Abstract
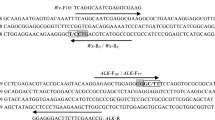
Improving grain quality is an important goal in breeding new elite rice varieties, requiring effective tools for the identification of target genotypes. Molecular marker-aided selection (MAS), combined with conventional breeding approaches, enables us to precisely identify the individual genotypes that are associated with different grain quality features, which can dramatically improve the breeding efficiency. However, to date, the number of molecular markers used in MAS for grain quality improvement is still somewhat limited. In this study, based on our previous study that rice grain quality is strongly associated with starch synthesis in the endosperm, we developed 51 gene-tagged molecular markers according to sequence variations in 18 starch synthesis-related genes from 16 typical rice cultivars. These markers can discriminate the different alleles among rice germplasms. These novel markers will provide effective tools in improving grain quality via the breeding new elite rice varieties.
Similar content being viewed by others
References
Mo H D. Quality improvement of rice grain in China (in Chinese). Sci Agri Sin, 1993, 26: 8–14
Cheng S H, Min S K. Rice varieties in China: Status and Prospects (in Chinese). China Rice, 2000, 1: 13–16
Li Z K. Strategies for molecular rice breeding in China (in Chinese). Mol Plant Breed, 2005, 3: 603–608
Feng J C. Application of Marker-assisted Selection in rice breeding (in Chinese). Chin Agric Sci Bull, 2006, 22: 43–47
Bernatzky R, Tanksley S D. Restriction fragments as molecular markers for germplasm evaluation and utilization. In: Brown A H D, Frandel O H, Marshall D R, et al, eds. The Use of Plant Genetic Resources. Cambridge: Cambridge University Press, 1989. 353–362
Juliano B O. Criteria and test for rice grain quality. In: Juliano B O, ed. Rice Chemistry and Technology. Minnesota: American Association of Cereal Chemists, 1985. 443–513
Myers A M, Morell M K, James M G, et al. Recent progress toward understanding biosynthesis of the amylopectin crystal. Plant Physiol, 2000, 122: 989–997
Smith A M. The biosynthesis of starch granules. Biomacromolecules, 2001, 2: 335–341
Nakamura Y. Towards a better understanding of the matabolic system for amylopectin biosynthesis in plants: Rice endosperm as a model tissue. Plant Cell Physiol, 2002, 43: 718–725
Martin C, Smith A M. Starch biosynthesis. Plant Cell, 1995, 7: 971–985
Ball S, Guan H P, James M, et al. From glycogen to amylopectin: A model for the biogenesis of the plant starch granule. Cell, 1996, 86: 349–352
James M G, Denyer K, Myers A M. Starch synthesis in the cereal endosperm. Curr Opin Plant Biol, 2003, 6: 215–222
Baba T, Nishihara M, Mizuno K, et al. Identification, cDNA cloning, and gene expression of soluble starch synthase in rice (Oryza sativa L.) immature seeds. Plant Physiol, 1993, 103: 565–573
Kawasaki T, Mizuno K, Baba T, et al. Molecular analysis of the gene encoding a rice starch branching enzyme. Mol Gen Genet, 1993, 237: 10–16
Kubo A, Fujita N, Harada K, et al. The starch-debranching enzymes isoamylase and pullulanase are both involved in amylopection biosynthesis in rice endosperm. Plant Physiol, 1999, 121: 399–409
Fujita N, Yoshida M, Asakura N, et al. Functional and characterization of starch synthase I using mutants in rice. Plant Physiol, 2006, 140: 1070–1084
Dian W, Jiang H, Wu P. Evolution and expression analysis of starch synthase III and IV in rice. J Exp Bot, 2005, 56: 623–632
Jiang H, Dian W, Liu F, et al. Molecular cloning and expression analysis of three genes encoding starch synthase II in rice. Planta, 2004, 218: 1062–1070
Nakamura Y, Yamanouchi H. Nucleotide sequence of a cDNA encoding starch-branching enzyme, or Q-enzyme I, from rice endosperm. Plant Physiol, 1992, 99: 1265–1266
Nakamura Y, Francisco P B Jr, Hosaka Y, et al. Essential amino acids of starch synthase IIa differentiate amylopectin structure and starch quality between japonica and indica rice varieties. Plant Mol Biol, 2005, 58: 213–227
Wang Z Y, Wu Z L, Xing Y Y, et al. Nucleotide sequence of rice Waxy gene. Nucleic Acids Res, 1990, 18: 5898
Wang Z Y, Zheng F Q, Shen G Z, et al. The amylose content in rice endosperm is related to the post-transcriptional regulation of the waxy gene. Plant J, 1995, 7: 613–622
Sano Y. Different regulation of Waxy gene expression in rice endosperm. Theor Appl Genet, 1984, 68: 4567–4573
Mikami I, Uwatoko N, Ikeda Y, et al. Allelic diversification at the wx locus in landraces of Asianrice. Theor Appl Genet, 2008, 116: 979–989
Gao Z Y, Zeng D L, Cui X, et al. Map-based cloning of the ALK gene, which controls the gelatinization temperature of rice. Sci China Ser C-Life Sci, 2003, 46: 661–668
Bao J S, Corke H, Sun M. Nucleotide diversity in starch synthase IIa and validation of single nucleotide polymorphisms in relation to starch gelatinization temperature and other physicochemical properties in rice (Oryza sativa L.). Theor Appl Genet, 2006, 113: 1171–1183
Han Y P, Xu M L, Liu X Y, et al. Genes coding for starch branching enzymes are major contribution to starch viscosity characteristics in waxy rice (Oryza sativa L.). Plant Sci, 2004, 166: 357–364
Tian Z X, Qian Q, Liu Q Q, et al. Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualityes. Proc Natl Acad Sci USA, 2009, 106: 21760–21765
Rogers S O, Bendich A J. Extraction of DNA from plant tissues. In: Gelvin S B, Schilperoort R A, eds. Plant Molecular Biology Manual. Boston: Kluwer Academis Publishers, 1988. A1–10
Wanchana S, Toojinda T, Tragoonrung S, et al. Duplicated coding sequence in the waxy allele of tropical glutinous rice (Oryza sativa L.). Plant Sci, 2003, 165: 1193–1199
Cai X L, Liu Q Q, Tang S Z, et al. Development of a molecular marker for screening the rice cultivars with intermediate amylose content in Oryza sativa subsp. indica (in Chinese). J Plant Physiol Mol Biol, 2002, 28: 137–144
He P, Li S G, Qian Q, et al. Genetic analysis of rice grain quality. Theor Appl Genet, 1999, 98: 502–508
Li J, Xiao J, Grandillo S, et al. QTL detection for rice grain quality traits using an interspecific backcross population derived from cultivated Asian (O. sativa L.) and African (O. glaberrima S.) rice. Genome, 2004, 47: 697–704
Lanceras J C, Huang Z L, Naivikul O, et al. Mapping of genes for cooking and eating qualities in Thai jasmine rice (KDML105). DNA Res, 2000, 7: 93–101
Umemoto T, Yano M, Satoh H, et al. Mapping of a gene responsible for the difference in amylopectin structure between japonica-type and indica-type rice varieties. Theor Appl Genet, 2002, 104: 1–8
Wan X Y, Wan J M, Su C C, et al. QTL detection for eating quality of cooked rice in a population of chromosome segment substitution lines. Theor Appl Genet, 2004, 110: 71–79
Septiningsih E M, Trijatmiko K R, Moeljopawiro S, et al. Identification of quantitative trait loci for grain quality in an advanced backcross population derived from the Oryza sativa variety IR64 and the wild relative O. irufipogon. Theor Appl Genet, 2003, 107: 1433–1441
Tan Y F, Li J X, Yu S B, et al. The three important traits for cooking and eating quality of rice grains are controlled by a single locus in an elite rice hybrid, Shanyou 63. Theor Appl Genet, 1999, 99: 642–648
Aluko G, Martinez C, Tohme J, et al. QTL mapping of grain quality traits from the interspecific cross Oryza sativa × O. glaberrima. Theor Appl Genet, 2004, 109: 630–639
Tanksley S D, Nelson J C. Advanced backcross QTL analysis: A method for the simultanouse discovery and transfer of valuable QTLs from unadapted germplasm into elite breedng lines. Theor Appl Genet, 1996, 92: 191–203
Author information
Authors and Affiliations
Corresponding authors
Additional information
These authors contributed equally to this work
About this article
Cite this article
Tian, Z., Yan, C., Qian, Q. et al. Development of gene-tagged molecular markers for starch synthesis-related genes in rice. Chin. Sci. Bull. 55, 3768–3777 (2010). https://doi.org/10.1007/s11434-010-4124-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11434-010-4124-1