whenever the ANOVA was significant, Holm-Sidak post hoc test was conducted for multiple comparisons of either level or layer. Statistical significance was set at P < 0.05.
3 Results
3.1 Validation studies using the software-implemented phantoms
3.1.1 Rotating phantom
Results for the rotating phantom are summarized in Table 1. The table contains the imposed peak rotation values (Degree) for five ωout [radians/frames] values, versus peak rotation measurements estimated by the LS-STE program. The velocity values for one cine are depicted in Fig. 3. The average errors of rotation between the measured values and the imposed values for the inner, middle, and outer layers are 7.5%, 2.9%, and 3.4%, respectively.
3.1.2 Rotating and shrinking/expanding phantom
The results are summarized in Table 2. The table contains the imposed peak rotation (Degrees) and peak circumferential strain (%) values versus the measurements obtained by the LS-STE program. The results show that the LS-STE program estimates the rotation and circumferential strain with rather small errors between the measured values and the imposed values (average rotation error—1.7%, average circumferential strain error—4.2%).
3.2 Validation studies using mechanical phantoms
3.2.1 Cylindrical rotating phantom
The maximal angle and velocity of rotation were imposed on the mechanical rotating phantom and its rotation was measured. Thirty four loops with different rotation values were scanned for the large phantom and 12 loops were scanned for the small phantom. Analysis was done by the LS-STE program and by the EchoPAC as described in Sect. 2. Although the inner layer was supposed to be static, it still rotated. However, the rotation of the inner layer was smaller than the rotation of the outer layer to create shear angle. The rotation of the inner layer was 2–6 [deg] and the rotation of the outer layer was 3–8 [deg]. Bland–Altman analysis for the rotation values of the human LV (A) and rat LV (B) phantoms are presented in Fig. 4. The results show a good agreement between the two methods.
Bland–Altman analysis between the LS-STE program with wide ROI and the EchoPAC applied to narrow ROIs for the mechanical cylindrical rotating phantom (LS-STE–STE): a results for the human LV phantom (degrees of freedom = 242) b results for the rat LV phantom (degrees of freedom = 12). The mean value and limits of agreement are presented
3.2.2 Shrinking/expanding phantom
The results obtained by the LS-STE program at three layers were compared to the strain values that were imposed by the computer controlled electrical motor. Analysis of 10 cines, where strain values of 10% were imposed, provided strain results of 9.8 ± 0.7% (6% error). Analysis of three cines with imposed strain values of 15% provided strain results of 14.7 ± 1.1% (6% error). Analysis of three cines with imposed strain values of 20% provided strain results of 19.8 ± 2.4% (10% error).
3.3 Animal studies
3.3.1 Peak rotation
Both the EchoPAC and the LS-STE program showed that the apex and base rotate in opposite directions as seen in the Bland–Altman analysis in Fig. 5. The two-way ANOVA indicated significant difference between the rotations at the different levels (P < 0.001). Significant difference in rotation values was found between the layers in the AP and MV levels (P < 0.001), but not in the PM level. Bland–Altman statistical analysis showed an agreement between the LS-STE program results and the manual narrow ROIs analyzed by the EchoPAC (Fig. 5).
Bland–Altman analysis comparing the rotation values calculated by the EchoPAC and by the LS-STE program (LS-STE–STE). The rotation values were evaluated for the inner half and outer half of the myocardium. The results are plotted for 50 rats and for three levels of short axis: AP apex, PM papillary muscles, and MV mitral valve. The mean value and limits of agreement are presented. The confidence interval is presented for the mean value and limits of agreement
3.3.2 Peak circumferential strain
The results were obtained for the three myocardial layers in the three short axis levels: AP, PM, and MV levels. The peak circumferential strain was found to be larger in the endocardium than in the epicardium (Fig. 6). The two-way ANOVA indicates significant difference between the peak circumferential strains in the different levels of the heart (P < 0.001) as well as the different layers (P < 0.001). Furthermore, significant difference in circumferential strain was found between the layers at each level (P < 0.0001), indicating that at each level each layer has its own discrete circumferential strain. Bland–Altman statistical analysis showed an agreement between the LS-STE program results and the manual narrow ROIs analyzed by the EchoPAC (Fig. 6).
Bland–Altman analysis comparing the circumferential strain values calculated by the EchoPAC and by the LS-STE program (LS-STE–STE). The values were evaluated for the inner half and outer half of the myocardium. The results are plotted for 50 rats and for 3 levels of short axis: AP apex, PM papillary muscles, and MV mitral valve. The mean value and limits of agreement are presented. The confidence interval is presented for the mean value and limits of agreement
4 Discussion
Transmural analysis of LV function provides new information, potentially of clinical significance, which is unattainable by visual inspection of ultrasound cines, or even by 2D strain analysis. Since pathologies induced in rats commonly serve as models of human pathologies, which may be studied under controlled conditions, studying the function of the rat LV is of high interest. Yet, transmural analysis of LV function of a rat is very challenging due to the small size of the heart and its high heart rate, which require excellent spatial and temporal resolutions. In this study, the analysis of transmural LV function was performed by enhancing a speckle tracking echocardiography (STE) commercial program (EchoPAC) to create a layer specific program (LS-STE), which provides clinically relevant information at a spatial resolution of three myocardial layers. Such an analysis was possible owing to the application of 3D wavelet de-noising to the myocardial velocities. Thus far, transmural myocardial strain analysis for small rodents was reported by Luo et al. [28], who utilized radio frequency (RF) data, instead of B-mode images, for the strain calculations. The advantage of the method presented here over the method proposed by Luo et al. is the utilization of an available and user-friendly commercial program (EchoPAC) along with a nearly real-time (2 s) and easy to use Matlab routine, where the utilization of RF data requires costly special equipment, an expert that will run it, and a longer post-processing (7 min).
The LS-STE method was validated by performing measurements of controlled movements, performed by software-implemented phantom models and by mechanical-phantom models, and was proved to provide circumferential strain and rotation values similar to the imposed values. Therefore, the results of the animal studies can be regarded as reliable. The results of the animal studies, presented here, demonstrate that the endocardium rotates significantly more than the epicardium and the endocardium produces larger circumferential strains than the epicardium.
The EchoPAC, which is able to produce strain parameters for the whole myocardium, was validated against sonomicrometry [1, 10] and tagged MRI [1, 10]. Recently, a validation of the segmental strain parameters was performed against histology on a rat model, thus it is possible to assume that the segmental strain measurements are reliable even for small animals as rats [35]. The rotation parameter of the STE program of the EchoPAC was validated against sonomicrometry [20] and tagged MRI [20, 34]. Therefore, another kind of validation of the LS-STE method was performed by applying narrow ROIs on the inner and outer regions of the short axis cross-sections, and measuring the rotation and circumferential strain for the two regions as performed by Hui et al. [21]. The results of the EchoPAC were compared to the results of the LS-STE program by averaging the results of the LS-STE program from three layers to two layers. Bland–Altman statistical analysis demonstrated an agreement between the two methods (Figs. 5, 6). It is important to mention that even though it is possible to apply transmural analysis by using two narrow ROIs as described by Hui et al. [21], such an analysis has some major limitations: (1) the tracking is applied to a narrow ROI, which contains small amount of strong reflectors and thus the strain parameters are less reliable a the tracking quality, defined by the EchPAC, is lower. (2) The choice of the ROI is user dependant. (3) The outer ROI has low tracking quality due to the fact that the epicardium looks blurred, and its borders are hard to track. However, the inner half has very clear borders, and the tracking quality is good. In this study, the inner half analysis had a reliable tracking quality, and it was not user dependant since we took the wide ROI of the LS-STE analysis and just made it narrower without any additional changes. The outer half had a low tracking quality and it was user dependant.
The ultrasound cines of the animal studies were performed by utilizing a VIVID i system and a pediatric probe, which enabled us to achieve clear short-axis images of the LV with high frame rate (225 frames per second). All 150 ultrasound cines were trackable by the EchoPAC as determined by the tracking quality index of the EchoPAC, and therefore it ensured that the animal tracking results were reliable.
An additional validation was conducted by operating the LS-STE method while choosing a narrow ROI, encompassing either the inner, middle, or outer layers of the rotating software-implemented phantom (Table 1) and of a mechanical cylindrical rotating phantom (Fig. 4). The peak rotation value for a narrow ROI was calculated by the EchoPAC and was compared to the results of the LS-STE program that used a wide ROI, which contained the three radial layers. The Bland–Altman analysis of the mechanical cylindrical rotating phantom, depicted in Fig. 4, shows a good agreement between the two methods. Although it is possible to perform a layer-specific analysis by applying the EchoPAC while using narrow ROIs [21], selecting three ROIs is user-dependant, and thus prone to intra-observer and inter-observer variability, due to different selections of the ROI.
4.1 Study limitations
The LS-STE method, like any other STE method, performs tracking of myocardial tissue analysis and thus depends on image quality. It requires images in which the whole cross-section of the myocardium is well seen, otherwise the tracking deteriorates. Additionally, the speckle tracking is performed for four layers of the myocardium, and the strain is calculated for three layers, of equal size. Since the actual width of each myocardial layer for a particular subject is unknown, and is probably different for each layer, the assumption that the layers have equal width produces an error. Another limitation, characteristic to any 2D method that acquires functional LV data, is the dependency on the choice of the angle of the scan plane. When tilting the ultrasound probe, the values of rotation at the different layers change due to the different orientations of the myocardial fibers in the different layers. This is probably the reason for the relatively high variance of the rotation measurements in the animal studies. Another limitation, also common to all 2D ultrasound imaging methods, is the out-of-plane motion of the heart, which occurs mostly when scanning at the basal short axis. Therefore, it is less accurate to track the basal short axis level. This limitation can be overcome by using a 3D ultrasound system. Moreover, the out-of-plane movement was not simulated in the phantoms, and thus the tracking error that occurs due to the out-of-plane movement was not handled in this study.
5 Conclusions
The measurement of LV rotation and circumferential strain at a resolution that allows discerning LV function at three myocardial layers is feasible on a rat model owing to the usage of 3D wavelet de-noising of the myocardial velocities. LV rotation and circumferential strain obtain higher values at the endocardial layer, and the values decrease towards the epicardium. This heterogeneity across the layers, of the peak LV rotation and the peak circumferential strain values, was found to be significant in normal LVs, as was measured in this study. The ability to measure myocardial function on a rat model at three layers may provide a useful experimental tool.
References
Amundsen BH, Helle-Valle T, Edvardsen T et al (2006) Noninvasive myocardial strain measurement by speckle tracking echocardiography validation against sonomicrometry and tagged magnetic resonance imaging. J Am Coll Cardiol 47:789–793
Arts T, Reneman RS (1989) Dynamics of left ventricular wall and mitral valve mechanics. J Biomech 22(3):261–271
Azhari H, Buchalter M, Sideman S et al (1992) A conical model to describe the nonuniformity of the left ventricular twisting motion. Ann Biomed Eng 20(2):149–165
Beyar R, Sideman S (1985) Effect of the twisting motion on the nonuniformities of transmyocardial fiber mechanics and energy demand. IEEE Trans Biomed Eng 32(10):764–769
Billingham ME, Mason JW, Bristow MR et al (1978) Anthracycline cardiomyopathy monitored by morphologic changes. Cancer Treat Rep 62(6):865–872
Bland JM, Altman DG (1986) Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1(8476):307–310
Bogaert J, Rademakers FE (2001) Regional nonuniformity of normal adult human left ventricle. Am J Physiol Heart Circ Physiol 280(2):H610–H620
Bovendeerd PH, Arts T, Huyghe JM et al (1992) Dependence of local left ventricular wall mechanics on myocardial fiber orientation: a model study. J Biomech 25(10):1129–1140
Buckberg GD (2002) Basic science review: the helix and the heart. J Thorac Cardiovasc Surg 124:863–883
Cho GY, Chan J, Leano R et al (2006) Comparison of two-dimensional speckle and tissue velocity based strain and validation with harmonic phase magnetic resonance imaging. Am J Cardiol 97:1661–1666
Clark NR, Reichek N, Bergey P et al (1991) Circumferential myocardial shortening in the normal human left ventricle. Assessment by magnetic resonance imaging using spatial modulation of magnetization. Circulation 84(1):67–74
Delhaas T, Kotte J, van der Toorn A et al (2004) Increase in left ventricular torsion-to-shortening ratio in children with valvular aortic stenosis. Magn Reson Med 51(1):135–139
Derumeaux G, Ovize M, Loufoua J et al (2000) Assessment of nonuniformity of transmural myocardial velocities by color-coded tissue Doppler imaging: characterization of normal, ischemic, and stunned myocardium. Circulation 101(12):1390–1395
Dong SJ, Hees PS, Huang WM et al (1999) Independent effects of preload, after-load, and contractility on left ventricular torsion. Am J Physiol Heart Circ Physiol 277:H1053–H1060
Donoho DL (1995) De-noising by soft-thresholding. IEEE Trans Inf Theory 41(3):613–627
Donoho DL, Johnstone IM (1994) Ideal spatial adaptation by wavelet shrinkage. Biometrika 84(3):425–455
Fuchs E, Muller MF, Oswald H et al (2004) Cardiac rotation and relaxation in patients with chronic heart failure. Eur J Heart Fail 6(6):715–722
Goffinet C, Chenot F, Robert A et al (2009) Assessment of subendocardial vs. subepicardial left ventricular rotation and twist using two-dimensional speckle tracking echocardiography: comparison with tagged cardiac magnetic resonance. Eur Heart J 30(5):608–617
Guccione JM, McCulloch AD, Waldman LK (1991) Passive material properties of intact ventricular myocardium determined from a cylindrical model. J Biomech Eng 113(1):42–55
Helle-Valle T, Crosby J, Edvardsen T et al (2005) New noninvasive method for assessment of left ventricular rotation: speckle tracking echocardiography. Circulation 112:3149–3156
Hui L, Pemberton J, Hickey E et al (2007) The contribution of left ventricular muscle bands to left ventricular rotation: assessment by a 2-dimensional speckle tracking method. J Am Soc Echocardiogr 20(5):486–491
Jensen JA (1996) Field: a program for simulating ultrasound systems. Med Biol Eng Comput 34:351–353
Jensen JA, Svendsen NB (1992) Calculation of pressure fields from arbitrarily shaped, apodized, and excited ultrasound transducers. IEEE Trans Ultrason Ferroelectr Freq Control 39:262–267
Kelly DJ, Zhang Y, Connelly K et al (2007) Tranilast attenuates diastolic dysfunction and structural injury in experimental diabetic cardiomyopathy. Am J Physiol Heart Circ Physiol 293(5):H2860–H2869
Kramer CM, Reichek N, Ferrari VA et al (1994) Regional heterogeneity of function in hypertrophic cardiomyopathy. Circulation 90(1):186–194
Leitman M, Lysyansky P, Sidenko S et al (2004) Two-dimensional strain—a novel software for real-time quantitative echocardiographic assessment of myocardial function. J Am Soc Echocardiogr 17(10):1021–1029
Lumens J, Delhaas T, Arts T et al (2006) Impaired subendocardial contractile myofiber function in asymptomatic aged humans, as detected using MRI. Am J Physiol Heart Circ Physiol 291(4):H1573–H1579
Luo J, Fujikura K, Homma S et al (2007) Myocardial elastography at both high temporal and spatial resolution for the detection of murine infarcts. Ultrasound Med Biol 33(8):1206–1223
Maier SE, Fischer SE, McKinnon GC et al (1992) Evaluation of left ventricular segmental wall motion in hypertrophic cardiomyopathy with myocardial tagging. Circulation 86:1919–1928
Mallat S (1989) A theory for multiresolution signal decomposition: the wavelet representation. IEEE Pattern Anal Mach Intell 11(7):674–693
Migrino RQ, Aggarwal D, Konorev E et al (2008) Early detection of doxorubicin cardiomyopathy using two-dimensional strain echocardiography. Ultrasound Med Biol 34(2):208–214
Moore CC, Logo-Olivieri CH, McVeigh ER et al (2000) Three-dimensional systolic strain patterns in the normal human left ventricle. Radiology 214:453–466
Nagel E, Stuber M, Lakatos M et al (2000) Cardiac rotation and relaxation after anterolateral myocardial infarction. Coron Artery Dis 11:261–267
Notomi Y, Lysyansky P, Setser RM et al (2005) Measurement of ventricular torsion by two-dimensional ultrasound speckle tracking imaging. J Am Coll Cardiol 45:2034–2041
Popović ZB, Benejam C, Bian J et al (2007) Speckle-tracking echocardiography correctly identifies segmental left ventricular dysfunction induced by scarring in a rat model of myocardial infarction. Am J Physiol Heart Circ Physiol 292(6):2809–2816
Rappaport D, Adam D, Lysyansky P et al (2006) Assessment of myocardial regional strain and strain rate by tissue tracking in B-mode echocardiograms. Ultrasound Med Biol 32(8):1181–1192
Reisner SA, Lysyansky P, Agmon Y et al (2004) Global longitudinal strain: a novel index of left ventricular systolic function. J Am Soc Echocardiogr 17(6):630–633
Sandstede JJ, Johnson T, Harre K et al (2002) Cardiac systolic rotation and contraction before and after valve replacement for aortic stenosis: a myocardial tagging study using MR imaging. Am J Roentgenol 178(4):953–958
Streeter DD, Spotniz HM, Patel DJ et al (1969) Fiber orientation in the canine left ventricle during diastole and systole. Circ Res 24:339–347
Torrent-Guasp F, Kocica MJ, Corno A et al (2005) Towards new understanding of the heart structure and function. Eur J Cardiothorac Surg 27:191–201
Veyrat C, Pellerin D, Larrazet F et al (2003) Clinical relevancy of the myocardial velocity gradient: limitations of a binary response. J Am Soc Echocardiogr 16(12):1217–1225
Young AA, Kramer CM, Ferrari VA et al (1994) Three-dimensional left ventricular deformation in hypertrophic cardiomyopathy. Circulation 90(2):854–867
Acknowledgments
This work was funded by The Chief Scientist, the Ministry of Industry and Commerce Magneton project, the Technion VP for Research and the Alfred Mann Institute at the Technion (AMIT).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bachner-Hinenzon, N., Ertracht, O., Lysiansky, M. et al. Layer-specific assessment of left ventricular function by utilizing wavelet de-noising: a validation study. Med Biol Eng Comput 49, 3–13 (2011). https://doi.org/10.1007/s11517-010-0662-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-010-0662-6