Abstract
Per- and polyfluoroalkyl substances (PFASs) have received increasing attention due to their widespread presence in diverse environments including wastewater treatment plants (WWTPs) and their potential adverse health effects. Perfluorooctanoic acid (PFOA) is one of the most detected forms of PFASs in WWTPs. However, there is still a paucity of knowledge about the effect of PFASs on microorganisms of the key component of WWTP, activated sludge. In this study, lab-scale microcosm experiments were established to evaluate the influences of PFOA on activated sludge microbes under aerobic and anaerobic conditions. The diversity, structure, and microbe-microbe interaction of microbial community were determined by 16S rRNA gene amplicon sequencing and co-occurrence network analysis. After 90 days of exposure to PFOA, activated sludge microbial richness decreased under both aerobic and anaerobic conditions. Specifically, under aerobic condition, Rhodopseudomonas (mean relative abundance 3.6%), Flavobacterium (2.4%), and Ignavibacterium (6.6%) were enriched in PFOA-spiked activated sludge compared with that in the unspiked sludge (2.6%, 0.1%, and 1.9%, respectively). By contrast, after 90 days of exposure to PFOA, Eubacterium (2.1%), Hyphomicrobium (1.8%), and Methyloversatilis (1.2%) were enriched under anaerobic condition, and more abundant than that in the control sludge (0.4%, 1.5%, and 0.6%, respectively). These genera were the potential PFOA-resistant members. In addition, Azospirillum and Sporomusa were the most connected taxa in PFOA-aerobic and PFOA-anaerobic networks, respectively. Prediction of the functional gene showed that PFOA inhibited some gene expression of sludge microbes, such as transcription, amino acid transport and metabolism, and energy production and conversion. In summary, continued exposure to PFOA induced substantial shifts of the sludge bacterial diversity and composition under both aerobic and anaerobic conditions.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Per- and polyfluoroalkyl substances (PFASs, CnF2n+1–R) are a group of ubiquitous synthetic compounds, which is used extensively in a wide range of industrial and consumer applications. Owing to the unique strong carbon–fluorine (C-F) bond of the molecule, PFASs display oil and water repellency, high chemical and thermal stability, and surfactant properties (Lehmler 2005; Lindstrom et al. 2011). Among them, perfluorooctanoic acid (PFOA) is one of the most widely detected groups of PFASs. However, considerable studies show that PFOA exerts toxic effects on animals and human, such as endocrine disruption, reproductive toxicity, developmental toxicity, immunotoxicity, and neurotoxicity (Mariussen 2012; Sun et al. 2016). To be worse, high concentrations of PFOA have been widely detected in various natural environments, including river, sediment (Wang et al. 2019), soil (Zareitalabad et al. 2013), and wastewater treatment plants (Higgins et al. 2005). Therefore, before designing any applicable removal protocols, it becomes more urgent for environmentalists to investigate and evaluate the impact of PFOA on these environments.
Among these impacted environments by PFOA, wastewater treatment plants (WWTPs) gained increasing attentions because of their pivotal function in modern cities to receive and treat municipal and industrial wastewater. Activated sludge is a major contributor of WWTP to pollutant removal (Jiang et al. 2008). However, activated sludge is likely to be affected by high concentration of PFOA. PFOA is ubiquitous in wastewater with various concentrations ranging from 58 to 1050 ng/L (Sinclair and Kannan 2006). Some wastewater, such as aqueous film forming foam (AFFF)–contaminated wastewater or specific industrial wastewater, contains high concentrations of PFASs up to milligram per liter (mg/L) level (Arias et al. 2015; Houtz et al. 2013). Due to the low lipophilicity and solubility of long-chain PFASs, PFOA is preferential partitioning in the sludge during wastewater treatment (Zhang et al. 2013). By getting into WWTPs via domestic and industrial wastewater, PFOA could remain and accumulate in these plants through the process of biological treatment attributed to its recalcitrance to biodegradation (Liou et al. 2010). Conventional water treatment processes are ineffective in removing PFOA (Abunada et al. 2020). Moreover, precursor biodegradation may lead to an extra addition of PFOA (Sinclair and Kannan 2006). Consequently, PFOA is usually dominant in wastewater and activated sludge of WWTPs, and cannot be efficiently removed by conventional wastewater treatment. Given the bioaccumulation and toxicity of PFOA in WWTPs, it is necessary to investigate the impact of PFOA on sludge microbiota.
The structure and function of activated sludge microbial community are important factors affecting the efficiency and stability of most WWTPs (Wang et al. 2014). Studies indicated that PFOA might influence the geochemical behaviors and activities in activated sludge. And PFOA might induce stress to inhibit the growth of specific microbiome (Nobels et al. 2010). For example, PFOA inhibited nitrification rate of activated sludge (Chen et al. 2020), and suppressed the acidification and methanation of activated sludge anaerobic digestion (Wang et al. 2021). However, to date, the research on effects of PFAS on microorganisms are mainly focused in soil and sediment with limited studies on activated sludge microbiota. Qiao et al. (2018) applied a greenhouse experiment to investigate the effects of PFASs on soil microbiota and found that PFAS pollution significantly disturbs the normal function of soil microorganisms. Zhang et al. (2017b) measured the effect of 6:2 FTOH on a sediment microbial community under aerobic condition and potential 6:2 FTOH degraders and tolerant strains were identified in the sediment samples. Moreover, previous studies primarily focused on the effects of PFAS on activated sludge under fixed redox condition (mostly under aerobic conditions), ignoring the fluctuating oxygen content during biological treatment processes (Kassab et al. 2010). Yu et al. (2018) set up a sequencing batch reactor system to monitor dynamics of microbial communities under aerobic condition. Yang et al. (2020) reported that PFOA inhibited the cyclic transformations of polyhydroxyalkanoates and glycogen by the batch experiments with PFOA and aerobic granular sludge. Therefore, a comparative analysis under diverse redox conditions is necessary; giving different oxygen contents may give rise to different effects on the fate of PFOA.
In the current study, microcosm experiment was designed to investigate the PFOA stress on microbiome in activated sludge. PFOA was selected as representative PFAS owing to its ubiquity. Aerobic and anaerobic culture conditions were set up to observe similarity and difference of microbiome affected by PFOA in activated sludge. The diversity, composition, and interactions of microbial communities of activated sludge were determined by the high-throughput sequencing and statistical analyses. The objective of the present study was to elucidate the impacts of PFOA on microbial communities of activated sludge under aerobic and anaerobic conditions.
Materials and methods
Sludge collection
Inoculum sludge was collected from a secondary sedimentation tank of a municipal waste water treatment plant (WWTP) located in Guangzhou, Guangdong province, China, whose mixed liquor suspended solid (MLSS) concentration was 14.7 g/L and mixed liquor volatile suspended solid (MLVSS) concentration was 6.3 g/L. This WWTP adopted traditional anaerobic-anoxic-aerobic processes. Collected sludge samples were stored in a cryogenic tank during transportation and then stored at 4℃ in the laboratory. The evenly mixed sludge samples were divided into two identical parts, one for molecular analysis and the other for microcosm establishment.
Chemicals and reagents
All the chemicals used in the experiments were of analytical grade. PFOA (CAS# 335–67-1) was purchased from Sigma-Aldrich (St. Louis, MO, USA). The PFOA stock solution consisted of PFOA dissolved in methanol (> 99.8%) with the final concentration of 100 mg/L. Mineral salt medium (MSM) was used to construct the microcosms. It is composed of Na2HPO4·12H2O (10.55 g/L), NH4Cl (0.3 g/L), MgCl2·6H2O (0.1 g/L), KH2PO4 (1.5 g/L), vitamin solution (1 mL/L), and trace element solution (1 mL/L) (Xu et al. 2021a).
Microcosm experimental design
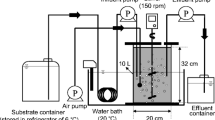
Aimed to evaluate the effects of PFOA on activated sludge under two distinct redox conditions, aerobic and anaerobic microcosms with activated sludge as inoculum were established respectively. Microcosms were constructed in triplicate for each treatment at different time points (30, 60, 90 days). To set up the aerobic microcosms, 50 mL activated sludge sample and 150 mL autoclaved MSM were first transferred into a clean sterile 250-mL serum bottle followed by 5 days of equilibration (Xu et al. 2021d). Two treatments were then established after equilibration by adding (1) 500 μL PFOA stock solution whose final concentration in the experiment was 0.25 mg/L (hereinafter, denoted as “PA_O” group); or (2) equal volume of methanol without PFOA (denoted as “Ct_O”) (Yu et al. 2018). To ensure adequate oxygen content, the microcosms were sealed with air permeable parafilm. All microcosms were then incubated at 25℃ in the dark (Li et al. 2021b).
The establishment and incubation of anaerobic microcosms were similar to those of aerobic microcosms, except for the addition of sodium nitrate (with a final concentration of 1000 mg/L) and nitrogen aeration for 30 min before sealing, in order to achieve anaerobic conditions. The serum bottles were sealed with butyl rubber septa and aluminum caps. Similar nomenclature was adopted in the two anaerobic treatments; i.e., group with PFOA was referred to “PA_N” and control group added with the same amount of methanol was referred to “Ct_N,” respectively.
After being set up, all microcosms were cultured continuously for 90 days. Another 500 μL PFOA or methanol stock solutions were kept being added to the corresponding microcosms every 15 days (totally 6 times) to mimic the accumulation process of PFOA in sludge. After 30, 60, and 90 days of incubation, sacrificial sampling was performed on triplicate cultures from each treatment before the next addition of PFOA/methanol. A total of 39 sludge samples (including three original sludge samples, representing cultures at the 0th day) were obtained for subsequent DNA extraction and 16S rRNA gene amplicon sequencing.
Illumina high-throughput sequencing
The extraction of genomic DNA from each activated sludge sample was carried out using the Powersoil DNA Isolation kit (MO BIO Laboratories, Inc., Carlsbad, CA) in accordance with the manufacturer’s protocol (Sun et al. 2020). The concentration of the extracted DNA was determined by Qubit (Invitrogen 3.0 Fluorometer) and the purity was determined by gel electrophoresis (Xu et al. 2021e). V3-V4 hypervariable regions of the bacterial 16S rRNA gene were amplified using the primers 338F (5ʹ-ACTCCTACGGGAGGCAGCA-3ʹ) and 806R (5ʹ-GGACTACHVGGGTWTCTAAT-3ʹ) (Kuczynski et al. 2011). PCR amplification was performed using the following thermal conditions: 60 s at 98 °C, 30 cycles of 10 s at 98 °C, 30 s at 50 °C, and 30 s at 72 °C, followed by a final extension step for 5 min at 72 °C. PCR products purified by gel extraction were sent for high-throughput sequencing on an Illumina MiSeq platform (Gao et al. 2021).
Bioinformatic and statistical analysis
After trimming of the barcodes and primers, generated raw sequences were merged and then filtered with USEARCH (v 10.0) (Xu et al. 2021d). UNOISE3 was applied to denoise the sequences retained after quality control and obtain the unique amplicon sequence variants (ASV). The ASVs were classified into taxonomy with the GreenGene database (v 13.5) (Xu et al. 2020). Functional gene prediction was performed with PICRUSt and annotated with the COG database (Langille et al. 2013).
Determination of alpha-diversity and beta-diversity was performed in R by “–alpha_div” and “–beta_div” function (Gao et al. 2021). MicrobiomeAnalyst platform was used for comprehensive statistical analysis, including analysis for the relative abundance (Xu et al. 2021b). Additionally, STAMP software was employed to analyze the difference in genus level with Welch’s t test among groups and filtered the genera with a p value exceeding 0.05 and effect sizes smaller than 0.1 (Parks et al. 2014). Co-occurrence networks was calculated with the R package “ggcorrplot” and was visualized by Gephi v 0.29 software (Sun et al. 2021; Xu et al. 2021c). The significance level of all tests was set at 0.05.
Quantitative PCR
Total bacteria biomass was estimated in this experiment by quantitative PCR targeting 16S rRNA using the primers 338F and 518R (Xu et al. 2020). The quantitative PCR reaction was performed in a volume of 20 μL containing 10 μL TB Green Premix Ex Taq (Tli RNaseH Plus) (Takara, Japan), 0.2 μL of each primer (10 pmol/μL), 1 μL of tenfold diluted template DNA (1–10 ng), and 8.6 μL of double-distilled H2O (ddH2O). The qPCR was performed on a CFX96 Real-Time System (Bio-Rad, USA) using the following thermal conditions: 15 min at 95 °C, 45 cycles of 15 s at 94 °C, 30 s at 55 °C, and 30 s at 72 °C, with a final extension step of 72 °C for 7 min. A standard curve was constructed using a serial of tenfold diluted known-copies plasmid solution harboring 16S rRNA genes (Li et al. 2021a). Each sample was quantified in triplicates.
Results
Richness of sludge microbial community affected by PFOA addition
Biomass and richness of microbial community under aerobic condition
As mentioned, total bacteria biomass of activated sludge was measured by qPCR of 16S rRNA gene on 30, 60, and 90 days (Fig. 1). Mean copy number of 16S rRNA gene in the PA_O treatment group was significantly lower than that with Ct_O treatment during matched incubation times. Specifically, Chao1 index was used to further determine the richness of sludge microbial communities (Fig. 2). At the first 30 days, PA_O sludge had a higher bacterial Chao1 index (1078.72) than Ct_O sludge (1038.43). However, over the course of incubation, the Chao1 value of PA_O sludge decreased gradually. Finally, the Chao1 index of bacteria in sludge treated with PFOA (1033.26) was relatively lower compared with that in control sludge (1125.07) after 90 days of incubation.
Biomass and richness of microbial community under anaerobic condition
Similar to aerobic incubation, final microbiome biomass of activated sludge was suppressed under exposure to PFOA under anaerobic condition according to the copy number of 16S rRNA gene. In detail, compared with Ct_N samples, abundance of 16S rRNA gene of PA_N samples was lower after 90 days of incubation. However, richness of activated sludge first raised promoted by coexistence with PFOA at the first 60 days, and then fell at the end of the 90-day anaerobic incubation. The mean value of Chao1 index in Ct samples was 864.71, while the value in PFOA samples was 659.23 on the 90th day.
Microbial community structure affected by PFOA
Structure of microbial community under aerobic condition
Initially in original activated sludge samples, Proteobacteria (45.8%, relative abundance), Nitrospirae (14.0%), Bacteroidetes (14.0%), and Actinobacteria (4.7%) were the predominant bacterial phyla. After 3 months of incubation, Firmicutes and Ignavibacteriae became the dominant phyla in control sludge.
The relative abundance of bacterial phyla of sewage sludge containing PFOA under oxic condition is shown in Fig. 3A. Proteobacteria, Bacteroidetes, Firmicutes, and Ignavibacteriae were the predominant bacterial phyla in both PFOA-added sludge and control sludge on the 90th day, accounting for more than 90% of the whole community. However, distinct structure of microbial community of the sludge amended with PFOA was observed. The long-term aerobic treatment of activated sludge with PFOA resulted in remarkable changes in microbial community composition. Proteobacteria, the most abundant phylum in the PFOA treatment (49.4%), was remarkably lower than that in the control (68.0%). In contrast, Ignavibacteriae and Bacteroidetes were more abundant in PFOA-added samples than in the control samples.
Exposure of PFOA over 90 days induced a shift of compositions of AS microbial community. “Inoculum” on the X-axis represents the original sludge at the 0th day, “Ct” is the control group without PFOA, and “PA” is the treatment group with PFOA. The bar charts showed the major phyla and genera under A, B aerobic and C, D anaerobic conditions, respectively
Genus level analysis of microbial community was further conducted. Top 20 bacterial genera in terms of the relative abundance are presented in Fig. 3B. Among them, Dokdonella and Xanthobacter were the abundant genera in all samples on the 90th day. But the addition of PFOA caused a decline of the abundance of Dokdonella, Xanthobacter, Azospirillum, and Pseudoxanthomonas after the 90-day incubation. Meanwhile, an obvious variation of Azospirillum was observed in PA_O samples. In the first 60 days, the abundance of Azospirillum in PA_O sludge samples was significantly higher than that in the Ct_O samples, accounting for more than 10%. However, by the 90th day, the abundance of Azospirillum decreased sharply and was even lower than that of the control group. Additionally, the relative abundances of Rhodopseudomona, Acetobacteriums, and Flavobacterium slightly increased in the samples adding with PFOA (3.6%, 3.3%, 2.4%) compared to control samples (2.6%, 0.3%, 0.1%). The proportion of Ignavibacterium was much higher in PFOA treatment (6.6%) than that in control samples (1.9%).
Structure of microbial community under anaerobic condition
The relative abundance of the sludge samples under anoxic condition is shown in Fig. 3C and D. Proteobacteria, Bacteroidetes, and Firmicutes were the dominant phyla in both groups under anaerobic condition. Bacteroidetes had a lower proportion in PFOA-added sludge (12.2%) than that in control treatment (24.3%) under anaerobic conditions on the 90th day. The relative abundance of Ignavibacteriae was decreased from 2.4% (Ct) to 1.0% (PFOA). The relative abundance of phylum Firmicutes remarkably increased in samples spiked with PFOA (58.8%) after 90 days of incubation, compared with the control sample (39.3%).
Genus level analysis in anaerobic samples was further performed (Fig. 3D). Sporomusa and Rhodopseudomonas consisted of a considerable proportion in the microbial community. The relative abundances of Sporomusa increased rapidly in sludge amended with PFOA, which was eventually 18.6% higher than that in control sludge. Additionally, Melioribacter, Acetobacterium, Oscillibacter, and Candidatus_Endomicrobium were slightly depressed over time treated with PFOA, whereas Hyphomicrobium, Methyloversatilis, and Eubacterium were promoted in the same group. For example, the final proportion of Eubacterium in PFOA-amended sludge (2.1%) was higher than that in the control sludge (0.4%).
Further genus level analysis by Welch’s t test
According to the abovementioned analysis, it is observed that the addition of PFOA resulted in the variations of the composition and diversity of community in sludge. In this study, the differential species were further identified and confirmed by Welch’s t test at the genus level (Parks et al. 2014).
Genera with significant difference under aerobic condition
Eleven significantly distinct bacteria genera were observed between the two groups under aerobic condition (Fig. 4A). The genera Azospirillum, Methylobacillus, Acetobacterium, Hydrogenophaga, Methyloversatilis, Gracilibacter, Fluviicola, and Brevundimonas demonstrated a relatively higher mean proportion in microbiota of PFOA-exposed sludge than that in Ct sludge. The mean proportions of Sporomusa, Ancylobacter, and Anaerovorax were lower in PFOA-amended samples.
Genera with significant difference under anaerobic condition
Genera significantly enriched in anaerobic condition are shown in Fig. 4B. A total of ten genera were identified with significant differences between PFOA-exposed sludge and Ct sludge. Sporomusa, of phylum Firmicutes, was the most different genus with the largest effect size, followed by Candidatus_Endomicrobium in Elusimicrobia. The relative abundance of Hyphomicrobium, Nitrospira, Acetoanaerobium, Anaerovorax, Proteiniphilum, Sedimentibacter, and Gp6 decreased slightly in PFOA-contaminated sludge. This indicates a growth inhibition of these microbes brought by the addition of PFOA. All of these 10 genera with significant difference could be applied as potential biomarkers responded to PFOA under anaerobic condition.
Sludge microbial interaction affected by PFOA
In this study, microbial co-occurrence networks were applied to investigate the interactions of sludge microbiota. According the experimental design, two co-occurrence networks, the “PFOA-aerobic network” and “PFOA-anaerobic network,” were constructed to investigate microbial interactions in activated sludge affected by the long-term exposure of PFOA under aerobic and anaerobic conditions, respectively (Fig. 5).
The co-occurrence networks of the AS microbiomes under the A PA_O treatments (n = 9) and B PA_N (n = 9) treatments. The color of most connected node (top 3) is marked using the assignment at the genus level. The size of each node was proportional to its connection number (mean degree). The links represent the significant and strong Spearman’s correlations (|R|> 0.85, p < 0.05)
Microbial interaction networks under aerobic condition
A total of 290 nodes and 6355 pair-wise correlations (3498 positive links and 2857 negative links) were obtained from the network. The majority of taxonomic nodes in PFOA-aerobic networks were mainly subordinate to Proteobacteria (58.63%), followed by Bacteroidetes (16.55%), Firmicutes (12.41%), and Nitrospirae (2.07%). The highly connected taxa at genera level can be characterized by calculating the network’s topological parameters such as high average degree (Banerjee et al. 2018). Accordingly, Azospirillum, Xanthobacter, and Methylophilus were identified as the most connected taxonomies in PA_O community due to their high mean degree in the PFOA-aerobic network. In addition, the ratio of the number of positive and negative correlated links in the networks was also calculated. In the PFOA-aerobic network, the ratio of positive links (~ 55%) was close to that of negative links (~ 45%).
Microbial interaction networks under anaerobic condition
The PFOA-anaerobic network was generated capturing 7716 pair-wise correlations (6328 positive links and 1388 negative links in Fig. 5B) among 219ASVs (nodes in Fig. 5B). The most taxonomic nodes in the network were assigned to Firmicutes (67.56%), followed by Bacteroidetes (18.48%), Proteobacteria (6.39%), and Nitrospirae (1.37%). Genera Sporomusa, Petrimonas, and Eubacterium were identified as the most connected members. In the PFOA-anaerobic network, the proportion of the positive links was much larger than the negative links, which was nearly four times larger.
Microbial functional performance in activated sludge affected by PFOA
Microbial community function analysis under aerobic condition
The prediction of functional genes of the activated sludge bacterial microbiota was performed using PICRUSt. Under aerobic condition (Fig. 6), compared with the control group, the exposure of activated sludge to PFOA for 90 days resulted in a relatively lower abundance of most functional genes, such as secondary metabolites biosynthesis transport and catabolism, transcription, amino acid transport and metabolism, chromatin structure and dynamics, energy production and conversion, inorganic ion transport and metabolism, and cell cycle control cell division chromosome partitioning. This lower abundance in PFOA-amended samples demonstrates that the exposure to PFOA might suppress the expression of microbial functional genes.
Microbial community function analysis under anaerobic condition
Similar to the samples under aerobic condition, PFOA also presented a certain inhibitory effect on some functional genes of the microbial community under anaerobic conditions (Fig. 6). But the abundance of many functional genes that was reduced at the end of the incubation with PFOA was different from that in aerobic condition in terms of metabolic pathways. The affected genes involved transcription, cell motility, energy production and conversion, inorganic ion transport and metabolism, signal transduction mechanisms, and chromatin structure and dynamics. Remarkably, at the 30th day, functional genes involved in cell motility, energy production and conversion, and amino acid transport and metabolism were facilitated, while, over the coming 60 days until the 90th day, abundance of these genes in PA_N gradually reduced and eventually lower than that in Ct_N. It might be due to the long-term accumulation of PFOA in activated sludge.
Discussion
Effect of PFOA on activated sludge bacterial richness
In our study, prolonged exposure to high concentrations of PFOA led to a decrease of activated sludge community richness under both aerobic and anaerobic conditions (Fig. 2). Similarly, Yu et al. (2018) also found a remarkable decrease of community richness in sludge exposed to PFOA. In soil environments, Senevirathna et al. (2022) observed similar patterns that high concentration of PFAS caused a reduction of the richness of microbial community. Bao et al. (2018) also indicated that perfluorooctanesulfonate (PFOS) lowered the microbial richness and diversity. These results were consistent with the current study.
However, the above related studies were all carried out under aerobic conditions. Few published results reveal the effects of PFOA on activated sludge microbial community richness under anaerobic condition. Our study found that the richness of PFOA-amended activated sludge decreased under anaerobic conditions. The anaerobic PFOA-treated samples had fewer copy numbers of the 16S rRNA gene than the control group, also supporting the current finding of reduced richness of activated sludge exposed to PFOA (Fig. 1).
In general, microbiota with higher richness are able to maintain a more stable ecology (Hooper and Macpherson 2010). And high richness implies a greater probability of functional redundancy, which can increase the community’s resilience to disturbance (Zhang et al. 2019). Therefore, our results indicate exposure to PFOA aerobically and anaerobically might both restrain the biomass growth and decrease the richness of activated sludge due to its cumulative toxicity, and might further influence the stableness of activated sludge bacteria consortia.
Effect of PFOA on bacterial community composition and structure in activated sludge
The addition of PFOA induced instinct changes of the composition of sludge microbial community under both aerobic and anaerobic conditions (Fig. 3). In our study, the high abundance of Proteobacteria, Bacteroidetes, and Firmicutes maintained over time of all groups regardless of aerobic and anaerobic conditions, indicating their tolerance to PFOA. Proteobacteria and Bacteroidetes have been found as the most abundant phyla in the aerobic sequencing batch reactor (SBR) continuously exposed to PFOA (Yu et al. 2018).
Under aerobic condition, Proteobacteria was the most abundant phylum in PFOA-spiked sludge, although its relative abundance was lower than in the control sludge. However, Senevirathna et al. (2022) reported that the relative abundance of Proteobacteria in PFAS-rich contaminated soil was higher than that in uncontaminated soil, which could be the result of different experimental subject and physicochemical parameters. Moreover, the abundance of some genus in PFOA-amended treatment increased with a variety of degrees, such as Ignavibacterium, Rhodopseudomona, and Flavobacterium. Enrichment of these taxonomies might be attributed to their preference, tolerance, or degradation of PFOA. Previous studies draw similar conclusions. Fluoroacetate dehalogenase (FAcD) RPA1163, a defluorinase, was already discovered from Rhodopseudomonas palustris CGA009, indicating that Rhodopseudomona might have the potential to defluorinate PFOA (Chan et al. 2011). Both Ignavibacterium and Flavobacterium possess dechlorinated potential. Ignavibacterium could enhance dechlorination of polychlorinated biphenyl (Yu et al. 2016). Flavobacterium was the classical aerobic pentachlorophenol (PCP)–degrading bacteria (Bosso and Cristinzio 2014). Since both fluorine and chlorine are halogen elements, they share similar physical and chemical properties, so it can be speculated that their metabolic pathways are likely to be similar (Neufeld and Wolfire 2009). Hence, Ignavibacterium and Flavobacterium might be resistant to PFOA, and more laboratory studies are needed in the future to investigate their potential for defluorination.
Under anaerobic condition (Fig. 3A), Firmicutes increased to be the most abundant phylum in PFOA-spiked sludge, suggesting that they have higher tolerance to the high concentration of PFOA. Firmicutes widely exists in the environment and has the ability to degrade a variety of organic pollutants (Li et al. 2021c; Wolfe et al. 2014; Zhang et al. 2018). The enrichment of genera Sporomusa, Hyphomicrobium, and Eubacterium also indicated their potential to tolerate or degrade PFOA. Sporomusa is known as an anaerobic methanol-utilizing bacterium (Ammam et al. 2016), and has been isolated from varied environments such as rice paddy soil, forest soil, sediment of river, and gut of termites (Balk et al. 2010). The high abundance of Sporomusa in sludge either with or without PFOA might be as a result of the addition of methanol in both groups. Besides, Sporomusa was reported as an acetogenic bacterium with the capacity to oxidize a wide range of organic substrates (Balk et al. 2010). Previous studies reported that Eubacterium was resistant to high concentrations of hexadecyltrimethylammonium bromide (CTAB) under anaerobic conditions (Keith and John 2001). Eubacterium processes the capacity to adapt to complex pollutant environments (Zhang et al. 2017a). The increasing number of Eubacterium in PA_N suggested their potential capability to resist the toxicity of PFOA under anaerobic condition.
PFOA usually played a dual role in the sewage sludge microbial community. At the same time, the inhibitory effect of PFOA on certain bacteria in the sludge community was also observed. Under aerobic condition, the relative abundance of genera Dokdonella, Azospirillum, Pseudoxanthomonas, and Xanthobacter was decreased with PFOA treatment compared with the Ct group. Among them, Dokdonella, Pseudoxanthomonas, and Xanthobacter show a substantial denitrification capacity during wastewater biological treatment (Long et al. 2019; Wang et al. 2018a; Zhang et al. 2020). Under anaerobic condition, Melioribacter, Acetobacterium, Oscillibacter, and Candidatus_Endomicrobium were suppressed over the incubation course with the presence of PFOA. Notably, Melioribacter is also a denitrifier in sludge (Han et al. 2021). Additionally, Acetobacterium and Oscillibacter are acid-producing bacteria. They play important roles in acidification process of activated sludge, which are capable of improving the degradation of organic matter in wastewater (Zhou et al. 2015). Thus, PFOA might affect the sludge performance of biological treatment due to its inhibitory effect on denitrification and acidification processes under both aerobic and anaerobic conditions.
The decline of these microorganisms suggests that they are vulnerable to PFOA’s toxicity. PFASs with long carbon chain may induce expression of several stress genes, resulting in membrane damage, oxidative damage, and DNA damage (Nobels et al. 2010). PICRUSt results in this study also supported that the majority of metabolic functions and gene expression was repressed in the PFOA group, such as nucleotide transport and metabolism, and secondary metabolite biosynthesis transport and catabolism (Fig. 6). These evidences could explain the decrease of this taxonomic richness. The community profiling results suggest the necessity of further investigations on physiology of the above taxonomies, which will help better assess their exact role in sludge communities exposed to PFOA.
Highly connected microbial taxonomies as responses to PFOA
Highly connected taxonomies play important roles for community structure and function (Banerjee et al. 2018). Azospirillum, Xanthobacter, and Methylophilus were identified as the most connected taxonomies in the PA_O community. The abundance variation of all these three microbes showed a rising trend at the first 60 days and then decreasing until the 90th day, which may be related to the overall gradual increase of PFOA concentration, ultimately exceeding their PFOA tolerance threshold. Azospirillum is known as typical associative N2-fixing bacteria (Steenhoudt and Vanderleyden 2000). It also has been reported to degrade a wide variety of organic pollutant such as polycyclic aromatic hydrocarbons (PAHs) and total petroleum hydrocarbons (TPHs) (Godini et al. 2019; Grobelak et al. 2020). In this study, Azospirillum was the genus with significant difference between PFOA-amended and unamended sludge samples; thus, the Azospirillum could be used as biomarkers to indicate the effect of PFOA. Xanthobacter was also recognized as nitrogen-fixing bacteria (Arzumanyan et al. 1997). Xanthobacter and Methylophilus processed the potential of dehalogenation and might play a key role in the PA_O community. Xanthobacter was reported to catalyze the conversion of halogenated hydrocarbons to their corresponding alcohols through hydrolytic haloalkane dehalogenase (Erable et al. 2005). Methylophilus was characterized as methylotrophic bacteria (Babbitt et al. 2009). Members of Methylophilus were identified to utilize dichloromethane as the sole carbon source with dichloromethane dehalogenase (Chaignaud et al. 2017).
Genera Sporomusa, Petrimonas, and Eubacterium were identified as the most connected taxonomies in the PFOA-anaerobic network (PA_N) by screening the bacteria with high average degree. Experiment results showed that the abundances of Sporomusa and Eubacterium were enriched in PFOA-exposed sludge. The enrichment of Sporomusa and Eubacterium was probably due to their potential to resist PFOA, which was discussed above. The abundance of Petrimonas was slightly enriched with PA_N treatment. As Petrimonas, a popular fermentative bacterium, was able to provide electron donor by degrading glucose for dichlorination (Wang et al. 2018b), Petrimonas in PA_N microbial consortium might exert assistant effects by providing extra nutrients.
When a large proportion of the members of a microbiome community are linked together by positive correlation, it tends to be unstable. This is because in such a community, positively correlated species respond to environmental fluctuations in the same way, leading to positive feedback. On the contrary, negative links representing competitive relationship may stabilize the community by suppressing the positive feedback loop (Coyte et al. 2015). In the PFOA-aerobic network (PA_O), the proportion of positive and negative correlation lines is closer, while in the PFOA-anaerobic network (PA_N), the ratio of negative correlations was much lower than that of positive one. Therefore, it can be inferred that the influence of PFOA on the stability of sludge microbial community under aerobic condition is relatively smaller than that under anaerobic condition.
The impact of PFOA on sludge function
Through functional prediction in the present study, it was found that PFOA exerted negative effects on the abundance of some specific genes at different oxygen levels. Similarly, previous studies had shown that PFASs with long carbon chain length inhibited the expression of genes responsible for defense mechanisms and cell wall membrane envelope biogenesis and also inhibited the enzyme activities of soil as well (Qiao et al. 2018). PFOA declined the translation of some mRNA and then further inhibited the transcription of the proteins (Choi et al. 2013).
In the current study, genes involved in transcription, amino acid transport and metabolism, energy production and conversion, chromatin structure and dynamics, inorganic ion transport and metabolism, and cell cycle control cell division chromosome partitioning were decreased in both aerobic and anaerobic conditions. Amino acid not only acts as the substrate to synthesize protein but also plays a regulatory role as signaling molecules (Sturme et al. 2002). Hence, it could be further speculated that cellular structure of bacterial cells may be damaged, resulting in the inactivation or death of some bacteria. In addition, the inhibition of amino acid transport and metabolism might further interrupt the synthesis and function of protein. The declined abundance of genes linked to energy production and conversion demonstrates that a wide range of microorganisms in activated sludge are susceptible to the selective pressure of PFOA and their ability to metabolize different energy sources was restrained (Milojevic and Weckwerth 2020). The results indicated that PFOA posed a potential threat to sludge function under both aerobic and anaerobic conditions. More evidences at microbial genomic and environmental metagenomic level are expected to uncover the effects of PFOA on AS microorganisms.
Conclusions
Both aerobic and anaerobic microcosm experiments were conducted to study the PFOA stress on microbiome in activated sludge. Substantial shifts in microbial community of activated sludge were observed, supported by the results of the appreciable variations of the structure, richness, and interaction of the bacterial community. After 90 days of coexistence with PFOA, sludge microbiota richness eventually decreased under both aerobic and anaerobic conditions compared with their counterparts amended with methanol only. Functional gene prediction indicated that the long-term exposure of PFOA may inhibit some sludge microbial metabolism processes in activated sludge microbiota including transcription, amino acid transport and metabolism, and energy production and conversion.
However, PFOA additions led to enrichment of potential PFOA-degrader or PFOA-resistant bacteria, such as Rhodopseudomonas, Flavobacterium, and Ignavibacterium under aerobic condition, while under anaerobic condition, the abundance of Eubacterium, Hyphomicrobium, and Methyloversatilis increased. The genera Azospirillum and Sporomusa were identified as the most connected nodes in the PFOA-aerobic and PFOA-anaerobic co-occurrence network, respectively.
Further studies are needed to investigate the potential risk of PFOA and identify the PFOA-degraders and their corresponding degrading mechanism. The bacteria with significant variation responding to the exposure of PFOA in this study are worthy studying as environmental biomarkers to PFOA and further to the PFAS group.
Data availability
The nucleic acid reads of 16S rRNA genes in the current study are available in the NCBI database with the project number PRJNA755805.
References
Abunada Z, Alazaiza MY, Bashir MJ (2020) An overview of per-and polyfluoroalkyl substances (PFAS) in the environment: source, fate, risk and regulations. Water 12:3590
Ammam F, Tremblay P-L, Lizak DM, Zhang T (2016) Effect of tungstate on acetate and ethanol production by the electrosynthetic bacterium Sporomusa ovata. Biotechnol Biofuels 9:163
Arias VA, Mallavarapu M, Naidu R (2015) Identification of the source of PFOS and PFOA contamination at a military air base site. Environ Monit Assess 187:8
Arzumanyan VG, Sakharova ZV, Panikov NS, Vshivtsev VS (1997) Growth and nitrogen-fixing activity of the batch culture of Xanthobacter autotrophicus at various concentrations of dissolved oxygen. Microbiology 66:627–630
Babbitt CW, Pacheco A, Lindner AS (2009) Methanol removal efficiency and bacterial diversity of an activated carbon biofilter. Bioresour Technol 100:6207–6216
Balk M, Mehboob F, van Gelder AH, Rijpstra WIC, Damste JSS, Stams AJM (2010) (Per)chlorate reduction by an acetogenic bacterium, Sporomusa sp., isolated from an underground gas storage. Appl Microbiol Biotechnol 88:595–603
Banerjee S, Schlaeppi K, van der Heijden MGA (2018) Keystone taxa as drivers of microbiome structure and functioning. Nat Rev Microbiol 16:567–576
Bao YX, Li BX, Xie SG, Huang J (2018) Vertical profiles of microbial communities in perfluoroalkyl substance-contaminated soils. Ann Microbiol 68:399–408
Bosso L, Cristinzio G (2014) A comprehensive overview of bacteria and fungi used for pentachlorophenol biodegradation. Rev Environ Sci Biotechnol 13:387–427
Chaignaud P, Maucourt B, Weiman M, Alberti A, Kolb S, Cruveiller S, Vuilleumier S, Bringel F (2017) Genomic and transcriptomic analysis of growth-supporting dehalogenation of chlorinated methanes in methylobacterium. Front Microbiol 8:1600
Chan PWY, Yakunin AF, Edwards EA, Pai EF (2011) Mapping the reaction coordinates of enzymatic defluorination. J Am Chem Soc 133:7461–7468
Chen H, Zou M, Zhou Y, Zeng L, Yang X (2020) Monitoring the nitrous oxide emissions and biological nutrient removal from wastewater treatment: impact of perfluorooctanoic acid. J Hazard Mater 402:123469
Choi SK, Kim JH, Park JK, Lee KM, Kim E, Jeon WB (2013) Cytotoxicity and inhibition of intercellular interaction in N2a neurospheroids by perfluorooctanoic acid and perfluorooctanesulfonic acid. Food Chem Toxicol 60:520–529
Coyte KZ, Schluter J, Foster KR (2015) The ecology of the microbiome: Networks, competition, and stability. Science 350:663–666
Erable B, Goubet I, Lamare S, Seltana A, Legoy MD, Maugard T (2005) Nonconventional hydrolytic dehalogenation of 1-chlorobutane by dehydrated bacteria in a continuous solid-gas biofilter. Biotechnol Bioeng 91:304–313
Gao P, Song B, Xu R, Sun X, Lin H, Xu F, Li B, Sun W (2021) Structure and variation of root-associated bacterial communities of Cyperus rotundus L. in the contaminated soils around Pb/Zn mine sites. Environ Sci Pollut Res 28:58523–58535
Godini K, Samarghandi MR, Tahmasebi H, Zarei O, Karimitabar Z, Yarahmadi Z, Arabestani MR (2019) Biochemical and molecular characterization of novel PAH-degrading bacteria isolated from polluted soil and sludge. Pet Sci Technol 37:1763–1769
Grobelak A, Worwag M, Grosser A (2020) Removal of total petroleum hydrocarbons from wastewater and sewage sludge generated in oil separators and evaluation of the process efficiency. Desalination Water Treat 199:205–211
Han F, Li X, Zhang M, Liu Z, Han Y, Li Q, Zhou W (2021) Solid-phase denitrification in high salinity and low-temperature wastewater treatment. Bioresour Technol 341:125801.
Higgins CP, Field JA, Criddle CS, Luthy RG (2005) Quantitative determination of perfluorochemicals in sediments and domestic sludge. Environ Sci Technol 39:3946–3956
Hooper LV, Macpherson AJ (2010) Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol 10:159–169
Houtz EF, Higgins CP, Field JA, Sedlak DL (2013) Persistence of perfluoroalkyl acid precursors in AFFF-impacted groundwater and soil. Environ Sci Technol 47:8187–8195
Jiang X, Mingchao MA, Jun LI, Anhuai LU, Zhong Z (2008) Bacterial diversity of active sludge in wastewater treatment plant. Earth Sci Front 15:163–168
Kassab G, Halalsheh M, Klapwijk A, Fayyad M, van Lier JB (2010) Sequential anaerobic-aerobic treatment for domestic wastewater - a review. Bioresour Technol 101:3299–3310
Keith RC, John GH (2001) Induction of a stress protein in Eubacterium biforme by the surfactant CTAB. Microb Ecol Health Dis 13:229–233
Kuczynski J, Stombaugh J, Walters WA, González A, Caporaso JG, Knight R (2011) Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Bioinformatics 10:Unit 10.7
Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Thurber RLV, Knight R, Beiko RG, Huttenhower C (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31, 814-+
Lehmler HJ (2005) Synthesis of environmentally relevant fluorinated surfactants - a review. Chemosphere 58:1471–1496
Li Y, Lin H, Gao P, Yang N, Xu R, Sun X, Li B, Xu F, Wang X, Song B, Sun W (2021) Variation in the diazotrophic community in a vertical soil profile contaminated with antimony and arsenic. Environ Pollut 291:118248
Li Y, Lin H, Gao P, Yang N, Xu R, Sun X, Li B, Xu F, Wang X, Song B, Sun W (2021b) Synergistic impacts of arsenic and antimony co-contamination on diazotrophic communities. Microb Ecol. https://doi.org/10.1007/s00248-021-01824-6
Li Y, Zhang M, Xu R, Lin H, Sun X, Xu F, Gao P, Kong T, Xiao E, Yang N, Sun W (2021) Arsenic and antimony co-contamination influences on soil microbial community composition and functions: relevance to arsenic resistance and carbon, nitrogen, and sulfur cycling. Environ Int 153:106522
Lindstrom AB, Strynar MJ, Libelo EL (2011) Polyfluorinated compounds: past, present, and future. Environ Sci Technol 45:7954–7961
Liou JSC, Szostek B, DeRito CM, Madsen EL (2010) Investigating the biodegradability of perfluorooctanoic acid. Chemosphere 80:176–183
Long X, Tang R, Zhan B, Fang Z, Xie C, Li Y (2019) Biological nitrogen removal in a flow-separating biochemical reactor with coral sand. Pol J Environ Stud 28:3767–3778
Mariussen E (2012) Neurotoxic effects of perfluoroalkylated compounds: mechanisms of action and environmental relevance. Arch Toxicol 86:1349–1367
Milojevic T, Weckwerth W (2020) Molecular mechanisms of microbial survivability in outer space: a systems biology approach. Front Microbiol 11:22
Neufeld DA, Wolfire MG (2009) The chemistry of interstellar molecules containing the halogen elements. Astrophys J 706:1594–1604
Nobels I, Dardenne F, De Coen W, Blust R (2010) Application of a multiple endpoint bacterial reporter assay to evaluate toxicological relevant endpoints of perfluorinated compounds with different functional groups and varying chain length. Toxicol in Vitro 24:1768–1774
Parks DH, Tyson GW, Hugenholtz P, Beiko RG (2014) STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30:3123–3124
Qiao W, Xie Z, Zhang Y, Liu X, Xie S, Huang J, Yu L (2018) Perfluoroalkyl substances (PFASs) influence the structure and function of soil bacterial community: greenhouse experiment. Sci Total Environ 642:1118–1126
Senevirathna STMLD, Krishna KCB, Mahinroosta R, Sathasivan A (2022) Comparative characterization of microbial communities that inhabit PFAS-rich contaminated sites: a case-control study. J Hazard Mater 423:126941
Sinclair E, Kannan K (2006) Mass loading and fate of perfluoroalkyl surfactants in wastewater treatment plants. Environ Sci Technol 40:1408–1414
Steenhoudt O, Vanderleyden J (2000) Azospirillum, a free-living nitrogen-fixing bacterium closely associated with grasses: genetic, biochemical and ecological aspects. FEMS Microbiol Rev 24:487–506
Sturme MHJ, Kleerebezem M, Nakayama J, Akkermans ADL, Vaughan EE, de Vos WM (2002) Cell to cell communication by autoinducing peptides in gram-positive bacteria. Antonie Van Leeuwenhoek 81:233–243
Sun WM, Sun XX, Li BQ, Xu R, Young LY, Dong YR, Zhang MM, Kong TL, Xiao EZ, Wang Q (2020) Bacterial response to sharp geochemical gradients caused by acid mine drainage intrusion in a terrace: relevance of C, N, and S cycling and metal resistance. Environ Int 138:12
Sun XX, Song BR, Xu R, Zhang MM, Gao P, Lin HZ, Sun WM (2021) Root-associated (rhizosphere and endosphere) microbiomes of the Miscanthus sinensis and their response to the heavy metal contamination. J Environ Sci (china) 104:387–398
Sun YJ, Wang TY, Peng XW, Wang P, Lu YL (2016) Bacterial community compositions in sediment polluted by perfluoroalkyl acids (PFAAs) using Illumina high-throughput sequencing. Environ Sci Pollut Res 23:10556–10565
Wang C, Wu L, Zhang YT, Wei W, Ni BJ (2021) Unravelling the impacts of perfluorooctanoic acid on anaerobic sludge digestion process. Sci Total Environ 796:149057
Wang H, Zhang W, Ye Y, He Q, Zhang S (2018a) Isolation and characterization of Pseudoxanthomonas sp. strain YP1 capable of denitrifying phosphorus removal (DPR). Geomicrobiol J 35:537–543
Wang J, Zhou BY, Ge RJ, Song TS, Yu JP, Xie JJ (2018b) Degradation characterization and pathway analysis of chlortetracycline and oxytetracycline in a microbial fuel cell. RSC Adv 8:28613–28624
Wang Q, Tsui MMP, Ruan Y, Lin H, Zhao Z, Ku JPH, Sun H, Lam PKS (2019) Occurrence and distribution of per- and polyfluoroalkyl substances (PFASs) in the seawater and sediment of the South China sea coastal region. Chemosphere 231:468–477
Wang XH, Xia Y, Wen XH, Yang YF, Zhou JZ (2014) Microbial community functional structures in wastewater treatment plants as characterized by GeoChip. PLoS ONE 9:10
Wolfe GV, Fitzhugh C, Almasary A, Green A, Bennett P, Wilson M, Siering P (2014) Microbial heterotrophic production in an oligotrophic acidic geothermal lake: responses to organic amendments and terrestrial plant litter. FEMS Microbiol Ecol 89:606–624
Xu R, Huang D, Sun X, Zhang M, Wang D, Yang Z, Jiang F, Gao P, Li B, Sun W, Villanueva L (2021a) Diversity and metabolic potentials of As(III)-oxidizing bacteria in activated sludge. Appl Environ Microbiol 87:e01769-e1821
Xu R, Li BQ, Xiao EZ, Young LY, Sun XX, Kong TL, Dong YR, Wang Q, Yang ZH, Chen L, Sun WM (2020) Uncovering microbial responses to sharp geochemical gradients in a terrace contaminated by acid mine drainage. Environ Pollut 261:114226
Xu R, Sun X, Häggblom MM, Dong Y, Zhang M, Yang Z, Xiao E, Xiao T, Gao P, Li B, Sun W (2021b) Metabolic potentials of members of the class Acidobacteriia in metal-contaminated soils revealed by metagenomic analysis. Environ Microbiol 24:803–818
Xu R, Tao W, Lin HZ, Huang DY, Su PZ, Gao P, Sun XX, Yang ZH, Sun WM (2021c) Effects of perfluorooctanoic acid (PFOA) and perfluorooctane sulfonic acid (PFOS) on soil microbial community. Microb Ecol 83:929–941
Xu R, Zhang M, Lin H, Gao P, Yang Z, Wang D, Sun X, Li B, Wang Q, Sun W (2021d) Response of soil protozoa to acid mine drainage in a contaminated terrace. J Hazard Mater 421:126790
Yang GJ, Zhang N, Yang JN, Fu QZ, Wang Y, Wang DB, Tang L, Xia JF, Liu XR, Li XM, Yang Q, Liu YW, Wang QL, Ni BJ (2020) Interaction between perfluorooctanoic acid and aerobic granular sludge. Water Res 169:11
Yu H, Feng CH, Liu XP, Yi XY, Ren Y, Wei CH (2016) Enhanced anaerobic dechlorination of polychlorinated biphenyl in sediments by bioanode stimulation. Environ Pollut 211:81–89
Yu XL, Nishimura F, Hidaka T (2018) Impact of long-term perfluorooctanoic acid (PFOA) exposure on activated sludge process. Water Air Soil Pollut 229:12
Zareitalabad P, Siemens J, Hamer M, Amelung W (2013) Perfluorooctanoic acid (PFOA) and perfluorooctanesulfonic acid (PFOS) in surface waters, sediments, soils and wastewater - a review on concentrations and distribution coefficients. Chemosphere 91:725–732
Zhang DQ, Zhang WL, Liang YN (2019) Distribution of eight perfluoroalkyl acids in plant-soil-water systems and their effect on the soil microbial community. Sci Total Environ 697:11
Zhang K, Liu YH, Luo HB, Chen Q, Zhu ZY, Chen W, Chen J, Ji L, Mo Y (2017a) Bacterial community dynamics and enhanced degradation of di-n-octyl phthalate (DOP) by corncob-sodium alginate immobilized bacteria. Geoderma 305:264–274
Zhang L, Fu G, Zhang Z (2020) Long-term stable and energy-neutral mixed biofilm electrode for complete nitrogen removal from high-salinity wastewater: mechanism and microbial community. Bioresour Technol 313:123660
Zhang LL, Li LJ, Pan XG, Shi Z, Feng XH, Gong B, Li J, Wang LS (2018) Enhanced growth and activities of the dominant functional microbiota of chicken manure composts in the presence of maize straw. Front Microbiol 9:11
Zhang S, Merino N, Wang N, Ruan T, Lu X (2017b) Impact of 6:2 fluorotelomer alcohol aerobic biotransformation on a sediment microbial community. Sci Total Environ 575:1361–1368
Zhang W, Zhang YT, Taniyasu S, Yeung LWY, Lam PKS, Wang JS, Li XH, Yamashita N, Dai JY (2013) Distribution and fate of perfluoroalkyl substances in municipal wastewater treatment plants in economically developed areas of China. Environ Pollut 176:10–17
Zhou A, Liu W, Varrone C, Wang Y, Wang A, Yue X (2015) Evaluation of surfactants on waste activated sludge fermentation by pyrosequencing analysis. Bioresour Technol 192:835–840
Funding
This work was supported by the Science and Technology Planning Project of Guangzhou (grant no. 202002020072), GDAS’ Project of Science and Technology Development (grant nos. 2020GDASYL-20200102015, 2020GDASYL-20200102018, 2020GDASYL-20200102014, 2022GDASZH-2022010106, and 2021GDASYL-20210302003), the National Natural Science Foundation of China (grant nos. 42007357, 52170035, and U20A20109), and Guangdong Basic and Applied Basic Research Foundation (grant no. 2021A1515011374).
Author information
Authors and Affiliations
Contributions
Duanyi Huang wrote the original draft, and Zhaohui Yang, Hanzhi Lin, and Weimin Sun contributed to the conception of the study. Rui Xu, Xiaoxu Sun, Yongbin Li, and Duanyi Huang performed the experiment and data analysis. Rui Xu, Hanzhi Lin, Enzong Xiao, Zhimin Xu, Qi Wang, and Gao Pin offered comments for revisions to the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Responsible Editor: Robert Duran
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Huang, D., Xu, R., Sun, X. et al. Effects of perfluorooctanoic acid (PFOA) on activated sludge microbial community under aerobic and anaerobic conditions. Environ Sci Pollut Res 29, 63379–63392 (2022). https://doi.org/10.1007/s11356-022-18841-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-022-18841-8