Abstract
Dietary restriction (DR) increases lifespan in a range of evolutionarily distinct species. The polyphenol resveratrol may be a dietary mimetic of some effects of DR. The pivotal role of the mammalian histone deacetylase (HDAC) Sirt1, and its homologue in other organisms, in mediating the effects of both DR and resveratrol on lifespan/ageing suggests it may be the common conduit through which these dietary interventions influence ageing. We propose the novel hypothesis that effects of DR relevant to lifespan extension include maintenance of DNA methylation patterns through Sirt1-mediated epigenetic effects, and proffer the view that dietary components, including resveratrol, may mimic these actions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In this article we discuss key findings that led to our hypothesis that epigenetic actions of Sirt1, culminating in the maintenance of DNA methylation integrity, contribute to the beneficial effects of dietary restriction (DR) on ageing and lifespan. We consider first the general phenomenon of increased lifespan/delayed ageing in response to DR and highlight evidence that Sirt1, or its homologue in other species, plays a pivotal role in this response. Speculative and proven epigenetic-independent actions of Sirt1 with likely relevance to lifespan extension are noted; discussion of recently emerging literature reporting effects of Sirt1 on epigenetic status forms a major focus of the article, since effects at this level are pivotal to our hypothesis. We then summarise some of the evidence that supports the general premise that diet is an important component in defining the ageing trajectory, then consider how ageing and changes to the epigenetic status of the genome may be interrelated and how environmental influences, with a particular emphasis on diet, may modulate epigenetic marking. Finally, we consider the likelihood that dietary or pharmaceutical interventions that mimic the effect of DR on ageing/lifespan may act through Srt1-mediated maintenance of epigenomic integrity and explore ideas for future research priorities in this area.
Lifespan extension in response to dietary restriction and the role of sirtuins
DR remains one of the most robust dietary interventions that has proved effective in increasing lifespan across evolutionarily distinct species, including yeast, Caenorhabditis elegans, Drosophila and rodents (Guarente and Picard 2005). The longevity effects of DR were observed first in rodents many decades ago (McCay et al. 1935), and these early observations have been substantiated by many later studies in which restriction of energy intake by 25–50% compared with ad libitum levels increased lifespan (e.g. Ross 1961; Weindruch and Walford 1982; Yu et al. 1982; Weindruch et al. 1986). For example, the longest-living 10% of mice fed a diet providing only 35% of the ad libitum intake but enriched with vitamins and minerals (to avoid deficiency) lived a remarkable average of 53 months, compared with 35 months for the longest-lived 10% of the control ad libitum-fed group (Weindruch et al. 1986). In rodents, an element of the longevity response to DR is a reduction in chronic diseases associated with ageing, including diabetes, atherosclerosis, cardiomyopathy, kidney disease, respiratory disease and cancer (Fontana and Klein 2007). However, DR appears also to extend lifespan through other mechanisms, acting on what might be considered as a “healthy ageing trajectory”; in a study in rats no evidence of organ pathology was detected at death in approximately one-third of animals (Shimokawa et al. 1993), and in young, apparently disease-free animals DR induced effects indicative of a biologically “younger” state (Fontana and Klein 2007). These effects included reduced production of reactive oxygen species, decreased plasma concentrations of inflammatory cytokines, increased expression of protein chaperones, including HSP70, and reduced cellular “debris” associated with ageing, including damaged proteins, oxidised lipids and advanced glycation end products (Fontana and Klein 2007). Other general metabolic and physiological effects of DR in mammals that may be linked to longer and healthier life include lower plasma concentrations of glucose, insulin, triglycerides and cholesterol along with increased insulin sensitivity and glucose tolerance (reviewed in Guarente and Picard 2005).
Debate continues about whether or not reduced adiposity contributes to the longevity response to DR. This premise is thrown into question by reports that in mice, without DR, exercise (on running wheels) to reduce body weight to the same level as in DR did not increase lifespan (Holloszy et al. 1985; Holloszy 1997) and that genetically obese ob/ob mice on DR lived longer than control lean mice despite maintaining body fat in excess of that of control animals (Harrison et al. 1984).
In the absence of validated surrogate biomarkers of ageing, studies in mammals of the longevity response to DR must rely, ultimately, on the measurement of lifespan as the primary measure of an effect on ageing. Thus, investigating the effects of DR in long-lived mammals, including primates and humans, offers particular challenges relating to the length of time over which experiments must be conducted. Data from such studies at this point are insufficient to support any conclusions concerning effects on lifespan as an end point; however, data from ongoing studies in rhesus monkeys demonstrate changes in metabolic and physiological parameters similar to many of those observed in response to DR in rodents, including reduced body weight and adiposity (Colman et al. 1999), reduced core body temperature (Lane et al. 1996) and resting energy expenditure (Blanc et al. 2003), reduced blood pressure (Lane et al. 1999), reduced plasma glucose and insulin concentrations (Lane et al. 1999; Gresl et al. 2001), increased insulin sensitivity (Lane et al. 1999; Gresl et al. 2001), decreased plasma levels of inflammatory mediators (Kim et al. 1997) and reduced levels of glycation end products in skeletal muscle (Zainal et al. 2000). Specific beneficial effects of DR may be restricted to particular windows of exposure, as indicated by contrasting effects of DR in rhesus monkeys on measures of T cell function; DR initiated during adolescence (3–5 years) delayed T cell senescence (Messaoudi et al. 2006) but when initiated either in juvenile (1–2 years) or old (> 15 years) males resulted in changes in the T cell population consistent with accelerated T cell senescence (Messaoudi et al. 2008). Various lines of epidemiological data demonstrate an association, but not causality, between DR in humans and longevity. Such evidence includes observations based on the inhabitants of Okinawa Island in Japan. A recent analysis indicated that, since 1949, the energy intake of individuals currently in their eighth decade of life was approximately 11% lower than recommended on the basis of energy balance calculation, provided through a diet rich in micronutrients and antioxidants, for the first half of adult life (Willcox et al. 2006). Survival curves based on data for 1995 show increases in both average and maximum lifespan compared with Japanese and United States populations, and data also reveal reduced mortality from age-related diseases (Willcox et al. 2006). These findings corroborate earlier observations concerning reduced energy intake coupled with health and longevity in this population (Kagawa 1978). Recent data based on a group of individuals (18–28 subjects) who have practiced DR (energy intake of a micronutrient-replete, protein-sufficient diet approximately 70% of age- and sex-matched individuals consuming a typical Western diet) with the aim of extending lifespan, reveal positive effects compared with age- and sex-matched control subjects in parameters including blood pressure, inflammatory markers, lipid profile, insulin sensitivity and diastolic function along with reduced body fat (Fontana et al. 2004; Meyer et al. 2006; Fontana and Klein 2007).
Replicative lifespan in the budding yeast Saccharomyces cerevisiae was extended by reducing glucose concentration in the medium from 2% to 0.5% (Lin et al. 2002) (now considered to represent conditions of moderate DR; Bishop and Guarente 2007), thereby establishing a model of DR in yeast on which numerous further studies have been conducted, resulting elucidation of some of the metabolic and genetic mediators of the response (reviewed in Bishop and Guarente 2007).
The NAD-dependent (class III) histone deactylase Sirt1 in mammals, and its ortholog in other species (sir2 in yeast), may be pivotal in mediating the effect of DR to increase lifespan. This premise is supported by observations including the abolition of lifespan extension by DR in mutants of yeast (Lin et al. 2002), C. elegans (Tissenbaum and Guarente 2001) and Drosophila (Rogina and Helfand 2004) that do not express sir2. In rats and mice, DR increased Sirt1 expression in several tissues, including liver (Cohen et al. 2004; Nisoli et al. 2005), and in human subjects DR increased Sirt1 mRNA in skeletal muscle (Civitarese et al. 2007). Other data show no apparent effect of DR on Sirt1 expression in mice (Barger et al. 2008) or tissue-specific responses, with increases in mouse muscle and white adipose tissue but a reduction in Sirt1 expression in response to DR in liver (Chen et al. 2008). Possible explanations for these discordant observations may include species-specific effects, differences in the age of animals and/or duration of DR or differences in sampling time-point, particularly in relation to time after daily feeding. A recent study demonstrated that several effects of DR in mice—including improved glucose tolerance, reduced fasting levels of insulin, glucose and cholesterol and reduced adiposity—were mimicked in a transgenic model in which Sirt1 was overexpressed in several tissues, including brown and white adipose tissue and brain (Bordone et al. 2007). It remains to be established if this genetic manipulation will increase lifespan.
Although classed as a histone deacetylase, Sirt1 deacetylates a broad range of substrates. Potential downstream targets for modification by Sirt1 with a potential role in modifying the ageing process are numerous, and include several key transcription factors involved in the regulation of multiple pathways linked to physiological and metabolic processes that are influenced by, or contribute to, ageing or that improve stress resistance. It is beyond the scope of this article to discuss, or even archive, all known cellular targets for Sirt1 action and their potential consequences for ageing/lifespan; for a more comprehensive overview the reader is referred to recent reviews (Chen and Guarente 2007; Guarente 2007). Here, we select some specific examples of Sirt1 downstream targets to illustrate the variety of mechanisms through which effects on lifespan may be mediated. Of particular note in this context are p53 and peroxisome proliferator-activated receptor gamma co-activator 1 alpha (PGC-1α). Deacetylation of p53 by Sirt1 may, arguably, have effects on lifespan by reducing cellular senescence, in which active (acetylated) p53 plays a key role (Oren 2003) and which may be causal in the ageing process (de Magalhaes and Faragher 2008). In agreement with this notion, overexpressed Sirt1 antagonised the acetylation of p53 induced by promyelocytic leukemia protein (PML) in primary mouse embryo fibroblasts and prevented PML-mediated premature cellular senescence (Langley et al. 2002) and in human breast cancer MCF-7 cells and lung cancer H1299 cells the Sirt1 inhibitor Sirtinol induced a senescence-like growth arrest (Ota et al. 2006). Observations consistent with a role for cellular senescence in ageing includes exponential accumulation of senescent cells in the skin of ageing baboons (Herbig et al. 2006); however, it should be noted that current data are insufficient to demonstrate a causal, rather than merely associative, relationship. PGC-1α is the major regulator of mitochondrial biogenesis (Wu et al. 1999) and activation of PGC-1α by Sirt1-mediated deacetylation may contribute to healthy ageing through the maintenance of mitochondrial energy production and the prevention of endogenous mitochondria-derived oxidative stress (Guarente 2007; Lopez-Lluch et al. 2008). PGC-1α is also a key regulator of gluconeogenesis (Schilling et al. 2006) and fatty acid oxidation (Vega et al. 2000), providing a link between Sirt1 activation and the stimulation of gluconeogenesis and fatty acid oxidation observed under DR. Effects of Sirt1 on lifespan may also be unrelated to deacetylation activity, such as regulation of insulin secretion through binding to and repressing the promoter of the mitochondrial uncoupling protein 2 gene (UCP2) and thus allowing an increase in cellular ATP levels after glucose stimulation so that insulin is released (Bordone et al. 2006). In general, the mechanisms through which effects of Sirt1 activation on its numerous downstream targets then translate into effects on ageing and/or lifespan remain unclear. In their recent review, Chen and Guarente note the apparent idiosyncratic causes of ageing across the diverse range of species in which Sirt1 (or its homologue) appears to play a pivotal role, and proffer the view that the dependency of Sirt1 on NAD+ for activity may form the basis of a pan-organismal link between Sirt1 and cellular energy status and, thus, lifespan (Chen and Guarente 2007).
Evidence for epigenetic effects of Sirt1
The abovementioned broad range of substrates for Sirt1-mediated deacetylation raises the question of to what extent histones are a major substrate target of Sirt1 and thus brings into question the likelihood that Sirt1 may influence epigenetic status.
Actions of the yeast Sirt1 homologue, sir2, point towards activity concerned with modifying epigenetic markings. Sir2 associated with the homologous mating type loci HML and HMR represses gene expression at these sites (Klar et al. 1979; Rine et al. 1979). Loss of this gene silencing effect results in sterility, which is a hallmark of ageing in yeast (Smeal et al. 1996). Sir2 also suppresses recombination at the ribosomal DNA locus, a process that, as cells age, leads to the accumulation of toxic extra-chromosomal ribosomal DNA circles (Sinclair and Guarente 1997).
In vitro, Sirt1 has the capacity to deacetylate histone substrates in an NAD+ -dependent manner, specifically H4-K16 and H3-K9 (Vaquero et al. 2004), and the same action in cells was indicated by hyperacetylation of histones at these sites under conditions of Sirt1 knockdown by siRNA (Vaquero et al. 2004). This same study demonstrated a role for Sirt1 in the establishment of repressive heterochromatin by targeting Sirt1 to an exogenous integrated luciferase reporter gene under the control of Gal4 binding sites by virtue of heterologous expression of a Gal4–Sirt1 fusion protein in 293F cells. Deacetylation of H4-K16 and H3-K9 in the region of the integrated promoter was detected by chromatin immunoprecipitation (ChIP) and was concurrent with reduced luciferase expression, loss of dimethylation at H3-K79, which is a mark of (active) euchromatin, increased trimethylation at H3-K9, characteristic of heterochromatin, and also recruitment of and deacetylation of histone H1 (Vaquero et al. 2004). Epigenetic regulation of endogenous gene targets by Sirt1was demonstrated in breast and colon cancer cell lines, in which Sirt1 was found to be associated with the promoters of tumour suppressor genes only in cell lines in which these genes were hypermethylated and aberrantly silenced, and not in cell lines lacking promoter hypermethylation and showing basal levels of gene expression. RNAi knockdown, pharmacological inhibition or expression of a dominant negative mutant of Sirt1 resulted in the reactivation of these aberrantly silenced, hypermethylated tumour suppressor genes (Pruitt et al. 2006). Perhaps surprisingly, this gene reactivation was not accompanied by any change in promoter methylation status for any of the genes studied, but was associated with increased acetylation of H4-K16 and H3-K9 in the promoter regions, consistent with Sirt1-mediated deacetylation at these loci contributing to gene repression (Pruitt et al. 2006). The basis of an effect of Sirt1 on H3-K9 trimethylation was shown, in a separate study, to be via a functional interaction between Sirt1 and the lysine methyltransferase SUV39H1, which was found to undergo activation coincident with NAD+-dependent, Sirt1-catalysed deacetylation (Vaquero et al. 2007).
The first, very recent, published evidence for an effect of Sirt1 on DNA methylation is based on effects at an exogenous, integrated E cadherin promoter, which contains a CpG island at which aberrant methylation in cancer is known to repress gene expression (O'Hagan et al. 2008). This elegant experimental design involved the inclusion of a rare restriction endonuclease cleavage site within the CpG island of the exogenous E cadherin promoter, which was upstream of a herpes simplex thymidine kinase (HSTK) reporter gene integrated into the MB-MDA-231 breast cancer cell line. The cognate restriction endonuclease was expressed under tetracycline induction from an exogenous transgene, so introducing a double-strand break. Analysis by chromatin immunoprecipitation (ChIP) demonstrated that induction of the double-strand break led to the accumulation at the break site of: (1) Sirt1, coincident with deacetylation of the Sirt1 substrate H4-K16; (2) the lysine methyltransferase EZH2, accompanied by the products of its activation—di- and tri-methylation at H3-K9 and trimethylation at H3-K27, all marks of repressive heterochromatin; (3) the DNA methyltransferases DNMT1 and DNMT3b. Whereas in the majority of cells HSTK reporter gene activity was preserved after double-strand break repair, conferring sensitivity to ganciclovir, approximately 1% of cells retained ganciclovir resistance and showed accrual of DNA methylation in the region around the break site, which spread over the CpG island with subsequent passages. Importantly, these epigenetic effects resulting from induction of the double-strand break were abrogated by Sirt1 knockdown. In relation to effects specifically on DNA methylation, Sirt1 knockdown did not have an effect on the frequency of ganciclovir resistance following induction of the double-strand break but, compared with control cells (no Sirt1 knockdown), a significantly smaller proportion of these resistant cells showed increased exogenous E cadherin promoter CpG island methylation. The authors, reasonably, interpret these observations as an indication that reduced Sirt1 recruited to the break site did not affect silencing but that Sirt1 was important in seeding DNA methylation. In agreement with such an interpretation, Sirt1 did not persist at the promoter in ganciclovir-resistant cells (with no Sirt1 knockdown) after 5 passages but DNMT1 and DNMT3b remained enriched, with DNMT1 (but not DNMT3b) persisting after 34–36 passages.
It remains important to establish if Sirt1 can affect methylation of endogenous targets, both within gene promoter regions and also at non-promoter elements. The latter include subtelomeric CpG islands, whose methylation may maintain telomere integrity (Gonzalo et al. 2006), along with repetitive sequences such as endogenous retroviral A-type particles (IAPs), centromeric repeats and short interspersed nuclear elements such as Alu repeats, which provide a readout of global DNA methylation (Gaudet et al. 2003; Rodriguez et al. 2008) so may be relevant to effects on genome stability (Eden et al. 2003; Gaudet et al. 2003) and, thus, to ageing.
Recent compelling evidence that Sirt1-influenced chromatin remodelling modulates chromatin stability and gene expression, and plays a role in ageing, centres on observations concerning Sirt1 redistribution in the genome under oxidative stress, coupled with expression changes in Sirt1-associated genes with ageing, and effects of Sirt1 expression on genome stability (Oberdoerffer et al. 2008). Sirt1 binding to major satellite repeats in ES cells was reduced by both oxidative stress and the Sirt1 inhibitor nicotinamide, coincident with increased histone H1-K26 acetylation at these sites (used as a readout of Sirt1 deaceteylase activity) and concomitant increased transcript expression. Moreover, the distribution of promoters associated with Sirt1, which corresponded with H1-K26 acetylation, was shifted under conditions of oxidative stress (as determined by microarray profiling) to result in a pattern of transcriptional change parallel to that observed in the ageing mouse brain, with release of Sirt1 corresponding with transcript de-repression. Importantly, these effects on transcript de-repression, both in ES cells and in the ageing mouse brain, were reversed by Sirt1 over-expression, an observation that has possible direct relevance to effects of dietary restriction via Sirt1 to counteract ageing; in particular these observations may be indicative of a role for “anti-ageing” Sirt1-mediated epigenetic effects specifically under conditions such as oxidative stress, which is generally considered as an ageing assault. Indicating a role for Sirt1 in the maintenance of chromatin stability, transgenic mice over-expressing Sirt1 in lymphoid tissues against a p53+/− background, which confers susceptibility to irradiation-induced tumorigenesis due to loss of heterozygosity at the p53 locus, showed reduced tumour-related deaths following irradiation. It will be particularly informative in the context of our current hypothesis that epigenetic effects of Sirt1, including the maintenance of DNA methylation patterns, contribute to the longevity response to DR to establish if such changes in chromatin association of Sirt1 are accompanied by changes to the DNA methylation status. Speculatively, observations that loss of Sirt1 binding was not associated with gene de-repression at all loci (Oberdoerffer et al. 2008) may be indicative of the requirement for other accompanying epigenetic changes, such as effects on DNA methylation.
Effects of diet on ageing
Evidence from observational studies supports the premise that the quality of the diet affects both length of life and risk of a wide range of age-related diseases. For example, adherence to a modified Mediterranean diet was associated with reduced mortality in healthy individuals across nine European countries aged 60 or older at recruitment into the prospective EPIC cohort (Trichopoulou et al. 2005). In other studies, associations between diet and age-related cognitive decline included: a protective effect of a high intake of mono-unsaturated fatty acids in an elderly population in Southern Italy; increased risk of Alzheimer disease associated with high dietary fat and energy intake and reduced risk associated with the consumption of white fish and cereals in Europe and North America; and a protective effect of moderate red wine consumption against Alzheimer disease and vascular dementia in a French cohort (Panza et al. 2004). Of course, a major limitation associated with such observational studies is the difficulty in correcting for all confounding variables, so it remains essential to establish unequivocally the effects of a broad range of specific nutrients, other dietary components and dietary patterns on ageing through rigorous intervention studies. As an early step in this direction, a meta-analysis of total mortality in randomised trials testing vitamin D supplementation concluded that supplementation at ordinary doses was associated with reduced total mortality rate; however, the authors note the importance of confirming the findings through population-based, placebo-controlled randomised trials with total mortality as the main end point (Autier and Gandini 2007).
Evidence that diet in general can affect the ageing trajectory raises the possibility that dietary mimetics of the response to DR, or pharmaceuticals designed rationally on the basis of naturally-occurring dietary compounds with desirable actions, may offer an alternative route to reap the benefits of DR.
Epigenetic changes in ageing
Ageing is accompanied by changes to the epigenetic status of the genome that are known to influence factors of likely relevance to ageing such as gene expression, genome stability and telomere length. Descriptions in the literature of ageing-related changes to the DNA methylation profile are relatively abundant and concordant, but reports of changes in histone markers with ageing are, in comparison, less numerous and reports of more global effects on chromatin organisation appear inconsistent.
Based on the observation that the total methyl-cytosine content of DNA of mammalian cells decreased with time in culture, Wilson and Jones (1983) proffered the view that this alteration in DNA methylation might explain reported changes in gene expression with age. Much of this loss of genomic methyl groups occurs in repeated sequences within DNA (Fraga et al. 2007)—especially within Alu sequences (Rodriguez et al. 2008), which are short (∼300 bp) interspersed nuclear elements (SINEs) making up about 10% of the mass of the human genome. Later studies have gone on to show that this age-related demethylation appears to be a passive process associated with attenuated activity of the maintenance DNA methylase DNMT1, which results from reduced expression (Lopatina et al. 2002; Casillas et al. 2003). Loss of DNA methylation may contribute to genomic instability (Eden et al. 2003; Gaudet et al. 2003) and loss of telomere integrity (Gonzalo et al. 2006), and so contribute to an ageing cellular phenotype through these processes. In contrast to a reduction in global DNA methylation, ageing is associated with site-specific DNA hypermethylation, notably within CpG islands within the promoters of house-keeping genes such as the estrogen receptor (ESR1) gene, which investigators have found to be hypermethylated with ageing in normal human colon (Issa et al. 1994) and prostate tissue (Kwabi-Addo et al. 2007). Other genes hypermethylated in CpG islands in samples of normal prostate tissue from older individuals were RARβ2, RASSF1A, GSTP1 and NKX2-5 (Kwabi-Addo et al. 2007). Other reports of promoter CpG hypermethylation point towards a role for gene-specific hypermethylation in ageing-related diseases. For example, hypermethylation of ESR1, MYOD and P16 exon 1 was observed in macroscopically normal epithelium from ulcerative colitis patients (Issa et al. 2001), and coronary atherosclerotic tissues had higher ESR2 methylation than apparently normal femoral artery tissue (Kim et al. 2007). The mechanism underlying the accrual of promoter CpG island methylation with ageing is poorly understood but observations that the expression and activity of the do novo DNA methylase DNMT3b increased with ageing in fibroblasts (Lopatina et al. 2002; Casillas et al. 2003) provide a plausible possible mechanism.
The increased mosaicism between cells in an ageing tissue resulting from stochastic methylation changes to the genome in individual cells may, speculatively, be a major factor in age-related decline in tissue function (reviewed in Mathers and Ford 2009).
The phenomenon of cell senescence, which may, debatably, be causal in the ageing process (de Magalhaes and Faragher 2008), is associated with changes to chromatin structure; indeed, the accumulation of so-called senescence-associated heterochromatic foci is becoming an accepted biomarker of cell senescence (Herbig et al. 2006; Narita et al. 2006). In contrast, however, and illustrating the abovementioned discordance in the literature concerning changes to gross chromatin structure with ageing, other investigators have reported an ageing-related progressive reduction in heterochromatin-like domains (e.g. Howard 1996; Villeponteau 1997). With respect to specific changes in histone marks with ageing, increased H4-K20 trimethylation was reported in rat kidney and liver in older animals (Sarg et al. 2002). As a second example, a progressive dephosphorylation of histone H1 somatic subtypes 4 and 5 (H1.4 and H1.5) was measured in peripheral blood lymphocytes when individuals of age 23–30 years, 38–50 years and 60–65 years were compared (Happel et al. 2008). The phenotypic consequences of these changes in histone markers, and the extent to which they may be causal in the ageing process, rather than the result of cellular ageing, are currently unclear.
Dietary and other environmental influences on epigenetic marking
Evidence that the epigenetic status of the genome can be modified through environmental exposures includes the observation that differences between monozygotic twins in DNA methylation and H3/H4 acetylation in lymphocytes, all of which were more marked in older compared with younger twin pairs, were greater in pairs who had different lifestyles and spent less of their lives together (Fraga et al. 2005). Diet is a particularly compelling candidate environmental exposure with large potential to influence epigenetic marking, and numerous studies provide proof of principle that diet can have epigenetic consequences. The observation that the susceptibility of rat liver chromatin to digestion by micrococcal nuclease was altered as a function of diet was an early indication of the potential of diet to modify gross chromatin structure (Castro and Sevall 1980). Intermediates in 1-carbon metabolism, specifically folate, choline, betaine and methionine, have been a focus for much of the research on effects of individual dietary components on DNA methylation. These dietary agents are likely to affect DNA methylation through effects on the supply of methyl groups [ultimately as S-adenosylmethionine (SAM), the methyl donor to cytosine in the reaction catalysed by the DNMTs]. Also, the literature on the potential role of bioactive dietary phytochemicals, principally polyphenols and isothiocyanates, in moderating DNA methylation is expanding rapidly.
Studies on the capacity of specific dietary agents to modify DNA methylation have used in vitro cell culture models, dietary intervention in rodents and also include observational studies in humans. The agouti variable yellow mouse model, which shows a shift in coat colour from yellow to pseudo-agouti in response to methylation of DNA at a specific locus (a retrotransposon insertion into the agouti gene; Michaud et al. 1994), provides a particularly convenient model for investigating the capacity of specific dietary agents to affect DNA methylation in vivo. Also, it is worth noting that much of the research on the epigenetic effects of dietary components, in particular methyl donors and bioactive polyphenols, has focussed on exposure in utero, often making use of the agouti variable yellow mouse model. The focus on exposure in utero has been driven largely by the, now compelling, evidence that nutritional stresses during gestation result in lifelong consequences for health (Fernandez-Twinn and Ozanne 2006), coupled with the apparent feasibility of epigenetic modification as a mechanism through which these in utero challenges might be recorded in a manner that will allow functional consequences to persist throughout the lifecourse. It remains to be established directly if epigenetic marking in utero, or indeed epigenetic effects of diet later in the lifecourse, has an effect on ageing per se. Table 1 lists selected examples of specific studies demonstrating effects of methyl donors and bioactive polyphenols on DNA methylation, along with other studies selected from the smaller body of literature providing evidence for effects on DNA methylation also of zinc, selenium, arsenic, vitamin A and alcohol. The authors have recently provided a more comprehensive review of such studies elsewhere (Mathers and Ford 2009).
Comparison of the main findings of different studies on the effects on DNA methylation of specific dietary agents reveals examples of single agents having opposing effects on DNA methylation (reviewed in Mathers and Ford 2009, and see also Table 1). There are many possible explanations for such apparent discordant findings including cell line/tissue-specific responses, interrogation of effects on DNA methylation at different sites in different studies, which differ in their response to the agent tested, influence of dose, exposure time and/or interactions with dietary and other factors and different effects on DNA methylation of parent compounds (generally tested in cell culture models) compared with the major in vivo metabolites.
Diet appears to influence epigenetic marking at the level of histone modification, as well as DNA methylation. The evidence for effects at this level is most robust for the short chain fatty acid butyrate, generated in the large bowel as a result of bacterial fermentation of dietary fibre, and organosulphur compounds, including the isothiocyanates. Proven ability to inhibit HDAC activity (Aviram et al. 1994; Dashwood et al. 2006; Myzak et al. 2006) provides a mechanistic basis for the histone-modifying activity of these compounds. Examples of other dietary agents or nutritional modifications that appear to or have the potential to influence histone modification include methyl donors, retinoic acid (the active metabolite of vitamin A), ethanol, and protein restriction in utero (reviewed in Mathers and Ford 2009).
The potential for dietary interventions that mimic DR to act through Sirt1-mediated effects on epigenetic marking
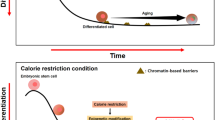
Knowledge that Sirt plays a pivotal role in the longevity response to DR and that its substrates for deacetylation include specific histone residues, in the context of observations that epigenetic marking alters with ageing and may be causal in the ageing process, led us to the hypothesis that, under conditions of DR, Sirt1 promotes longevity in part through epigenetic effects, including the maintenance of DNA methylation integrity, and that these can be mimicked by specific dietary components (Fig. 1).
A hypothesis concerning potential effects of dietary restriction on epigenetic marking. Dietary restriction (DR; reduced energy intake) or DR-mimetic compounds, including dietary components, increase expression and/or activity of the histone deacetylase Sirt1, leading to increased histone deacetylation and, thereby, to increased DNA methylation
The effects of DR on epigenetic marking appear little studied. However, the limited published evidence supports the premise that DR affects epigenetic marking, in particular DNA methylation. Transient global DNA hypomethylation in liver and suppression of age-dependent changes in methylation of the c-myc oncogene were observed in mice in response to DR (Miyamura et al. 1993). In another study, DR in rats led to hypermethylation of the c-Ha-ras oncogene in pancreatic acinar cells (Hass et al. 1993).
The plant polyphenol resveratrol is emerging as a potential mimetic of DR. Dietary resveratrol protected mice against diet-induced obesity and insulin resistance and induced other metabolic and physiological effects associated with longer lifespan (Baur et al. 2006; Lagouge et al. 2006). Resveratrol treatment increased PGC-1α deactylation in multiple tissues, consistent with Sirt1 activation, and resveratrol failed to induce deacetylation of PGC-1α in Sirt1−/− mouse embryonic fibroblasts, further supporting the notion that Sirt1 is a key mediator of the effects of DR on lifespan/ageing. Evidence concerning Sirt1 activation by resveratrol remains equivocal, however. In particular, the ability of resveratrol to activate Sirt1 was reported to be substrate-dependent (Borra et al. 2005). Compelling data for an effect of resveratrol on gene expression to mimic changes seen in DR and to reverse changes seen in ageing were obtained by Prolla and co-workers through comparison of the transcriptomic profiles of mice fed resveratrol with those of CR mice; genes expressed differentially in response to either intervention in a pattern that opposed ageing-related changes showed a remarkable level of overlap between the two treatments (Barger et al. 2008). Moreover, genes involved in chromatin remodelling were identified as among those showing such changes in expression, highlighting the likelihood that effects of DR are likely to include alterations in chromatin structure and revealing that effects may be through mechanisms less direct than Sirt1-mediated effects on epigenetic marking. Indeed, it should be noted that Prolla and co-workers did not observe any increase in Sirt1 expression in either their DR mice or in the animals fed resveratrol (Barger et al. 2008). As already noted, experiments in which Sirt1 was overexpressed in mice against a p53+/− background indicted a role for Sirt1 in the maintenance of chromosome stability (as indicated by the reduced incidence of death from irradiation-induced tumours; Oberdoerffer et al. 2008). Of particular relevance to the current argument is that administration of resveratrol to the p53+/− background strain had a similar effect to Sirt1 overexpression, consistent with resveratrol having epigenetic effects through activation of endogenous Sirt1 (Oberdoerffer et al. 2008). Our own preliminary data, based on overexpression of Sirt1 in the human intestinal Caco-2 cell line, indicate that resveratrol and Sirt1 together increase DNA methylation at LINE1 elements, a surrogate for global DNA methylation (Yang et al. 2004; L.J.I., L.A.W., D.F. unpublished data), commensurate with a mechanism involving Sirt1 activation by resveratrol.
The limited bioavailability of resveratrol, contrasting with some of its major metabolites (Walle et al. 2004), indicates that physiological effects result principally from the action of metabolites rather than from the parent compound itself. This reasoning supports the view that natural or synthetic derivatives of resveratrol, or even other polyphenolic compounds, may prove better mimetics than resveratrol of the beneficial response to DR. In view of our hypothesis that epigenetic effects mediated via Sirt1 contribute to the longevity response to DR, recent evidence that the soyabean isoflavone daidzein activated recombinant Sirt1 in vitro and increased mitochondrial biogenesis in renal proximal tubular cells, in concert with deacetylation of the Sirt1 substrate PGC1α, (Rasbach and Schnellmann 2008) is of particular interest. Given the fairly robust evidence for the effects of isoflavones on DNA methylation (see Mathers and Ford 2009 for a review, and also above and Table 1), we proffer the suggestion that isoflavones may mimic aspects of the longevity response to DR that are mediated through epigenetic actions. Indeed, our preliminary data indicate an effect of daidzein on DNA methylation similar to that observed using resveratrol in the same cell culture model (Caco-2 cells overexpressing Sirt1; L.J.I., L.A.W., D.F. unpublished data).
The observed effects of DR on gene expression per se do not lend direct support to the hypothesis that a component of the response is mediated through epigenetic effects, but they would certainly be an expectation if DR-mediated effects of relevance to extending healthy lifespan included epigenetic actions. Consistent with (although by no means demonstrating) epigenetic effects, many published studies report effects of DR on gene expression (e.g. Sreekumar et al. 2002; Dhahbi et al. 2004; Higami et al. 2004; Park and Prolla 2005).
Future directions and conclusions
As noted, our preliminary, unpublished data reveal that resveratrol and Sirt1 interact to influence epigenetic factors, including DNA methylation, in our experimental cell culture model. Priorities for future research in the area should include investigating the extent to which such effects are observed under conditions of DR in vivo or with other dietary agents or resveratrol derivatives both in vivo and in cell culture models. Future work should also locate specific genomic sites at which these effects are manifest, achievable through approaches such as ChIP or selective immunoprecipitation of methylated versus unmethylated DNA followed by microarray hybridisation. Mapping such sites to loci whose epigenetic marking changes in the opposite direction in ageing tissues would evaluate the contribution of epigenetic changes induced by DR/DR-mimetics to counteracting the epigenetic effects of ageing. Such approaches may also reveal specific gene targets for further research whose epigenetic modification by Sirt1 and/or resveratrol or other agents may potentially contribute to healthy ageing/longevity. Studies of epigenetic status and how this is affected by ageing and by specific dietary or pharmacological interventions, including DR and administration of test DR-mimetic agents, in mouse models in which Sirt1 expression is knocked out or increased (Cheng et al. 2003; Bordone et al. 2007; Sequeira et al. 2008), and in long-lived strains such as the Ames dwarf mouse (Bartke and Brown-Borg 2004) are likely to be particularly illuminating in the context of the current hypothesis.
In conclusion, one can propose a feasible mechanism through which dietary agents acting through Sirt1-mediated epigenetic modification, including DNA methylation, may mimic some aspects of the response to DR. Emerging evidence, from laboratories including our own, supports the premise that Sirt1 and also resveratrol affect epigenetic marking, highlighting the need for a detailed investigation of these effects in the context of changes to the epigenetic status of ageing cells.
References
Autier P, Gandini S (2007) Vitamin D supplementation and total mortality: a meta-analysis of randomized controlled trials. Arch Intern Med 167:1730–1777
Aviram A, Zimrah Y, Shaklai M et al (1994) Comparison between the effect of butyric acid and its prodrug pivaloyloxymethylbutyrate on histones hyperacetylation in an HL-60 leukemic cell line. Int J Cancer 56:906–909
Barger JL, Kayo T, Vann JM et al (2008) A low dose of dietary resveratrol partially mimics caloric restriction and retards aging parameters in mice. PLoS ONE 3:e2264
Bartke A, Brown-Borg H (2004) Life extension in the dwarf mouse. Curr Top Dev Biol 63:189–225
Baur JA, Pearson KJ, Price NL et al (2006) Resveratrol improves health and survival of mice on a high-calorie diet. Nature 444:337–342
Beach RS, Gershwin ME, Hurley LS (1982) Gestational zinc deprivation in mice: persistence of immunodeficiency for three generations. Science 218:469–471
Bishop NA, Guarente L (2007) Genetic links between diet and lifespan: shared mechanisms from yeast to humans. Nat Rev Genet 8:835–844
Blanc S, Schoeller D, Kemnitz J et al (2003) Energy expenditure of rhesus monkeys subjected to 11 years of dietary restriction. J Clin Endocrinol Metab 88:16–23
Bonsch D, Lenz B, Reulbach U et al (2004) Homocysteine associated genomic DNA hypermethylation in patients with chronic alcoholism. J Neural Transm 111:1611–1616
Bordone L, Motta MC, Picard F et al (2006) Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol 4:e31
Bordone L, Cohen D, Robinson A et al (2007) SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell 6:759–767
Borra MT, Smith BC, Denu JM (2005) Mechanism of human SIRT1 activation by resveratrol. J Biol Chem 280:17187–17195
Burdge GC, Slater-Jefferies J, Torrens C et al (2007) Dietary protein restriction of pregnant rats in the F0 generation induces altered methylation of hepatic gene promoters in the adult male offspring in the F1 and F2 generations. Br J Nutr 97:435–439
Casillas MA Jr, Lopatina N, Andrews LG et al (2003) Transcriptional control of the DNA methyltransferases is altered in aging and neoplastically-transformed human fibroblasts. Mol Cell Biochem 252:33–43
Castro CE, Sevall JS (1980) Alteration of higher order structure of rat liver chromatin by dietary composition. J Nutr 110:105–116
Chen D, Guarente L (2007) SIR2: a potential target for calorie restriction mimetics. Trends Mol Med 13:64–71
Chen D, Bruno J, Easlon E et al (2008) Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev 22:1753–1757
Cheng HL, Mostoslavsky R, Saito S et al (2003) Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci USA 100:10794–10799
Choi SW, Stickel F, Baik HW et al (1999) Chronic alcohol consumption induces genomic but not p53-specific DNA hypomethylation in rat colon. J Nutr 129:1945–1950
Civitarese AE, Carling S, Heilbronn LK et al (2007) Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS Med 4:e76
Cohen HY, Miller C, Bitterman KJ et al (2004) Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science 305:390–392
Colman RJ, Ramsey JJ, Roecker EB et al (1999) Body fat distribution with long-term dietary restriction in adult male rhesus macaques. J Gerontol A Biol Sci Med Sci 54:B283–B290
Dashwood RH, Myzak MC, Ho E (2006) Dietary HDAC inhibitors: time to rethink weak ligands in cancer chemoprevention? Carcinogenesis 27:344–349
Davis CD, Uthus EO (2002) Dietary selenite and azadeoxycytidine treatments affect dimethylhydrazine-induced aberrant crypt formation in rat colon and DNA methylation in HT-29 cells. J Nutr 132:292–297
Davis CD, Uthus EO (2003) Dietary folate and selenium affect dimethylhydrazine-induced aberrant crypt formation, global DNA methylation and one-carbon metabolism in rats. J Nutr 133:2907–2914
Davis CD, Uthus EO, Finley JW (2000) Dietary selenium and arsenic affect DNA methylation in vitro in Caco-2 cells and in vivo in rat liver and colon. J Nutr 130:2903–2909
de Magalhaes JP, Faragher RG (2008) Cell divisions and mammalian aging: integrative biology insights from genes that regulate longevity. Bioessays 30:567–578
Dhahbi JM, Kim HJ, Mote PL et al (2004) Temporal linkage between the phenotypic and genomic responses to caloric restriction. Proc Natl Acad Sci USA 101:5524–5529
Di Croce L, Raker VA, Corsaro M et al (2002) Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science 295:1079–1082
Dolinoy DC, Weidman JR, Waterland RA et al (2006) Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect 114:567–572
Dolinoy DC, Huang D, Jirtle RL (2007) Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci USA 104:13056–13061
Eden A, Gaudet F, Waghmare A et al (2003) Chromosomal instability and tumors promoted by DNA hypomethylation. Science 300:455
Fang MZ, Chen D, Sun Y et al (2005) Reversal of hypermethylation and reactivation of p16INK4a, RARbeta, and MGMT genes by genistein and other isoflavones from soy. Clin Cancer Res 11:7033–7041
Fernandez-Twinn DS, Ozanne SE (2006) Mechanisms by which poor early growth programs type-2 diabetes, obesity and the metabolic syndrome. Physiol Behav 88:234–243
Fontana L, Klein S (2007) Aging, adiposity, and calorie restriction. JAMA 297:986–994
Fontana L, Meyer TE, Klein S et al (2004) Long-term calorie restriction is highly effective in reducing the risk for atherosclerosis in humans. Proc Natl Acad Sci USA 101:6659–6663
Fraga MF, Ballestar E, Paz MF et al (2005) Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA 102:10604–10609
Fraga MF, Agrelo R, Esteller M (2007) Cross-talk between aging and cancer: the epigenetic language. Ann N Y Acad Sci 1100:60–74
Gaudet F, Hodgson JG, Eden A et al (2003) Induction of tumors in mice by genomic hypomethylation. Science 300:489–492
Gonzalo S, Jaco I, Fraga MF et al (2006) DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol 8:416–424
Gresl TA, Colman RJ, Roecker EB et al (2001) Dietary restriction and glucose regulation in aging rhesus monkeys: a follow-up report at 8.5 yr. Am J Physiol Endocrinol Metab 281:E757–E765
Guarente L (2007) Sirtuins in aging and disease. Cold Spring Harb Symp Quant Biol 72:483–488
Guarente L, Picard F (2005) Calorie restriction—the SIR2 connection. Cell 120:473–482
Happel N, Doenecke D, Sekeri-Pataryas KE et al (2008) H1 histone subtype constitution and phosphorylation state of the ageing cell system of human peripheral blood lymphocytes. Exp Gerontol 43:184–199
Harrison DE, Archer JR, Astle CM (1984) Effects of food restriction on aging: separation of food intake and adiposity. Proc Natl Acad Sci USA 81:1835–1838
Hass BS, Hart RW, Lu MH et al (1993) Effects of caloric restriction in animals on cellular function, oncogene expression, and DNA methylation in vitro. Mutat Res 295:281–289
Herbig U, Ferreira M, Condel L et al (2006) Cellular senescence in aging primates. Science 311:1257
Higami Y, Pugh TD, Page GP et al (2004) Adipose tissue energy metabolism: altered gene expression profile of mice subjected to long-term caloric restriction. Faseb J 18:415–417
Holloszy JO (1997) Mortality rate and longevity of food-restricted exercising male rats: a reevaluation. J Appl Physiol 82:399–403
Holloszy JO, Smith EK, Vining M et al (1985) Effect of voluntary exercise on longevity of rats. J Appl Physiol 59:826–831
Howard BH (1996) Replicative senescence: considerations relating to the stability of heterochromatin domains. Exp Gerontol 31:281–293
Ishida E, Nakamura M, Ikuta M et al (2005) Promotor hypermethylation of p14ARF is a key alteration for progression of oral squamous cell carcinoma. Oral Oncol 41:614–622
Issa JP, Ottaviano YL, Celano P et al (1994) Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet 7:536–540
Issa JP, Ahuja N, Toyota M et al (2001) Accelerated age-related CpG island methylation in ulcerative colitis. Cancer Res 61:3573–3577
Jacob RA, Gretz DM, Taylor PC et al (1998) Moderate folate depletion increases plasma homocysteine and decreases lymphocyte DNA methylation in postmenopausal women. J Nutr 128:1204–1212
Kagawa Y (1978) Impact of Westernization on the nutrition of Japanese: changes in physique, cancer, longevity and centenarians. Prev Med 7:205–217
Kim MJ, Aiken JM, Havighurst T et al (1997) Adult-onset energy restriction of rhesus monkeys attenuates oxidative stress-induced cytokine expression by peripheral blood mononuclear cells. J Nutr 127:2293–2301
Kim J, Kim JY, Song KS et al (2007) Epigenetic changes in estrogen receptor beta gene in atherosclerotic cardiovascular tissues and in-vitro vascular senescence. Biochim Biophys Acta 1772:72–80
Klar AJ, Fogel S, Macleod K (1979) MAR1—a regulator of the HMa and HMalpha loci in Saccharomyces cerevisiae. Genetics 93:37–50
Kwabi-Addo B, Chung W, Shen L et al (2007) Age-related DNA methylation changes in normal human prostate tissues. Clin Cancer Res 13:3796–3802
Lagouge M, Argmann C, Gerhart-Hines Z et al (2006) Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell 127:1109–1122
Lane MA, Baer DJ, Rumpler WV et al (1996) Calorie restriction lowers body temperature in rhesus monkeys, consistent with a postulated anti-aging mechanism in rodents. Proc Natl Acad Sci USA 93:4159–4164
Lane MA, Ingram DK, Roth GS (1999) Calorie restriction in nonhuman primates: effects on diabetes and cardiovascular disease risk. Toxicol Sci 52:41–48
Langley E, Pearson M, Faretta M et al (2002) Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J 21:2383–2396
Lee WJ, Shim JY, Zhu BT (2005) Mechanisms for the inhibition of DNA methyltransferases by tea catechins and bioflavonoids. Mol Pharmacol 68:1018–1030
Lillycrop KA, Phillips ES, Jackson AA et al (2005) Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J Nutr 135:1382–1386
Lillycrop KA, Slater-Jefferies JL, Hanson MA et al (2007) Induction of altered epigenetic regulation of the hepatic glucocorticoid receptor in the offspring of rats fed a protein-restricted diet during pregnancy suggests that reduced DNA methyltransferase-1 expression is involved in impaired DNA methylation and changes in histone modifications. Br J Nutr 97:1064–1073
Lin SJ, Kaeberlein M, Andalis AA et al (2002) Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature 418:344–348
Lopatina N, Haskell JF, Andrews LG et al (2002) Differential maintenance and de novo methylating activity by three DNA methyltransferases in aging and immortalized fibroblasts. J Cell Biochem 84:324–334
Lopez-Lluch G, Irusta PM, Navas P et al (2008) Mitochondrial biogenesis and healthy aging. Exp Gerontol 43:813–819
Marutha Ravindran CR, Ticku MK (2004) Changes in methylation pattern of NMDA receptor NR2B gene in cortical neurons after chronic ethanol treatment in mice. Brain Res Mol Brain Res 121:19–27
Mathers JC, Ford D (2009) Nutrition, epigenetics and aging. In: Friso S, Choi SW (eds) Nutrients and epigenetics. CRC, Boca Raton, pp 175–206
McCay CM, Crowell MF, Maynard LA (1935) The effect of retarded growth upon the length of life span and upon the ultimate body size. J Nutr 10:63–79
Messaoudi I, Warner J, Fischer M et al (2006) Delay of T cell senescence by caloric restriction in aged long-lived nonhuman primates. Proc Natl Acad Sci USA 103:19448–19453
Messaoudi I, Fischer M, Warner J et al (2008) Optimal window of caloric restriction onset limits its beneficial impact on T-cell senescence in primates. Aging Cell 7:908–919
Meyer TE, Kovacs SJ, Ehsani AA et al (2006) Long-term caloric restriction ameliorates the decline in diastolic function in humans. J Am Coll Cardiol 47:398–402
Michaud EJ, van Vugt MJ, Bultman SJ et al (1994) Differential expression of a new dominant agouti allele (Aiapy) is correlated with methylation state and is influenced by parental lineage. Genes Dev 8:1463–1472
Miyamura Y, Tawa R, Koizumi A et al (1993) Effects of energy restriction on age-associated changes of DNA methylation in mouse liver. Mutat Res 295:63–69
Moreno FS, SW T, Naves MM et al (2002) Inhibitory effects of beta-carotene and vitamin a during the progression phase of hepatocarcinogenesis involve inhibition of cell proliferation but not alterations in DNA methylation. Nutr Cancer 44:80–88
Myzak MC, Ho E, Dashwood RH (2006) Dietary agents as histone deacetylase inhibitors. Mol Carcinog 45:443–446
Narita M, Narita M, Krizhanovsky V et al (2006) A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell 126:503–514
Nisoli E, Tonello C, Cardile A et al (2005) Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science 310:314–317
Oberdoerffer P, Michan S, McVay M et al (2008) SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell 135:907–918
O'Hagan HM, Mohammad HP, Baylin SB (2008) Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet 4:e1000155
Olthof MR, Hollman PC, Zock PL et al (2001) Consumption of high doses of chlorogenic acid, present in coffee, or of black tea increases plasma total homocysteine concentrations in humans. Am J Clin Nutr 73:532–538
Oren M (2003) Decision making by p53: life, death and cancer. Cell Death Differ 10:431–442
Ota H, Tokunaga E, Chang K et al (2006) Sirt1 inhibitor, Sirtinol, induces senescence-like growth arrest with attenuated Ras-MAPK signaling in human cancer cells. Oncogene 25:176–185
Panza F, Solfrizzi V, Colacicco AM et al (2004) Mediterranean diet and cognitive decline. Public Health Nutr 7:959–963
Park SK, Prolla TA (2005) Gene expression profiling studies of aging in cardiac and skeletal muscles. Cardiovasc Res 66:205–212
Pruitt K, Zinn RL, Ohm JE et al (2006) Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet 2:e40
Pufulete M, Al-Ghnaniem R, Leather AJ et al (2003) Folate status, genomic DNA hypomethylation, and risk of colorectal adenoma and cancer: a case control study. Gastroenterology 124:1240–1248
Pufulete M, Al-Ghnaniem R, Rennie JA et al (2005) Influence of folate status on genomic DNA methylation in colonic mucosa of subjects without colorectal adenoma or cancer. Br J Cancer 92:838–842
Rampersaud GC, Kauwell GP, Hutson AD et al (2000) Genomic DNA methylation decreases in response to moderate folate depletion in elderly women. Am J Clin Nutr 72:998–1003
Rasbach KA, Schnellmann RG (2008) Isoflavones promote mitochondrial biogenesis. J Pharmacol Exp Ther 325:536–543
Rine J, Strathern JN, Hicks JB et al (1979) A suppressor of mating-type locus mutations in Saccharomyces cerevisiae: evidence for and identification of cryptic mating-type loci. Genetics 93:877–901
Rodriguez J, Vives L, Jorda M et al (2008) Genome-wide tracking of unmethylated DNA Alu repeats in normal and cancer cells. Nucleic Acids Res 36:770–784
Rogina B, Helfand SL (2004) Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci USA 101:15998–16003
Ross MH (1961) Length of life and nutrition in the rat. J Nutr 75:197–210
Rowling MJ, McMullen MH, Schalinske KL (2002) Vitamin A and its derivatives induce hepatic glycine N-methyltransferase and hypomethylation of DNA in rats. J Nutr 132:365–369
Sarg B, Koutzamani E, Helliger W et al (2002) Postsynthetic trimethylation of histone H4 at lysine 20 in mammalian tissues is associated with aging. J Biol Chem 277:39195–39201
Schilling MM, Oeser JK, Boustead JN et al (2006) Gluconeogenesis: re-evaluating the FOXO1-PGC-1alpha connection. Nature 443:E10–E11
Sequeira J, Boily G, Bazinet S et al (2008) sirt1-null mice develop an autoimmune-like condition. Exp Cell Res 314:3069–3074
Shimokawa I, Higami Y, Hubbard GB et al (1993) Diet and the suitability of the male Fischer 344 rat as a model for aging research. J Gerontol 48:B27–B32
Sinclair DA, Guarente L (1997) Extrachromosomal rDNA circles–a cause of aging in yeast. Cell 91:1033–1042
Sirchia SM, Ren M, Pili R et al (2002) Endogenous reactivation of the RARbeta2 tumor suppressor gene epigenetically silenced in breast cancer. Cancer Res 62:2455–2461
Smeal T, Claus J, Kennedy B et al (1996) Loss of transcriptional silencing causes sterility in old mother cells of S. cerevisiae. Cell 84:633–642
Sreekumar R, Unnikrishnan J, Fu A et al (2002) Effects of caloric restriction on mitochondrial function and gene transcripts in rat muscle. Am J Physiol Endocrinol Metab 283:E38–E43
Stempak JM, Sohn KJ, Chiang EP et al (2005) Cell and stage of transformation-specific effects of folate deficiency on methionine cycle intermediates and DNA methylation in an in vitro model. Carcinogenesis 26:981–990
Tissenbaum HA, Guarente L (2001) Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410:227–230
Trichopoulou A, Orfanos P, Norat T et al (2005) Modified Mediterranean diet and survival: EPIC-elderly prospective cohort study. BMJ 330:991
Vaquero A, Scher M, Lee D et al (2004) Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell 16:93–105
Vaquero A, Scher M, Erdjument-Bromage H et al (2007) SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature 450:440–444
Vega RB, Huss JM, Kelly DP (2000) The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol Cell Biol 20:1868–1876
Villeponteau B (1997) The heterochromatin loss model of aging. Exp Gerontol 32:383–394
Walle T, Hsieh F, DeLegge MH et al (2004) High absorption but very low bioavailability of oral resveratrol in humans. Drug Metab Dispos 32:1377–1382
Wallwork JC, Duerre JA (1985) Effect of zinc deficiency on methionine metabolism, methylation reactions and protein synthesis in isolated perfused rat liver. J Nutr 115:252–262
Wasson GR, McGlynn AP, McNulty H et al (2006) Global DNA and p53 region-specific hypomethylation in human colonic cells is induced by folate depletion and reversed by folate supplementation. J Nutr 136:2748–2753
Waterland RA, Jirtle RL (2003) Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol 23:5293–5300
Waterland RA, Travisano M, Tahiliani KG (2007) Diet-induced hypermethylation at agouti viable yellow is not inherited transgenerationally through the female. Faseb J 21:3380–3385
Weindruch R, Walford RL (1982) Dietary restriction in mice beginning at 1 year of age: effect on life-span and spontaneous cancer incidence. Science 215:1415–1418
Weindruch R, Walford RL, Fligiel S et al (1986) The retardation of aging in mice by dietary restriction: longevity, cancer, immunity and lifetime energy intake. J Nutr 116:641–654
Willcox DC, Willcox BJ, Todoriki H et al (2006) Caloric restriction and human longevity: what can we learn from the Okinawans? Biogerontology 7:173–177
Wilson VL, Jones PA (1983) DNA methylation decreases in aging but not in immortal cells. Science 220:1055–1057
Wu Z, Puigserver P, Andersson U et al (1999) Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98:115–124
Yang AS, Estecio MR, Doshi K et al (2004) A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res 32:e38
Yu BP, Masoro EJ, Murata I et al (1982) Life span study of SPF Fischer 344 male rats fed ad libitum or restricted diets: longevity, growth, lean body mass and disease. J Gerontol 37:130–141
Zainal TA, Oberley TD, Allison DB et al (2000) Caloric restriction of rhesus monkeys lowers oxidative damage in skeletal muscle. Faseb J 14:1825–1836
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Wakeling, L.A., Ions, L.J. & Ford, D. Could Sirt1-mediated epigenetic effects contribute to the longevity response to dietary restriction and be mimicked by other dietary interventions?. AGE 31, 327–341 (2009). https://doi.org/10.1007/s11357-009-9104-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11357-009-9104-5