Abstract
Purpose
Marine environments, especially sediments, are rich sources of actinomycetes that provide many bioactive compounds, primarily antibiotics. The goal of this study was to investigate the diversity of cultivable actinomycetes and their potential to produce antibiotics from sediments collected from the coastal zones of Turkey.
Materials and methods
Thirty sediment samples were collected from nine different coastal sites in three seas surrounding the Anatolian Peninsula of Turkey. Of the samples, 6 were collected from one site in the Black Sea, 18 from seven sites in the Aegean Sea, and 6 from one site in the Mediterranean Sea. Strains of pure actinomycetes were isolated by modified actinomycetes isolation agar (MAIA), M1 agar, M6 agar, and modified R2A agar. Ethyl acetate extracts and fermentation broths were used for the evaluation of antimicrobial activity against antibiotic resistant test microorganisms. The identification of the isolates was undertaken by 16S rRNA gene sequencing.
Results and discussion
A total of 261 strains of actinomycetes were isolated, of which 66 (25 %) were active against at least one antibiotic-resistant microorganism. Sixty-five of the actinomycetes isolates with antimicrobial activity were Streptomyces spp. and one was Nocardia sp., which implied that genus Streptomyces was predominant. Whereas MAIA agar was the best medium to recover actinomycetes, M6 agar was superior to others for the isolation of antibiotic-producing strains.
Conclusions
Extensive screening of the extracts from the 261 isolates for antimicrobial activities revealed considerable potential to produce antibiotics. These findings imply that actinomycetes from marine sediments of the Anatolian Peninsula coasts have potential for the discovery of novel bioactive compounds.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Actinomycetes (Actinomycetales; Actinomyces) is derived from the Greek word “actis” for beam and “mykes” for mucus, fungus. Actinomycetes are Gram-positive bacteria that are mostly aerobic and mycelial, and are characterized by high guanine–cytosine (GC) contents. Actinomycetes are morphologically rod-shaped and their colonies form fungus-like branched networks of hyphae (Holt 1994). These bacteria play vital roles in turnover of organic matter and the formation of humus by decomposing recalcitrant organic materials such as keratin, lignocelluloses, and chitin (Goodfellow and Williams 1983; McCarthy and Williams 1992). Actinomycetes also produce volatile substances such as geosmin that give soils their characteristic “wet earthy scent” (Wilkins 1996). Actinomycetes of the genus Streptomyces are known to produce commercial antimicrobial compounds. For instance, nearly 80 % of the antimicrobial compounds that have been characterized are derived from Streptomyces spp. (Jensen et al. 2005a; Bull and Stach 2007).
Due to the rapid emergence of fatal antibiotic-resistant pathogens, it is necessary to continue the search for novel antibiotics. Despite considerable efforts to chemically synthesis new antimicrobial compounds, nature still remains the richest and the most versatile source for novel antibiotics (Bredholdt et al. 2008). During the second half of the last century, the abundant soil bacteria, actinomycetes, have been extensively screened to discover new bioactive molecules (Anzai et al. 2008). To date, most of the bioactive compounds have been derived from terrestrial actinomycetes. However, in the recent years, the chance to discover novel bioactive compounds from terrestrial actinomycetes has reduced significantly (Alvan et al. 2011). Therefore, it is necessary to search for novel bioactive compounds from diverse sources such as marine environments.
Previously, a bacterial strain isolated from a marine environment was thought to be a function of terrestrial contamination because these environments were regarded as very salty and nutritionally inadequate to support bacterial diversity. However, recent research has revealed that some actinomycetes taxa are natural or well-adapted inhabitants of marine environments (Williams 2008). Such marine-derived actinomycetes have been shown to have great potential in the synthesis of diverse bioactive metabolites such as antibiotics and anticancer agents with unusual structures and properties (Jensen et al. 2005a). Sediments are the most common source of marine-derived actinomycetes; other sources include plankton, weeds, stones, algae, shellfish, mangroves, sea grass, coral reefs, fjords, and sponges (Mukku et al. 2000; Pukall et al. 2001; Kim et al. 2005; You et al. 2005; Anzai et al. 2008; Bredholdt et al. 2008; Eccleston et al. 2008).
The seas that surround the Anatolian Peninsula of Turkey have a high biodiversity. For instance, the coastal waters of the Aegean Sea are one of the global biodiversity hotspots (Meyers et al. 2000). However, despite the rich marine biodiversity in Turkey, only a few studies have been performed to screen for bioactive compound in actinomycetes (Hames-Kocabas and Uzel 2007). Therefore, the aim of this study was to screen for cultivable actinomycetes isolates in the coastal sediments of Turkey and to evaluate the antimicrobial activities of compounds present in these isolates.
2 Material and methods
2.1 Sediment samples
Nine biologically rich areas, determined based on biodiversity, along the coastal zone of Turkey were selected for sediment sampling (Fig. 1). Sediment samples were obtained by SCUBA diving and a total of 30 samples were collected during marine expeditions in 2009 and 2010 (Table 1). Six samples were collected from one site in the Black Sea, 18 samples from seven sites in the Aegean Sea, and 6 samples from one site in the Mediterranean Sea. Sediment samples were taken from different depths ranging from 0.5 to 35 m after removal of the 3–5 cm surface layer using a sterile spoon and placed into sterile 50-ml plastic falcon tubes by the diver with the aid of a sterile scoop and the container was immediately sealed. The diver was positioned downstream of the sample site and paid attention so as not to disturb the fine sediment.
2.2 Isolation of actinomycetes
Four different media were used for the isolation of actinomycetes from sediment samples: (1) modified actinomycetes isolation agar (MAIA) (Difco); (2) modified R2A agar (MR2AA) (Difco); (3) M1 agar: 10.0 g soluble starch, 4.0 g yeast extract, 2.0 g peptone, 20 g agar, and 1,000 ml natural aged seawater; and (4) M6 agar: 4.0 g beef extract, 4.0 g peptone, 1.0 g yeast extract, 10.0 g glucose, 20.0 g NaCl, 20.0 g agar, and 1,000 ml distilled water. MAIA was prepared according to the manufacturers’ instructions by using 50 % (v/v) natural filtered and aged seawater. All media were supplemented with nystatin (50 μg ml−1) and nalidixic acid (20 μg ml−1) in order to suppress fungi and fast-growing Gram-negative bacteria, respectively, in isolation plates.
Sediment samples were brought to the laboratory in cold boxes and processed in two different ways (Mincer et al. 2002; Jensen et al. 2005a). Ten grams of wet sediment sample were added to 90 ml sterile natural aged seawater and homogenized for 2 h at 100 rpm at room temperature. After serial dilutions, 200 μl aliquots were plated onto the isolation media in triplicate. The sediment samples were air-dried aseptically in a laminar flow hood and homogenized by mixing with a sterile spatula. After separation of large granules, a small round foam plug (2 cm diameter) was pressed onto the dried sample and stamped onto the surface of the agar by gently tapping the swab onto the surface of the agar. A continuous stamping process with the same swab gave the desired serial dilution effect. The plates were incubated at 28 °C for up to 4 weeks, and the isolates were purified in their original isolation media by repeated streaking. The seawater requirement of the purified isolates was determined by using the same isolation media as those free of seawater or NaCl.
2.3 Fermentation and antimicrobial activity screening
Fermentations of the purified strains were performed in 250-ml flasks containing 50-ml media. Both versions of the original isolation media were used for fermentation experiments. Selective antibiotic supplements were also omitted from media formulations. Fermentation media were inoculated with 2.5 % (v/v) activated strains and the flasks were incubated by shaking (150 rpm) at 28 °C for 7–10 days (depending on the average cell density). The fermentation broths were separated from the cells by centrifugation and filtration and divided into two portions. One part of the fermentation broths was extracted by half volume of ethyl acetate twice. The combined extracts were evaporated under vacuum (Heidolph, Germany) at temperatures of ≤40 °C. The extracts were dissolved in 300 μl EtOAc. Both cell-free fermentation broths and Ethyl acetate (EtOAc) extracts were used for assessing antimicrobial activity.
Disk diffusion assays were used to determine the antimicrobial activities of cell-free fermentation broth and solvent extracts (CLSI 2007). Enteropathogenic Escherichia coli 0157:H7 (RSKK 234), methicillin-resistant Staphylococcus aureus ATCC 43300, vancomycin-resistant Enterococcus faecium DSMZ 13590, Candida albicans DSMZ 5817 (antifungal agents testing strain), and Pseudomonas aeruginosa ATCC 27853 were used as test microorganisms. Active cultures for experiments were prepared by transferring a loopful of cells from the stock cultures to Mueller–Hinton agar (MHA) for bacteria and Sabouraud dextrose agar (SDA) for C. albicans that were incubated for 24 h at 37 and 25 °C, respectively. The cultures were diluted with sterile water to obtain optical densities corresponding to 0.5 McFarland, and they were spread on MHA and SDA in Petri dishes. Sterile paper disks (6 mm in diameter) impregnated with 30 μl of the fermentation broth in parts were placed on the inoculated plates and allowed to diffuse for 2 h at 4 °C. In the case of EtOAc extracts, disks are impregnated with 30 μl of the EtOAc extracts in parts then allowed to dry for 30 min. After, they were placed on the previously inoculated plates and allowed to diffuse for an additional 2 h at 4 °C. All plates were incubated at 37 °C for 24 h. The diameters of the inhibition zones were measured in millimeters.
2.4 DNA extraction and PCR amplifications
Total genomic DNA was extracted from the marine-derived actinomycetes isolates by FastDNA™ 2-ml SPIN Kit for Soil (MP). All polymerase chain reactions (PCR) were performed with the G-Storm® PCR system (Applied Biosystems). The PCR amplifications were carried out in 50 μl reaction volumes containing 10 mM PCR buffer, 1.5 mM MgCl2, 0.4 mM dNTP, 0.2 mM primer, 1.25 U of Taq DNA polymerase (Go Taq® Hot Start Polymerase), and 20–50 ng of genomic DNA template. The primers used for the amplification of the 16S rDNA were FC27 (5′-AGAGTTTGATCCTGGCTCAG-3′) and RC1492 (5′-TACGGCTACCTTGTTACGACTT-3′). The PCR conditions for 16S rDNA reactions were: one cycle of denaturation for 5 min at 94 °C; 30 amplification cycles consisting of denaturation (94 °C for 30 s), primer annealing (56 °C for 40 s), and primer extension (72 °C for 90 s); and a final extension of 5 min at 72 °C. The 16S rDNA amplicons were cleaned with a GeneJET™ PCR Purification Kit (Fermentas) according to manufacturers’ recommendations.
2.5 Phylogenetic analysis
Sequencing of the purified PCR products was performed bidirectionally by an ABI 3730xl automated sequencer (GATC-Biotech, Germany). The sequencing reads were used for phylogenetic analyses. The nucleotide sequences of reference species were downloaded from NCBI Gene Bank. Nucleotide–nucleotide Basic Local Alignment Search Tool (BLAST) analysis was performed for isolates using 16S rDNA sequence data. The evolutionary history was inferred using the neighbor-joining method (Saitou and Nei 1987). The evolutionary distances were computed using the maximum composite likelihood method (Tamura et al. 2004) and are in units of the number of base substitutions per site. All positions containing gaps and missing data were eliminated from the dataset (complete deletion option). There were a total of 215 positions in the final dataset. Phylogenetic analyses were conducted in MEGA4 (Tamura et al. 2007).
3 Results
A total of 261 actinomycetes strains were isolated from 30 sediment samples by the four different isolation media. All of the isolates showed similar growth patterns on seawater-free media, which indicated that the isolates are not strictly dependent on seawater.
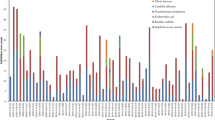
The distribution of the marine-derived isolates according to the locations (see Fig. 1) was as follows: Bodrum/Muğla, 23 (8.8 %); Saros/Çanakkale, 13 (4.9 %); Çanakkale, 26 (9.9 %); Kekova, 46 (17.6 %); Marmaris/Muğla, 48 (18.3 %); Didim/Aydın, 29 (11.1 %); Artvin, 28 (10.7 %); Yuvarlakçay/Muğla, 27 (10.3 %); and Azmak/Muğla, 21 (8.0 %). For each sediment sample, an average of 3.8–27 morphologically different colonies was obtained. Generally, estuarine sediments produced more diverse colonies than marine sediments. The average number of isolates per estuarine sediment was 12.5; the marine sediments had an average of 6.8 per sediment. Additionally, the distribution of the total isolates according to the isolation media revealed that the performance of the isolation media as follows: 46 isolates from M1 (17.6 %), 74 from M6 (28.4 %), 72 from MR2AA (27.6 %), and 69 from MAIA (26.4 %; Fig. 2a). The distribution of the active isolates is shown in Table 1. When subjected to the antimicrobial activity test, 25.3 % (66/261) of the actinomycetes strains were active against at least one test microorganism (Table 2). However, when considering the performance of the media in regard to the recovery of active isolates, different results were obtained: 7 from M1 (10.6 %), 32 from M6 (48.5 %), 16 from MR2AA (24.2 %), and 11 from MAIA (16.7 %; see Fig. 2b). In general, 68.2 % of the marine isolates were active against at least one test microorganism, whereas 31.8 % of the estuarine isolates were active.
Distribution of marine derived actinomycetes isolates: a number of total isolates according to the used culture media and b number of active isolates according to the isolation media. The four different media were: (1) modified actinomycetes isolation agar (MAIA), (2) modified R2A agar (MR2AA), (3) M1 agar, and (4) M6 agar (see text for details)
The isolates were sequenced for their 16S rDNA genes following PCR amplification and purification. Gene bank accession numbers were as follows: JX051234, JX051235, JX051236, JX051237, JX051238, JX051239, JX051240, JX051241, JX051242, JX051243, JX051244, JX051245, JX051246, JX051247, JX051248, JX051249, JX051250, JX051251, JX051252, JX051253, JX051254, JX051255, JX051256, JX051257, JX051258, JX051259, JX051260, JX051261, JX051262, JX051263, JX051264, JX051265, JX051266, JX051267, JX051268, JX051269, JX051271, JX051272, JX051273, JX051274, JX051275, JX051276, JX051277, JX051278, JX051279, JX051280, JX051281, JX051289, JX051290, JX051291, JX051292, JX051293, JX051295, JX051296, JX051297, JX051298, JX051301, JX051302, JX051303, JX051304, JX051305, JX051306, JX051308, JX051309, JX051310, and JX051311. 16S rDNA sequence data were used for BLAST analysis and construction of a phylogenetic tree (Fig. 3).
Evolutionary relationships of taxa. The optimal tree with the sum of branch length = 1.64455477 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree
4 Discussion
In recent years, marine sediments have become an attractive source of novel isolates of actinomycetes. Due to the wide diversity of bioactive natural products that have been isolated from these microorganisms, and their subsequent application in various biotechnological fields (Bérdy 2005), understanding the distribution of these microorganisms in marine environments has proven to be valuable, from both a scientific and an economical point of view (Fenical 2006).
Different pretreatment methods such as drying in SpeedVac (30 °C, 16 h), dry heat (120 °C, 60 min), phenol (1.5 %, 30 min at 30 °C), dry heat and phenol, and dry heat and benzethonium chloride (0.02 %, 30 min at 30 °C) prior to inoculation can be used to increase the relative numbers of actinomycetes (Bredholdt et al. 2008). For instance, Jensen et al. (1991) heated the sample dilutions in a water bath (50 °C, 60 min) to reduce the number of unicellular bacteria. Eccleston et al. (2008) used a dry heat treatment at 55 °C for 30 min to air dry sediment samples, followed by microwave irradiation (at 80 W for 30 s). Bredholdt et al. (2007) also used physical factors for the pretreatment of sediment samples, such as: UV irradiation (effective for the isolation of Nocardiopsis, Nocardia, and Pseudonocardia spp.), super-high frequency radiation (efficient for the isolation of Streptosporangium and Rhodococcus species), and extremely high frequency radiation (mostly favorable for Nocardiopsis, Nocardia, and Streptosporangium spp.). In the present study, we directly inoculated the wet sediment samples after serial dilution in sterile seawater and used stamping from air-dried sediments. Many different modified culture media can be used with or without specific antibiotic supplements in addition to the media originally used for terrestrial actinomycetes to isolate actinomycetes from sediments and other marine sources (Mincer et al. 2002; Jensen et al. 2005b; Hames-Kocabas and Uzel 2012). In this study, four different media were used for isolation. According to the recovery rate of actinomycetes strains from sediment samples, MR2AA and M6 were the best media (see Fig. 2a). However, when the isolation rates of the bioactive isolates were compared, M6 showed a superior performance (see Fig. 2b). Given that the aim of the study was to investigate phylogenetic diversity, MR2AA, M6, and MAIA might be useful since the number of cultured isolates was the highest among the other culture media. However, the percentage of active isolates from the M6 medium was found to be significantly higher among other isolation media. This phenomenon may be attributed to differences between the compositions of the two media in question. Higher amounts of complex nitrogen sources, such as beef extract and peptone, and the presence of glucose as carbon sources in M6—rather than low concentrations of yeast extract, proteose peptone, casein hydrolysate, glucose, and soluble starch that were found in MR2AA—may preferably promote the growth of antibiotic-producing strains.
In order to determine the antimicrobial activity of the marine-derived isolates, the fermentation broths and prepared EtOAc extracts were used. The reasons for employing this method were: (1) to prevent cytotoxic interactions observed in the cross-streak method between the tested actinomycete strain and the reference microorganism; (2) to hit the minor compounds produced by the actinomycete strains with high antimicrobial activity, but in very low concentrations; and (3) the high salt concentration in isolation media may negatively interfere with the growth of test bacteria. After obtaining EtOAc extracts via partitioning, the lower water phases were also screened for antimicrobial activity to avoid missing the water-soluble bioactive secondary metabolites (see Table 2). Inspection of the activity results revealed that the EtOAc extracts had higher activity compared with the water phases, probably due to a higher concentration of antimicrobial metabolites. The thin layer chromatography profiles and antimicrobial activity results of the isolates seems quite encouraging for large-scale fermentations and bioactivity guided isolation and characterization of the active compounds.
Of the five test microorganisms, P. aeruginosa was the most resistant. Only three Streptomyces strains isolated from Bordum and Artvin showed activity against P. aeruginosa. A total of 21, 25, 36, and 51 isolates were active against E. coli, E. faecium, S. aureus, and C. albicans, respectively. Both fermentation broth and EtOAc extracts of the Streptomyces 7CM26 isolated from Kekova/Antalya were active against all test bacteria with the higher inhibition zones. This strain appears to be a promising candidate to isolate more bioactive metabolites in future. Fifty-one isolates showed activity against C. albicans, implying that this test organism is the most sensitive to the isolated bioactive (antifungal) compounds. Interestingly, 21 of the 51 active isolates, including the single Nocardia strain, showed activity against C. albicans only.
The members of genus Streptomyces were found to be the dominating cultivable actinomycetes from the coastal sediments of Turkey. All the isolates found belong to Streptomyces except one Nocardia strain according to the 16SrDNA sequence results. This Nocardia strain was isolated from Marmaris/Mugla estuarine sediment. Moran et al. (1995), using 16S rRNA probes, reported that the first indigenous Streptomyces populations were a part of the marine bacterial community, and the first widespread distribution and persistent actinomycetes occurrence in the marine environment was reported by Mincer et al. (2002). The marine-derived actinomycetes strains isolated in the current study did not contain any obligate marine actinomycetes either from estuarine or marine sediments. This is probably due to the nature of the sediments and indigenous microflora. The samples were taken from areas relatively close to land, and were probably dominated by fast growing halotolerant strains. Consistent with previous results (e.g., Jensen et al. 1991), it is suggested that sampling from deeper zones and far from the coast is required to isolate obligate marine actinomycetes, where the numbers of streptomycetes decrease with depth and distance from the shore (Bredholdt et al. 2008). Taking samples far from the coastal areas is important to avoid isolating terrestrial actinomycetes as possible contaminants from runoff.
5 Conclusions
Based on evaluations of the numbers and antibiotic-producing potentials of the isolates in defined experimental conditions, the estuarine sediments produced actinomycetes isolates almost twice as often as marine samples. However, the marine isolates were found to be twofold more active than the estuarine isolates. These rates possibly reflect the fact that coastal marine sediments harbor more active strains than estuarine sediments. Taken together, the data presented in this study appear to suggest that the coastal sediments of Turkey may provide a good source of actinomycetes strains that could produce potent antimicrobial compounds.
References
Anzai K, Nakashima T, Kuwahara N, Suzuki R, Ohfuku Y, Takeshita S, Ando K (2008) Actinomycete bacteria isolated from the sediments at coastal and offshore area of Nagasaki Prefecture, Japan: diversity and biological activity. J Biosci Bioeng 106:215–217
Alvan G, Edlund C, Heddini A (2011) The global need for effective antibiotics—a summary of plenary presentations. Drug Resist Update 14:70–76
Bérdy J (2005) Bioactive microbial metabolites. J Antibiot 58:1–26
Bredholdt H, Galatenko OA, Engelhardt K, Tjaervik E, Terekhova LP, Zotchev SB (2007) Rare actinomycete bacteria from the shallow water sediments of the Trondheim fjord, Norway: isolation, diversity and biological activity. Environ Microbiol 9:2756–2764
Bredholdt H, Tjaervik E, Johnsen G, Zotchev SB (2008) Actinomycetes from sediments in the Trondheim fjord, Norway: diversity and biological activity. Mar Drugs 6:12–24
Bull AT, Stach JEM (2007) Marine actinobacteria: new opportunities for natural product search and discovery. Trends Microbiol 15:491–499
CLSI (Clinical and Laboratory Standards) (2007) Performance standards for antimicrobial susceptibility testing. 17th Informational Supplement, M100-S17, 27:1
Eccleston GP, Brooks PR, Kurtböke DI (2008) The occurrence of bioactive Micromonosporae in aquatic habitats of the Sunshine Coast in Australia. Mar Drugs 6:243–261
Fenical W (2006) Developing a new resource for drug discovery: marine actinomycete bacteria. Nature 2:666–673
Goodfellow M, Williams ST (1983) Ecology of actinomycetes. Annu Rev Microbiol 37:189–216
Hames-Kocabas EE, Uzel A (2007) Alkaline protease production by an actinomycete MA1-1 isolated from marine sediments. Ann Microbiol 57:71–75
Hames-Kocabas EE, Uzel A (2012) Isolation strategies of marine-derived actinomycetes from sponge and sediment samples. J Microbiol Meth 88:342–347
Holt JG (ed) (1994) Bergey’s manual of determinative bacteriology, 9th edn. Williams & Wilkins, New York. ISBN 0-683-00603-7
Jensen PR, Dwight R, Fenical W (1991) Distribution of actinomycetes in near-shore tropical marine sediments. Appl Environ Microbiol 57:1102–1108
Jensen PR, Gontang E, Mafnas C, Mincer TJ, Fenical W (2005a) Culturable marine actinomycete diversity from tropical Pacific Ocean sediments. Environ Microbiol 7:1039–1048
Jensen PR, Mincer TJ, Williams PG, Fenical W (2005b) Marine actinomycete diversity and natural product discovery. Antonie Leeuwenhoek 87:43–48
Kim TK, Garson MJ, Fuerst JA (2005) Marine actinomycetes related to the “Salinispora” group from the Great Barrier Reef sponge Pseudoceratina clavata. Environ Microbiol 7:509–519
McCarthy AJ, Williams ST (1992) Actinomycetes as agents of biodegradation in the environment—a review. Gene 115:189–192
Meyers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403:853–858
Mincer TJ, Jensen PR, Kauffman CA, Fenical W (2002) Widespread and persistent populations of a major new marine actinomycete taxon in ocean sediments. Appl Environ Microbiol 68:5005–5011
Moran MA, Rutherford LT, Hodson RE (1995) Evidence for indigenous streptomyces populations in a marine environment determined with a 16S rRNA probe. Appl Environ Microbiol 61:3695–3700
Mukku VJ, Speitling M, Laatsch H, Helmke E (2000) New butenolides from two marine Streptomycetes. J Nat Prod 63:1570–1572
Pukall R, Kramer I, Rohde M, Stackebrandt E (2001) Microbial diversity of cultivatable bacteria associated with the North Sea bryozoan Flustra foliacea. Syst Appl Microbiol 24:623–633
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbor-joining method. PNAS (USA) 101:11030–11035
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis [MEGA] software version 4.0. Mol Biol Evol 24:1596–1599
Wilkins K (1996) Volatile metabolites from actinomycetes. Chemosphere 32:1427–1434
Williams PG (2008) Panning for chemical gold: marine bacteria as a source of new therapeutics. Trends Biotechnol 27:45–52
You JL, Cao LX, Liu GF, Zhou SN, Tan HM, Lin YC (2005) Isolation and characterization of actinomycetes antagonistic to pathogenic Vibrio spp. from nearshore marine sediments. World J Microb Biot 21:679–682
Acknowledgments
This study was supported financially by the Scientific and Technological Research Council of Turkey (TÜBİTAK, SBAG-109S361) and Ege University Research Foundation (2009 Fen 061). We would also like to thank Associate Professor İkbal Agah Ince for his valuable help with the sequences and M. Baki Yokeş for providing the sediment samples.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Sabine Ulrike Gerbersdorf
Rights and permissions
About this article
Cite this article
Özcan, K., Aksoy, S.Ç., Kalkan, O. et al. Diversity and antibiotic-producing potential of cultivable marine-derived actinomycetes from coastal sediments of Turkey. J Soils Sediments 13, 1493–1501 (2013). https://doi.org/10.1007/s11368-013-0734-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11368-013-0734-y