Abstract
Silver nanoparticles were fabricated in the presence and absence of light with silver nitrate and aqueous extract of Prunus cerasifera leaf via facile and one-pot green method. P. cerasifera leaf extract reduced and stabilized the nanoparticles with phytometabolites expunging the need for addition of external reducing agents. Optimized silver nanoparticle syntheses was done with variations in leaf extract concentration, time, temperature, and molarity for deciphering the photocatalytic, antifungal, and antibacterial potential of synthesized nanoparticles. Optical, compositional, and morphological analyses of the synthesized nanoparticles were done by UV-visible spectrometry (UV-Vis), Fourier transform infrared spectroscopy (FTIR), atomic force microscopy (AFM), scanning electron microscopy (SEM), and X-ray diffraction (XRD). Formation of silver nanoparticles was confirmed firstly through UV-Vis by exhibition of peaks with 400–450 nm. FTIR confirmed the presence of major organic groups responsible for reduction of nanoparticles. AFM confirmed the spherical morphology of the synthesized nanoparticles with remarkable dispersion without any agglomeration. Phytochemical analysis for P. cerasifera leaf metabolites was done by GC-MS. Spherical nanoparticles having a size range of 57–144 nm were obtained with face-centered cubic crystals. The average crystallite size obtained from XRD spectra was 2.34 nm. Enhanced photocatalytic first-order kinetics were obtained for persistent organic pollutants, i.e., crystal violet, methylene blue, and malachite green (R2 = 0.99, 0.99, 0.98) in less than 15 min. Biomedical and agricultural significance as an antibiotic drug and utilization as a fungicides substitute was explored against nine resistant microbes. Statistically significant variations were analyzed via one-way analysis of variance (ANOVA) and Kruskal-Wallis test and specific multi comparison tests. Active to highly active inhibition zones manifested the use of biogenic silver nanoparticles as potential candidate for applications in biological arenas and as environmental remediators.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Nanotechnology and biology when unified, known as nanobiotechnology is a significant field associated with the fabrication of bioinspired and sustainable technology through which nanoscale materials are developed for different applications. Currently, nanotechnology has been greatly employed for balancing the ecological functioning via removal of obnoxious chemicals invading environment and deteriorating its quality explored in different investigations (Shaheen et al. 2016; Zahra et al. 2017; Amjad et al. 2019; Ali et al. 2019). Nanomaterials with desired characteristics and eco-friendly nature have brought a nanorevolution. However, such revolution is also marked by some concerns from environmentalists when the mode of synthesis is only limited to chemico-physical modes particularly aimed at medical applications (Nayak et al. 2019; Ahmad and Jaffri 2018b; Jaffri and Ahmad 2018c). Among different metallic nanoparticles synthesized, silver nanoparticles are of prime significance for their utilization in a myriad of medical, industrial, and eco-remediation purposes (Qasim Nasar et al. 2019). Variety of physicochemical methods employing synthetic reducing agents and organic solvents with elevated toxicities have been used for silver nanoparticle (SNP) synthesis. Such methods have detrimental impacts on environment despite the fact that they produce nanoparticles with higher purity indices (Jaffri and Ahmad 2018d). Physical synthetic modes are known for utilization of deposition of material in gaseous phase, laser burning, grinding in a mechanical mode, or ionic sputtering. In such methods, toxic substances are released into the environment in addition to the production of large-sized nanoparticles, e.g., 90–110 nm ranging SNPs were synthesized by DC magnetron sputtering by using anodic alumina template in nanoporous form (Gohil et al. 2007). Physical synthetic routes have a requirement for the elevated reaction conditions that not only employ heavier capital costs but also need installation of advanced machineries. Similarly, chemical modes cannot be completed without addition of the environmentally detrimental and expensive reducing agents. In case of chemical methods, solution left from reaction is usually environmentally obnoxious, harmful, or possess an elevated acidity or alkalinity, e.g., 10–250 nm SNPs were synthesized using poly[N-vinylpyrolidone] (PVP) as stabilizer in addition to two other chemical reducing agents, i.e., ethylene glycol and glucose. Compulsorily, the residual substances in form of PVP, glycol, and glucose were released to the ecosphere having considerable harmfulness towards environment (Samadi et al. 2010). Nevertheless, current silver nanoparticles are used in different applications particularly at commercialized scales; the fabrication modes are majorly physical and chemicals ones.
Marked toxicities incurred to environment due to these modes have recently attracted the scientific community to synthesize silver nanoparticles using biological entities in compliance with the principles of the green chemistry. Particularly, these challenges of toxicity and cost have now been efficiently addressed via use of plant biochemicals as a green alternative to the synthetic reducing and capping agents. Phytosynthesis is congruent with major principles of green chemistry for selection of highly abundant and nontoxic reducing agents (Ismail et al. 2019; Heydari et al. 2019), where plant material has been frequently utilized for various traditional and industrial purposed in form of food, pharmacy, textiles, perfumes, additives, etc. Plant material has recently found an interesting application in the nanotechnology for meeting the sustainability needs by acting as an alternative to synthetic reducing and stabilizing agents (Shaheen and Ahmad 2019; Bartosiak et al. 2019; Ahluwalia et al. 2018; Jaffri and Ahmad 2018a). Particularly, silver nanoparticle synthesis using different plant parts has been an attractive route due to its rapidity, ecological integrity, pathogenicity free, lower capital costs, and above all, single-step method in which plant phytochemicals are used for reduction and stabilization. Botanical extracts used for reduction and stabilization of nanoparticles and ensures the prevention of agglomeration (Gulbagca et al. 2019; Hussain et al. 2019).
Current environmental situation is comparatively worse than previous eras when human was not dependent upon synthetic commodities and environmental integrity was not challenged. Among different environmental challenges being faced by humans, the contamination of water bodies due to industrial discharge is most formidable due to complexity of the matrix. Such contaminants are mostly persistent in nature and do not degrade easily. Persistent organic pollutants are not easily degraded and mineralized by conventional methods, i.e., osmotic, coagulative, filtration, etc. due to their intricate ringed structure and elevated stability. Thus, the transformation of such obnoxious chemicals must be done through an efficient method. Water contaminants fall in different categories but textile dyes are known for making a major portion of the polluted matrix. World Bank reports are indicative of the fact that up to 20% water contamination is attributed to textile industry discharge. Textile industry releases poorly degradable substances, e.g., dye stuff and detergents. Variety of physical, chemical, biological, and combination of these methods have been devised for such contaminants, but cost-effectiveness and stability of the remediation technology remains an area of growing research.
Degradation of different contaminants through utilization of solar light and a nanocatalyst in a photolytic assay has been envisaged as an effective management strategy since few decades. In this regard, different metal and metal oxide nanoparticles have been utilized for degradation of different dyes. SNPs are efficient green photocatalysts which can degrade a variety of such persistent chemicals via series of chemo catalytic reactions. Usually, ultraviolet (UV) radiations are made use of in photo-degradative pathways but there are some concerns regarding the practical and environmental aspects in terms of commercialization since the solar energy region occupied by UV is only 4% in form of renewable resource and only rest of the 43% is visible light. Such drawback can be overcome by development of such photocatalysts that are highly acquiescent to the visible light so that photocatalysis will be efficient and rapid. Furthermore, photocatalysts are frequently used due to their simple fabrication modes, negligible cost, and regeneration of the photocatalysts. Such aspects are further made desirable by utilization of nanoparticles as nanophotocatalysts. In this way, variety of organic dyes can be degraded in a quick and efficient way since there is an annual production of 450,000 t, out of which major chunk (up to 11%) is lost during manufacturing and application modes. Remediation of such organic dyes is incumbent due to their inherent toxicity towards environment and carcinogenicity towards living organisms.
Among different dyes used in textile industries and other biomedical purposes, some dyes are particularly toxic due to their physicochemical properties. Crystal violet (CV), methylene blue (MB), and malachite green (MG) are one of those dyes. Both CV and MB dyes are used for coloring purposes while MG is used for extermination of different parasites, protozoans, and fungal cultures present in water which is a matrix for fish breeding. Despite higher industrial utilization, these synthetic dyes are highly harmful. CV, a tri-phenylmethane dye (Mittal et al. 2010), is known as an eye irritant and possess hazardous nature. Prolonged exposure to CV is associated with the corneal and conjunctival damage in addition to possibility of permanent blindness. Furthermore, the severe cases are also marked by renal and respiratory failure. Despite carcinogenicity associated with CV, it has been frequently used as a biological stain and as a dermatological agent in addition to its textile-based applications. MB is a cationic dye and has an essential toxicity towards human beings and causes different health issues, e.g., visual impairment, methemoglobinemia, vomiting, and nausea (Nafea and Hassan 2019).
Photocatalytic degradation of MB is essential for abatement of its toxicity in aquatic media (Recio-Sánchez et al. 2019). Antiparasitic, antifungal, and antiprotozoan aspects of MG are well established, but MG has been heavily criticized for its toxicity and causing different mammalian impairments, e.g., mutational, formative aberrations and organ damage (Jindal and Sinha 2019; Li et al. 2019). MG bioaccumulation can be due to the human exposure by ingesting the MG contaminated fish or it can also occur due to workplace exposure to dye or aquaculture industry. Furthermore, aquatic microbial communities are also adversely affected due to MG. Different parameters play an important role in the toxicity enhancement of MG as a result of exposure, e.g., time, temperature, and MG quantity. Since the reduction, product of MG is leuco-malachite green (LeucoMG) which is a highly toxic daughter product and poses challenges in elimination (Zhou et al. 2019).
In addition to this environmental remediation, SNPs possess inherent therapeutic potential. Numerous germicidal drugs have been found ineffective due to development of resistance among microbes widely distributed. Despite higher advances done in research and remedy of such diseases, such microbes are responsible for elevating death rate by 25% on a global scale (Fears and ter Meulen 2014). Thus, SNPs synthesized via novel routes have been found as potential candidate (Jaffri and Ahmad 2017). The inherent antimicrobial potential of phytosynthetic Ag NPs is augmented due to presence of plants phytochemicals which are themselves reservoirs for antimicrobial components (Mutua et al. 2017). The resistance exhibited by different microbial strains towards drugs, e.g., penicillin and tetracycline, is easily combated by silver nanoparticles for comparatively expanded time durations. Particularly, smaller size ranges obtained by greener synthesis has enabled the nanoparticles to better invade the microbial cellular membrane leading to cytoplasmic nanoparticle aggregation and disruption of enzyme-driven activity. This activity leads to the ultimate cell death by reactive oxygen species (ROS) regeneration (Ferreira et al. 2019). For such advantageous features, silver nanoparticles have been frequently employed in different biomedical products.
P. cerasifera is a wild as well as domesticated shrub found in European and Central Asia. The conspicuous antimicrobial and antioxidant activity of P. cerasifera leaves makes them riveting reducing agents to be used for SNP fabrication. Despite the fact that P. cerasifera possess such a unique blend of phytochemicals and other auspicious aspects, they have not been used on industrial scale for the welfare of human beings. Parachinar region of Pakistan is highly abundant in wild variety of P. cerasifera which can be used for large-scale production of SNPs in a highly sustainable mode. Studies with leaves of P. cerasifera have never been reported expressing the underutilization and scarcity of information about such a prodigious blend of reducing phytochemicals. Thus, present investigation has been designed to employ this stupendous blend of reducing chemicals in P. cerasifera for the synthesis of silver nanoparticles for first time in a cost-effective, facile, and sustainable synthetic mode. Current work is novel and different from all the reported biogenically synthesized nanoparticles especially silver, in selection of the P. cerasifera leaves for the first time, its utilization in exploration of the antimicrobial potential and photocatalysis. Synthetic mode comprised of fabrication of dark prunosilver nanoparticles (DPSNPs) and light prunosilver nanoparticles (LPSNPs) at variable optimization levels with P. cerasifera leaf extract (LE). Synthesized nanoparticles were characterized via ultraviolet–visible spectroscopy (UV-Vis), Fourier transform infrared spectroscopy (FTIR), scanning electron microscope (SEM), atomic force microscopy (AFM), and X-ray powder diffraction (XRD). P. cerasifera leaf phytometabolites were analyzed by gas chromatography–mass spectrometry (GC-MS). Furthermore, SNPs were applied for the photocatalysis of persistent organic pollutants, i.e., crystal violet (CV (C25N3H30Cl)), methylene blue (MB (C16H18ClN3S)) and malachite green (MG (C23H25ClN2)) in addition to inhibition of nine antibiotic resistant pathogens, i.e., X. citri (XC), P. syringae (PS), A. niger (AN), A. flavus (AF), A. fumigatus (AFG), A. terreus (AT), P. chrysogenum (PC), F. solani (FS), and L. theobromae (LT).
Materials and methods
Materials
Silver nitrate, potassium bromide, methanol, acetone, dichloromethane, ethyl acetate, and dyes, i.e., CV, MB, and MG were purchased from Sigma-Aldrich Chemicals (USA). Nutrient agar (NA) and potato dextrose agar (PDA) were purchased from Merck (Germany). Deionized water (DW) and methanol were used for preparation of all solutions. All apparatus was sterilized and oven dried prior to experimentation. All chemicals used for experimentation were of analytical grade and were employed without any further purification.
P. cerasifera leaf extraction
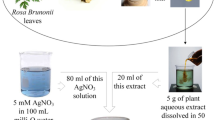
Fresh and healthy leaves of P. cerasifera were sampled in July 2016 from Alizai region of Parachinar valley, upper Kurram agency, Pakistan (Fig. 1). Leaves were washed with tap water and deionized water for removal of any possible impurities or dust particles being deposited on it. Leaves were spread on clean sheets in air and shade dried followed by drying in oven at 60 °C for 3 h for complete moisture removal. Leaves were finely powdered with help of pestle and mortar and were passed through sieve for obtaining complete powder. Leaf aqueous extract was prepared by boiling of 25 g of leaf powder in 250 mL DW in Erlenmeyer flask at 60 °C for 10 min. Suspension was stirred vigorously for 400 rpm. Leaf extract was sonicated at 20-kHz frequency with 10 s pulses. Sonication duration was 30 min using ultrasonic liquid processor (Sonics Vibra-Cell, USA). The resultant mixture was exposed to centrifugation at 8000 rpm for 25 min for complete removal of insoluble impurities. Extract was filtrated with Whatman No. 1 filter paper twice and stored at 4 °C for future use as a reducing agent in SNP optimized syntheses.
Syntheses and optimization
Green synthesis of silver nanoparticles was done by utilization of P. cerasifera leaf extract as a reducing cum stabilizing agent. Syntheses were done in both presence and absence of light to comprehend the impact and interplay of light in silver nanoparticle production. For both DPSNPs and LPSNPs, same optimization parameters were adopted for comparison. Optimized syntheses of DPSNPs and LPSNPs were done for LE conc., time of incubation, temperature, and AgNO3 solution molarity (Fig. 2). For LE concentration optimization, different concentrations were treated with AgNO3 1 mM solution. Addition of LE to AgNO3 was followed by magnetic stirring at 300 rpm in dark ambiance for DPSNPs, and same method was followed for fabrication of LPSNPs in sunlight. LE conc. variation comprised of 100, 150, 200, 250, and 300 mL in 500 mL of 1 mM AgNO3 solution at 60 °C. Incubation time required for SNP synthesis was also observed for time. Three hundred milliliters of LE in 500 mL of 1 mM AgNO3 solution was given incubation times of 30, 60, 90, 120, and 150 min at 60 °C. Temperature variation was also observed for 300 mL of LE in 500 mL of 1 mM AgNO3 solution at 40, 60, 80, 100, and 120 °C. AgNO3 solution molarity variation comprised of 300 mL of LE in 500 mL of 0.02, 0.05, 0.2, 0.5, and 1 mM. For LPSNPs, no additional temperature was provided for variation of LE conc., time of incubation, and AgNO3 solution molarity; however, the temperatures were varied for third parameter. Optimized batches for dark and light syntheses expressed successful SNP formation in all cases exhibited via reaction mixture color transformation. Solution color changed from yellow to dark brown was indicative of SNP formation which was monitored via UV–Vis Spectrophotometer (1602, Biomedical Services, Madrid, Spain). Synthesized nanoparticles were also sonicated for 10 min and then centrifuged at 8000 rpm for 20 min. It was followed by supernatant throwing away for collection of SNP pellet. SNP pellet was subjected to thrice sterile double-distilled water (SDDW) washing and dried for 24 h at 60 °C.
Characterization of SNPs
SNPs synthesized with P. cerasifera LE confirmation was done via a variety of spectroscopic and imaging techniques. SNPs were analyzed initially upon color transformation via UV-Vis spectrophotometer in the range of 200–800 nm with a scan rate 2 nm/s. SNPs (0.5 mL) were diluted with 2.5 mL of DW. SNPs were centrifuged at 6000 rpm for 10 min (C0060-230V, Labnet International, Inc. USA). Supernatant was discarded and SNPs were dried and stored for further analysis. SNPs were analyzed for functional groups via FTIR spectrophotometer (8400, Shimadzu, Japan) between wavenumber 4000 and 400 cm−1 after palletization of SNPs with overnight dried KBr. Crystalline features of SNPs stabilized with P. cerasifera LE was analyzed via Bruker AXS D-8 powder X-ray diffractometer (Shimadzu, Kyoto, Japan), operated at 40 kV, 20 mA, with CuKα radiation λ = 1.5406 Å. Morphological and size ranges were analyzed via scanning electron microscopy (SEM JOEL JSM-6490, Germany) and atomic force microscopy (AFM, Park systems, Suwon, Korea). Furthermore, P. cerasifera LE was analyzed for chemical composition via GC-MS (QP5050, Shimadzu, Japan) by extraction with methanol, acetone, dichloromethane, and ethyl acetate for obtaining maximum yield.
SNP-driven nanophotocatalysis
Nanophotocatalytic potential of the SNPs was analyzed against three dyes. All the degradative experiments were performed in triplicate, and mean of all the results have been shown. Prior to nanophotocatalysis, experiments were carried out for getting the optimized conditions and concentrations of dyes and SNP photocatalyst amount was kept be fixed for all three dyes. Photocatalysis was done by mixing 0.50 mL (0.01 M) of CV, MB, and MG; 1.68 mL DW; 1 mL of NaBH4 (0.05 M); and 0.02 mL (50 mg/mL) of SNPs kept in dark with 30-min constant magnetic stirring for establishment of adsorption-desorption equilibrium between dye molecules and photocatalyst surface. Control setup was not added with any quantity of SNPs. Both assemblies were kept in the sunlight for 20 min on a clear sunny day from 12: 30–12: 50 PM with sunlight having an average intensity 70–74 Klux (LT300, Extech, UK). Sunlight exposure triggered gradual decolorization in experimental setup while no change was observed for control set up. UV-Vis was used in range of 200–700 nm (scan rate 2 min/s) to evaluate the time dependent degradative response of dyes towards SNP nanophotocatalyst. Five milliliters of solution was withdrawn and analyzed at different time intervals, and overlays were plotted for alleviating absorbance (Ahmad and Jaffri 2018a). Degradation percentages were calculated by:
where Ao is the absorbance of dye before irradiation and At is the absorbance of dye after irradiation for time interval t.
Antimicrobial assay
Antimicrobial potential of DPSNPs and LPSNPs was evaluated against X. citri, P. syringae, A. niger, A. flavus, A. fumigatus, A. terreus, P. chrysogenum, F. solani, and L. theobromae by standard Kirby–Bauer disc diffusion assay. Fungal and bacterial strains were obtained from Mycology lab, Fatima Jinnah Women University, Rawalpindi, Pakistan. Microbial strains with known pathological history were used in the present research have been isolated from soils of different regions. DPSNP and LPSNP stock solution were prepared by dissolution of the nanoparticles in methanol for obtaining final concentration of 100 mg/mL and 50 μL of the volumes 2000, 4000, 6000, and 10,000 μg/mL were used for disc loading. Stock solution was sonicated for 30 min and used instantaneously for assays. SNP inhibition was compared with AgNO3 salt solution, standard antibiotic drugs ampicillin and amphotericin B for bacterial and fungal strains, respectively. Antibacterial assay was performed by swabbing the bacterial inocula onto presterilized petri dish containing NA after standardization to a density corresponding to BaSO4 standard of 0.5 McFarland units. Sterilized and round 0.45 cm disc loaded with 50 μL of the 10,000 μg/mL of ampicillin, 10,000 μg/mL of LE and 2000, 4000, 6000, and 10,000 μg/mL DPSNPs and LPSNPs, respectively, were inserted into inoculated plates and incubated for 24 h at 37 °C in incubator (Sanyo MR-153, GeminiBV, Netherlands) followed by measurement of zones of inhibition (ZOIs) in mm after 24 h. Antifungal assay involved the production of fresh fungal mycelia on PDA plates at 25 ± 1 °C followed by incubation for 5 days. Sterilized and round 0.45 cm disc loaded with 50 μL of the 10,000 μg/mL of Amphotericin B, 10,000 μg/mL of LE and 2000, 4000, 6000, and 10,000 μg/mL DPSNPs and LPSNPs, respectively, were inserted into inoculated plates and incubated for 24 h at 28 °C followed by measurement of ZOIs in mm after 48 h (Jaffri and Ahmad 2018b).
Statistical analysis
All assays were performed in triplicate, and final values obtained for zone of inhibition (mm) were mentioned as means and standard deviations. P > 0.05 were considered statistically significant. Inhibition was statistically evaluated by one-way analysis of variance (ANOVA) and Kruskal-Wallis test. Values were compared for Tukey HSD, Scheffé, Bonferroni, and Holm tests for confirming the specificity of statistical significance among different groups.
Results and discussions
SNP green synthesis
Current study is the first elucidation of the biogenic synthesis of SNPs in presence and absence of light by utilization of the reducing phytometabolites of the leafy parts of P. cerasifera. Phytometabolites not only reduced the silver salt to SNPs but also acted as a capping agent nullifying the need for any external agents. P. cerasifera, an angiospermic shrub found in Central Asia, has an abundant blend of phytochemicals with reducing and stabilizing potential. P. cerasifera exceeds high phenol and antioxidants containing fruits like strawberries, apples, and oranges because P. cerasifera has an elevated quantity of anthocyanins and phenols (Huo et al. 2019). Furthermore, the presence of organic acids, carbohydrates, aromatic and fibrous chemicals as expressed by P. cerasifera leaf GC-MS profile makes it a potential candidate for bioprospecting in SNP synthesis via facile and one-pot method. Though, several studies did molecular characterization of P. cerasifera leaf specifically; however, its potential in biogenic synthesis has never been reported with leaf. Thus, signifying the underutilization of such a miraculous blend of reducing phytometabolites. Relatively higher abundance of P. cerasifera can be attributed to the rapid growth and adaptability of P. cerasifera towards extreme environmental conditions. Thus, P. cerasifera leaf metabolites vanishes the need for chemical reducing agents that not only pose challenges to environmental integrity but are also not favorable in terms of cost benefit analysis. In contrast to these chemical reducing agents, P. cerasifera leaves that are abundantly available can reduce SNPs in short duration unlike microbe-mediated biogenic synthesis that needs culture maintenance. Thus, the choice of P. cerasifera for SNPs production can be scaled up on commercial level. Present study has been undertaken to synthesize SNPs utilizing the phytochemicals of P. cerasifera leaf extract in dark as well as sunlight. P. cerasifera leaf stabilized SNPs were also used for photocatalysis and inhibition of resistant human and plant pathogens.
Characterization of SNPs
UV-Vis
P. cerasifera leaves are a blend of phytochemicals with reducing nature. Such phytochemicals can be extracted to be used for green synthesis of SNPs in a facile and greener way. Green synthesis with P. cerasifera LE was done in different variations for obtaining the optimized reaction conditions in dark and light. Optimization was carried out for exploring the effect of LE conc. reacting with silver salt molar solution, time of incubation, temperature, and molarity of the AgNO3 solution on SNP synthesis in dark as well as light conditions. The reducing and capping potential of P. cerasifera leaf was proved by synthesis of SNPs with addition of only AgNO3 by varying experimental conditions without any addition of environmentally perilous chemicals. Addition of AgNO3 to LE and consequent SNP formation was primarily monitored by color change of reaction mixture from light yellow to brown (Alavi and Karimi 2017) being intensified as the reaction time proceeded. Color transformation is governed by the surface plasmon resonance (SPR) excitation for metal nanoparticles (Samari et al. 2019; Gavamukulya et al. 2019; Qais et al. 2019). SPR peaks (Table 1) for DPSNPs and LPSNPs show different peaks with variable shapes for all optimization parameters. All these variation produced variable SPR peaks when analyzed on UV-Vis after visual detection of color transformation from yellow to dark brown. SNP colloidal solution color transformation arise due to SPR excitation with SNPs as SNPs possess free electron which gives SPR absorption band by the dint of collective oscillations of SNPs’ electron with light waves in a resonating behavior.
Higher stability and negligible agglomeration was found in SNPs colloidal solution over a prolonged period. UV-Vis spectra was measured at different LE conc. with absorbance elevating for increasing concentration. Thus, in both cases, i.e., DPSNPs and LPSNPs, the highest production was obtained for 350 mL in 500 mL of 1 mM AgNO3 solution giving λmax at 433 and 441 nm, respectively (Fig. 3a, b). Similar highest trend was also observed for time and temperature; however, DPSNPs at 0.2, 0.5 mM and 0.75, 1 mM AgNO3 expressed similar SPR peaks signifying DPSNPs production feasibility at both molarities. LPSNPs expressed dissimilar behavior for molarity by giving highest SPR (463 nm) at 1 mM of AgNO3 (Fig. 3c–h). The incremental absorbance intensity in all optimization parameters is indicative of the reduction of Ag+ to Ag0 in the presence of P. cerasifera leaf metabolites. Sharp peaks for LPSNPs (Fig. 3b, d, f, h) in comparison to DPSNPs were indicative of relatively smaller sizes, as confirmed by SEM (Fig. 3b, d, f). While the magnitude of SNPs production was comprehended from increasing absorbance, i.e., upon highest LE conc., time, temperature, and molarity the highest SNP production was obtained. Such an augmented production in case of highest LE conc. can be attributed to the presence of larger phytometabolites available to reduce silver ions to SNPs (Moteriya and Chanda 2017). The simultaneous successful production of LPSNPs in sunlight with additional temperature (except in temperature variation case), suggests the sustainability of photo exposed SNPs to be more favorable over DPSNPs which needs maintenance in dark environments (Thatikayala et al. 2019).
UV-Vis spectra for P. cerasifera leaf reduced and stabilized silver nanoparticles optimized for different parameters (a, b) leaf extract variation for DPSNPs and LPSNPs (c, d) time variation for DPSNPs and LPSNPs (e, f) temperature variation for DPSNPs and LPSNPs (g, h) AgNO3 variation for DPSNPs and LPSNPs
FTIR
SNPs coating with P. cerasifera functional groups was analyzed via FTIR (Fig. 4a, b). LE exhibited major peaks at 874, 1020, 1415, 1637, and 3308 cm−1 which corresponded to N–H bend of primary/secondary amines, C–N stretch of aliphatic amines, C–C stretch of aromatics, N–H bend of primary amines, and O–H stretch or H–bonded stretch of alcohols or phenols, respectively. IR bands 1269, 1635, 2934, and 3447 cm−1 for DPSNPs arise from C–N stretch of aromatic amines, N–H stretch of primary amines, O–H stretch of carboxylic acids, and O–H stretch, H–bonded stretch of alcohols or phenols, respectively. LPSNPs also exhibited peaks nearly similar to DPSNPs but varied in sharpness and intensity. Such results are indicative of the richness of P. cerasifera leaves in phenolic content like other prunus plants (Stierlin et al. 2019; Smanalieva et al. 2019; Cerri et al. 2019). LPSNP IR bands at 1117, 1269, 1634, 2920, and 3458 cm−1 arise from C–N stretch of aliphatic amines, N–O symmetric stretch of nitro compounds, N–H bend of primary amines, C–H stretch of alkanes and O–H stretch, H–bonded stretch of alcohols or phenols, respectively. Although there were peak shifts with disappearance of several peaks and origination of new peaks but the resemblance of LE, DPSNPs, and LPSNPs for IR bands corresponding to amines, aromatics, alcohols, and phenols are suggestive of SNPs being reduced in presence of LE without any external chemical input. P. cerasifera metabolites simultaneously reduced and stabilized the SNPs, thus showing the transformative and multifunctional role. Phenolic, alcoholic, and proteinaceous phytochemicals present in P. cerasifera leaf extract explored via FTIR can be considered as possible reducing agents for SNPs synthesis.
GC-MS
The promising functional groups in LE involved in reductions of SNPs were further extended to phytochemical analysis of LE via optimized GC-MS parameters (Table 2). Major GC-MS peaks were detected in LE at 4.02 (2-heptadecanone), 6.11 ((e)-2-methyl-2-butenoic acid), 7.76 (benzoic acid, 1,3-dioxan-5-yl ester), 8.88 (benzydamine), 12.67 ((s)-(+)-1-(2-pyrrolidinylmethyl)-pyrrolidine), 15.87 (α-terpineol), 17.41 (hexadecanoic acid), 18.78 (phytol), 19.25 (1,10,13-hexadecatriene, 7,8-epoxy), and 26.87 min (3-cyclohexene-1-methanamine, α,α,4-trimethyl-). The presence of such an array of phytochemicals possessing antioxidant potential exerted influential impacts of SNP synthesis. Furthermore, the presence of phytochemicals of reducing nature were also expressed by GC-MS of LE signifying the duality in terms of reduction and capping of SNPs. Absence of agglomeration and the intactness of colloidal and dried SNPs at room temperature for extended durations was due to this capping role of LE making SNPs further sustainable and economical. The higher surface to volume ratio of SNPs enabled the surface adsorption of strong ligating agents, e.g., α-terpineol, hexadecanoic acid, phytol, 1,10,13-hexadecatriene, 7,8-epoxy, and 3-cyclohexene-1-methanamine, α,α,4-trimethyl- most probably by establishment of interaction via π-electrons. The presence of P. cerasifera phytometabolites not only caused the synthesis of SNPs in nanoregime but also influenced their shape (Batool et al. 2019; Rolim et al. 2019; Aisida et al. 2019).
AFM and SEM
SNPs were analyzed for morphological features via AFM and the microstructural analysis was done by SEM (Figs. 5 and 6). AFM of SNPs exceeded in giving a deeper insights of the morphology in qualitative manner for its resolution in both lateral and vertical nanometer range. Figure 5 shows the lateral and 3D micrograph for phytosynthetic SNPs to be of spherical shape with well-dispersed assemblage without any agglomeration. Presence of spherical shaped individual BSNPs were indicative of smooth surface morphology and homogeneity. BSNP topographical features were further confirmed via SEM that exhibited variable size ranges (Table 1) with lower size ranges predominantly obtained for LPSNPs. However, synthesized SNPs in agreement with AFM exhibited spherical shapes.
SEM micrographs for silver nanoparticles reduced by P. cerasifera leaf extract at different optimization parameters: a LE conc. (300 mL) variation DPSNPs, b conc. (300 mL) variation LPSNPs, c time (150 min) variation DPSNPs, d time (150 min) variation for LPSNPs, e temperature (120 °C) variation DPSNPs, and f temperature (120 °C) variation LPSNPs
XRD
SNPs were analyzed for their crystallographic parameters via XRD (Fig. 7). The diffraction pattern for SNPs showed peaks at 38.09°, 44.16°, 64.40°, and 77.29° which can be indexed to (111), (200), (220), and (222) planes of face-centered cubic crystal structure (Shelar et al. 2019; Roy et al. 2019). Diffraction peaks were matched with standard pattern of JCPDS card no. 04-0783. Pure crystalline nature of SNPs was revealed with an average crystallite size of 2.34 nm calculated by Scherer equation. Pure crystallinity of SNPs without any impurities was achieved signifying the role of P. cerasifera phytochemicals (Varshney et al. 2010). SNPs’ spherical shapes with lower size ranges and FCC crystal of 2.34 nm size when employed for chemo photocatalytic degradation and antimicrobial degradation exhibited remarkable results.
SNP nanophotocatalytic potential
Waste water pollution is not only a challenging task in terms of remediation but they also pose environmentally pernicious impacts on the directly exposed biota till the clean-up methodologies are applied. Despite advances, the conventional methods of ultrafiltration, adsorption and electrical treatments etc. persistent organic pollutants remain a challenge. Though metallic silver is a poor catalyst but SNPs in this case can be of great significance due to smaller sizes and an elevated size to volume ratio for toxic dyes comprising waste waters. The time-dependent photocatalytic activity of SNPs was analyzed against persistent pollutants under direct solar irradiance. SNPs triggered photocatalysis of CV, MB, and MG (Fig. 8) in less than 15 min in direct solar irradiation signifies the role of SNPs as efficient and sustainable environmental remediation tools. Control setup without SNP addition expressed no changes in color upon exposure to solar irradiance, thus expressing negligible self-catalytic behavior and exhibiting persistence in the aquatic media. However, SNPs treated experimental setup comprising three dyes expressed degradation firstly identified by conversion to achromatized forms followed by an alleviation in UV-Vis spectral absorbance (Fig. 9a–c). Degradation percentages expressed the rapid photocatalysis of MG with 85% degradation in 8 min with complete achromatization. However, CV and MB were degraded and converted to their leuco forms at 12 and 14 min, respectively (Fig. 9d–f). Furthermore, photocatalytic reaction rates were pseudo-first order for CV, MB, and MG (R2 = 0.99, 0.99, 0.98) (Fig. 9g–i).
UV-Vis spectra for photodegradation triggered by SNPs in direct solar irradiance. a–c Crystal violet, methylene blue, and malachite green. d–f Pie charts showing percent degradation at different time intervals for crystal violet, methylene blue, and malachite green. g–i Reaction kinetics for photodegradation of crystal violet, methylene blue, and malachite green
The photocatalytic potential of silver nanoparticles has been frequently investigated. Biogenic silver nanoparticles have been employed for the degradation of various dyes (Bhuyan et al. 2017; Elemike et al. 2017; Roy et al. 2015; Karthik et al. 2017; Saraswathi et al. 2017; Miri et al. 2018) (Table 3), but no reports are available on the P. cerasifera leaf stabilized SNPs in this regard. Pie chart (Fig. 9d–f) shows the gradual achromatization of CV after SNP treatment. Such shorter time duration explains the ease for transfer of electrons between valence and conduction band for photocatalysis by SNPs. Ninety-six percent MB degradation was achieved in 14 min which can attributed to the smaller size, polydispersity, and enhanced light absorption characteristic of SNPs. MG degradation with SNPs in 8 min is suggestive of the catalytic role of SNPs and its conversion to commercial scale. Such photodegrading is caused due to excitation of SPR upon exposure of SNPs to visible light, this gives rise to the oscillation of charge density followed by propagation between Ag+ and dielectric medium. Linear positive correlation between time and achromatization of CV, MB, and MG in accordance with ln At/Ao was revealed by kinetic studies is due to “electron relay effect.” Further studies on P. cersiafera stabilized SNPs with different quantities of photocatalyst, pH, and kinetic parameters will further strengthen the pragmatic grounds for use of SNPs in dye degradation at commercial scale.
Antimicrobial activity
Resistance towards multiple drugs exhibited by pathogenic bacteria and fungi has posed challenges to public health and agricultural production. Focus has been shifted towards the formulation of novel antimicrobial drugs comprising SNPs that can be effective for inhibition of resistant microorganisms. The enthralling antimicrobial effects towards such resistant microbes has not been previously reported with P. cerasifera leaf stabilized SNPs. SNPs in combination with other nanostructured materials retain its antimicrobial potential; however, in the present investigation, this potential was further enhanced due to phytometabolites. Plant pathogens have been destroying valuable crops since many centuries; such bacterial and fungal strains can be potentially inhibited by utilization of the biogenic SNPs. Smaller sized SNPs are particularly beneficial in the effective treatment of the bacterial or fungal pathogenicity in addition to their development into an antiviral medications (Sharma et al. 2019; Kailasa et al. 2019).
Current era is marked by an elevated demand for higher scale production of SNPs for greater utilization. Silver being an inherent antimicrobial agent has been engineered at nanoscale by use of P. cerasifera phytochemicals in the present investigation. SNPs were investigated for antimicrobial inhibition against nine human and plant pathogenic strains, i.e., X. citri, P. syringae, A. niger, A. flavus, A. fumigatus, A. terreus, P. chrysogenum, F. solani, and L. theobromae in a dose-dependent manner and compared with standard antibiotics (Table 4). Studied pathogens have been found resistant towards a myriad of antibiotics. SNPs were found effective against all nine microbes even at 2000 μg/mL volume, and clearance zones were formed. However, the susceptibilities of each microbes varied possibly due to their variable response towards LE metabolites. Anticipatedly, the increase in SNPs dose increased the clearance zones. Furthermore, SNPs astonishingly exceeded the standard drugs for all nine microbes. In case of bacterial strains, XC was more inhibited as compare to PS. For fungal pathogens, SNPs expressed highest clearance zones for AN and PC; however, AT was least inhibited. The degree of inhibition expressed by SNPs (10,000 μg/mL dose) against fungal pathogens was as follows: AN (25 ± 0.01) = PC (25 ± 0.01) > LT (23 ± 0.01) > AF (22 ± 0.01) = FS (22 ± 0.06) > AFG (20 ± 0.01) > AT (17 ± 0.05).
SNPs were found as effective therapeutic agents against nine tested microbes. Higher inhibition zones produced by SNPs revealed the antimicrobial potential of silver to be enhanced when converted to nanoform with higher surface to volume ratio. Such remarkable activity can be attributed to the antimicrobial potential of P. cerasifera LE (Table 3) which in turn increased the SNPs’ induced clearance zones due to synergistic effects. SNPs inhibited the growth of nine microbes in a dose-dependent manner and highest activity was obtained at 10,000 μg/mL and > 18 mm except AT showing active ZOI but not highly active as remaining eight microorganisms. Zones of inhibition are categorized as inactive (< 9 mm), fractionally active (9–12 mm), active (13–18 mm), and highly active (> 18 mm) (Mariselvam et al. 2014). Considerable variation was observed among all strains towards standard, LE, DPSNPs, and LPSNPs which arises due to the response of each microorganism towards P. cerasifera leaf capped SNPs. Bacterial inhibition by SNPs was due to SNPs’ penetration in bacterial cell wall and thus inducing bacteriostatic. Having crossed the bacterial cellular permeability, SNPs induce leakage of microbial cell material and consequently enter inner membrane for ROS production. These all events cause microbial cell growth inhibition. Microbial cell inevitable disintegration and death also arise due to SNP treatment after toppling of membrane and other components.
SNP-induced extirpation of fungal strains was done because P. cerasifera-mediated SNPs have not been reported for fungicidal action so far. Biogenic SNPs have been employed for fungal inhibition for fewer strains; however, P. cerasifera-based SNPs exceeded the previous studies by producing highly active ZOIs. Additionally, SNPs obliterative mechanism has been explored because multidrug resistance issue in case of fungal strains is not as precarious matter as in case of bacterial strains; however, the scarcity of drugs available to fight human and plant pathogenic fungal strains suggests the development of new effective antifungal formulations (Bocate et al. 2019). Furthermore, the use of antifungal drugs, e.g., amphotericin B and azoles (fluconazole or ketoconazole) for treatment of human fungal infections can be highly obnoxious to the major urinary and metabolic organs like kidney and liver. Thus, SNP-based formulations which do not pose any harm to human cells but effectively destruct the fungal cell are appropriate candidate for biomedical purposes. In addition to the clinical significance, the highly active ZOI produced for phytopathogens approves the role of SNPs in wiping out vicious agricultural pathogens. Thus, SNPs can be used as green fungicides as a sustainable alternative to chemical pesticides with known environmental toxicity.
Statistical evaluation
The statistical significance of enhanced antimicrobial activity of SNPs exceeding standard drugs was evaluated by one-way ANOVA and Kruskal-Wallis test (P value less than 0.05 was considered significant) (Table 5). Larger Kruskal-Wallis statistic (H = 17.50) reflects the statistical divergence among standard drug, LE, DPSNPs, and LPSNPs in agreement with ANOVA. Though such variance was indicated by ANOVA and Kruskal-Wallis test in general however the statistical significance of individual treatments was not expressed. Thus, it was evaluated by four additional tests namely Tukey HSD, Scheffé, Bonferroni, and Holm tests (P < 0.05) (Table 6). Assessment of inequality and multiple group specific comparison was done by these tests. Practical effectiveness of SNPs greatly depended upon the use of statistical tests for inhibition. Variations calculated for all cases are significant except between DPSNPs vs LPSNPs (P values: Tukey HSD 0.67, Scheffé 0.75, Bonferroni 1.69, and Holm 0.28) since both SNPs were synthesized at similar conditions with P. cerasifera phytometabolite reduction cum stabilization; thus, they both exhibited higher clearance zones nullifying the divergence in values of inhibition. In this case, the null hypothesis is acceptable for nearly equal ZOIs. Highly significant statistical differences were obtained for inhibition by standard vs DPSNPs, LE vs DPSNPs, and LE vs LPSNPs (P < 0.01).
Conclusions
Silver nanoparticles were fabricated for the first time using the reducing agents found in P. cerasifera leaf extract in conformity with all principles of green chemistry. LE conc., time, temperature, molarity, presence and absence of light exerted influential impacts on fabrication process yielding variable SPR peaks, functional group and size rages for spherical silver nanoparticles. Crystalline nature without any impurity with an average crystallite size 2.34 nm is indicative of the role of P. cerasifera metabolites in reduction and capping of synthesized nanoparticles. Persistent organic pollutants crystal violet, methylene blue, and malachite green were sustainably photodegraded with silver nanoparticles with pseudo-first-order kinetics (R2 = 0.99, 0.99, 0.98) in less than 15 min. Furthermore, SNPs can be efficiently used for obliteration of the bacterial and fungal cell growth by interfering with cellular functions due to inherent toxicity which was enhanced due to phytochemicals capping. Thus, P. cerasifera mediated silver nanoparticles are concluded to be potential photocatalytic and antimicrobial bullets for environmental, agricultural, and biomedical platforms under nanobiotechnology arena.
References
Ahluwalia V, Elumalai S, Kumar V, Kumar S, Sangwan RS (2018) Nano silver particle synthesis using Swertia paniculata herbal extract and its antimicrobial activity. Microb Pathog 114:402
Ahmad KS, Jaffri SB (2018a) Carpogenic ZnO nanoparticles: amplified nanophotocatalytic and antimicrobial action. IET Nanobiotechnol. https://doi.org/10.1049/iet-nbt.2018.5006
Ahmad KS, Jaffri SB (2018b) Phytosynthetic Ag doped ZnO nanoparticles: semiconducting green remediators. Open Chem 16(1):556
Aisida SO, Ugwu K, Akpa PA, Nwanya AC, Ejikeme PM, Botha S, Ahmad I, Maaza M, Ezema FI (2019) Biogenic synthesis and antibacterial activity of controlled silver nanoparticles using an extract of Gongronema latifolium. Mat Chem Phy 237:121859
Alavi M, Karimi N (2017) Characterization, antibacterial, total antioxidant, scavenging, reducing power and ion chelating activities of green synthesized silver, copper and titanium dioxide nanoparticles using Artemisia haussknechtii leaf extract. Artif Cells Nanomed Biotechnol 16
Ali M, Jaffri SB, Ahmad KS, Iqbal S (2019) Sorptive interactions of fungicidal 2-(4'-Thiazolyl) Benzimidazole with soils of divergent physicochemical composition. J Int Econ Envir Geol 10:97–104
Amjad I, Javaid M, Ikhlaq K, Gul S, Jaffri SB, Ahmad KS (2019) Adsorption-desorption mechanism of synthesized Benzimidazole based fungicide 2-(3’-Pyridyl) on selected soil minerals. J Int Econ Envir Geol 10:38–44
Bartosiak M, Giersz J, Jankowski K (2019) Analytical monitoring of selenium nanoparticles green synthesis using photochemical vapor generation coupled with MIP-OES and UV–Vis spectrophotometry. Microchem J 145:1169
Batool S, Hussain Z, Niazi MB, Liaqat U, Afzal M (2019) Biogenic synthesis of silver nanoparticles and evaluation of physical and antimicrobial properties of Ag/PVA/starch nanocomposites hydrogel membranes for wound dressing application. J Drug Del Sci Tech 52:403
Bhuyan B, Paul A, Paul B, Dhar SS, Dutta P (2017) Paederia foetida Linn. promoted biogenic gold and silver nanoparticles: synthesis, characterization, photocatalytic and in vitro efficacy against clinically isolated pathogens. J Photochem Photobiol B 173:210
Bocate KP, Reis GF, de Souza PC, Junior AG, Durán N, Nakazato G, Furlaneto MC, de Almeida RS, Panagio LA (2019) Antifungal activity of silver nanoparticles and simvastatin against toxigenic species of Aspergillus. Int J Food Microb 291:79
Cerri M, Rosati A, Famiani F, Reale L (2019) Fruit size in different plum species (genus Prunus L.) is determined by post-bloom developmental processes and not by ovary characteristics at anthesis. Sci Horti 255:1
Elemike EE, Onwudiwe DC, Ekennia AC, Ehiri RC, Nnaji NJ (2017) Phytosynthesis of silver nanoparticles using aqueous leaf extracts of Lippia citriodora: antimicrobial, larvicidal and photocatalytic evaluations. Mater Sci Eng C 75:980
Fears R, ter Meulen V (2014) What do we need to do to tackle antimicrobial resistance? Lancet Glob Health 2(1):e11
Ferreira LA, dos Reis SB, da Silva ED, Cadore S, da Silva BJ, Durán N, de Jesus MB (2019) Thiol-antioxidants interfere with assessing silver nanoparticle cytotoxicity. Nanomed: Nanotech Biol Med 1:102130
Gavamukulya Y, Maina EN, Meroka AM, Madivoli ES, El-Shemy HA, Wamunyokoli F, Magoma G (2019) Green synthesis and characterization of highly stable silver nanoparticles from ethanolic extracts of fruits of Annona muricata. J Inorg Org Polymers Mat 1:1–2
Gohil S, Chandra R, Chalke B, Bose S, Ayyub P (2007) Sputter deposition of self-organized nanoclusters through porous anodic alumina templates. J Nanosci Nanotech 7:641
Gulbagca F, Ozdemir S, Gulcan M, Sen F (2019) Synthesis and characterization of Rosa canina-mediated biogenic silver nanoparticles for anti-oxidant, antibacterial, antifungal, and DNA cleavage activities. Heliyon 5:e02980
Heydari R, Koudehi MF, Pourmortazavi SM (2019) Antibacterial activity of Fe3O4/Cu nanocomposite: green synthesis using Carum carvi L. Seeds Aqueous Extract ChemistrySelect 4:531
Huo Y, Yan M, Zhao X, Zhu Z, Yuan Z (2019) The complete chloroplast genome sequence of Prunus cerasifera Ehrh.‘Pissardii’(Rosaceae). Mitochondrial DNA Part B 4(2):3744
Hussain A, Oves M, Alajmi MF, Hussain I, Amir S, Ahmed J, Rehman MT, El-Seedi HR, Ali I (2019) Biogenesis of ZnO nanoparticles using Pandanus odorifer leaf extract: anticancer and antimicrobial activities. RSC Adv 9:15357
Ismail M, Gul S, Khan MI, Khan MA, Asiri AM, Khan SB (2019) Green synthesis of zerovalent copper nanoparticles for efficient reduction of toxic azo dyes congo red and methyl orange. Green processing and synthesis 8:–135
Jaffri SB, Ahmad KS (2017) Augmented photocatalytic, antibacterial and antifungal activity of prunosynthetic silver nanoparticles. Artif Cells Nanomed Biotechnol 46:127-137
Jaffri SB, Ahmad KS (2018a) Prunus cerasifera Ehrh. fabricated ZnO nano falcates and its photocatalytic and dose dependent in vitro bio-activity. Open Chem 16(1):141
Jaffri SB, Ahmad KS (2018b) Neoteric environmental detoxification of organic pollutants and pathogenic microbes via green synthesized ZnO nanoparticles. Envir Technol, (just-accepted), 1-42
Jaffri SB, Ahmad KS (2018c) Foliar-mediated Ag: ZnO nanophotocatalysts: green synthesis, characterization, pollutants degradation, and in vitro biocidal activity. Green Process Synthesis 1-11
Jaffri SB, Ahmad KS (2018d) Phytofunctionalized silver nanoparticles: green biomaterial for biomedical and environmental applications. Rev Inorg Chem 38(3):127
Jindal R, Sinha R (2019) Malachite green induced ultrastructural corneal lesions in Cyprinus carpio and its amelioration using Emblica officinalis. Bull Envir Contam Toxic 102(3):377
Kailasa SK, Park TJ, Rohit JV, Koduru JR (2019) Antimicrobial activity of silver nanoparticles. In Nanoparticles in pharmacotherapy (pp. 461-484). William Andrew publishing
Karthik R, Govindasamy M, Chen SM, Cheng YH, Muthukrishnan P, Padmavathy S, Elangovan A (2017) Biosynthesis of silver nanoparticles by using Camellia japonica leaf extract for the electrocatalytic reduction of nitrobenzene and photocatalytic degradation of eosin-Y. J Photochem Photobiol B 170:164
Li T, Tian D, Zhu Z, Jin W, Wu S, Li H (2019) The gut microbiota: a new perspective on the toxicity of malachite green (MG). App Micro Biotech 103(23–24):9723
Mariselvam R, Ranjitsingh AJ, Nanthini AU, Kalirajan K, Padmalatha C, Selvakumar PM (2014) Green synthesis of silver nanoparticles from the extract of the inflorescence of Cocos nucifera (Family: Arecaceae) for enhanced antibacterial activity. Spectrochim Acta A 129:537
Miri A, Vahed HOS, Sarani M (2018) Biosynthesis of silver nanoparticles and their role in photocatalytic degradation of methylene blue dye. Res Chem Inter:1–9
Mittal A, Mittal J, Malviya A, Kaur D, Gupta VK (2010) Adsorption of hazardous dye crystal violet from wastewater by waste materials. J Colloid Interface Sci 343:463e473
Moteriya P, Chanda S (2017) Synthesis and characterization of silver nanoparticles using Caesalpinia pulcherrima flower extract and assessment of their in vitro antimicrobial, antioxidant, cytotoxic, and genotoxic activities. Artif Cells Nanomed Biotechnol 45(8):1556–1567
Mutua JK, Imathiu S, Owino W (2017) Evaluation of the proximate composition, antioxidant potential, and antimicrobial activity of mango seed kernel extracts. Food Science Nutri 5(2):349
Nafea OE, Hassan HA (2019) Comparative effectiveness of methylene blue versus intravenous lipid emulsion in a rodent model of amlodipine toxicity. Clinical Toxicol 1:1–6
Nayak S, Sajankila SP, Rao CV, Hegde AR, Mutalik S (2019) Biogenic synthesis of silver nanoparticles using Jatropha curcas seed cake extract and characterization: evaluation of its antibacterial activity. Energy Sourc Part A 1-9
Qais FA, Shafiq A, Khan HM, Husain FM, Khan RA, Alenazi B, Alsalme A, Ahmad I (2019) Antibacterial effect of silver nanoparticles synthesized using Murraya koenigii (L.) against multidrug-resistant pathogens. Bioinorg Chem App 1:1–10
Qasim Nasar M, Zohra T, Khalil AT, Saqib S, Ayaz M, Ahmad A, Shinwari ZK (2019) Seripheidium quettense mediated green synthesis of biogenic silver nanoparticles and their theranostic applications. Green Chem Lett Rev 12:310
Recio-Sánchez G, Tighe-Neira R, Alvarado C, Inostroza-Blancheteau C, Benito N, García-Rodríguez A, Marcos R, Pesenti H, Carmona ER (2019) Assessing the effectiveness of green synthetized silver nanoparticles with Cryptocarya alba extracts for remotion of the organic pollutant methylene blue dye. Envir Sci Poll Res 26(15):15115
Rolim WR, Pelegrino MT, de Araújo LB, Ferraz LS, Costa FN, Bernardes JS, Rodigues T, Brocchi M, Seabra AB (2019) Green tea extract mediated biogenic synthesis of silver nanoparticles: characterization, cytotoxicity evaluation and antibacterial activity. Appl Surf Sci 463:66
Roy K, Sarkar CK, Ghosh CK (2015) Photocatalytic activity of biogenic silver nanoparticles synthesized using potato (Solanum tuberosum) infusion. Spectrochim Acta A 146:286
Roy K, Ghosh CK, Sarkar CK (2019) Rapid detection of hazardous H2O2 by biogenic copper nanoparticles synthesized using Eichhornia crassipes extract. Microsys Tech 25(5):1699
Samadi N, Hosseini SV, Fazeli A, Fazeli MR (2010) Synthesis and antimicrobial effects of silver nanoparticles produced by chemical reduction method. DARU J Pharma Sci 18:168
Samari F, Parkhari P, Eftekhar E, Mohseni F, Yousefinejad S (2019) Antioxidant, cytotoxic and catalytic degradation efficiency of controllable phyto-synthesised silver nanoparticles with high stability using Cordia myxa extract. J Exp Nanosci 14:141–159
Saraswathi VS, Kamarudheen N, BhaskaraRao KV, Santhakumar K (2017) Phytoremediation of dyes using Lagerstroemia speciosa mediated silver nanoparticles and its biofilm activity against clinical strains Pseudomonas aeruginosa. J Photochem Photobiol B 168:107
Shaheen I, Ahmad KS (2019) Chromatographic identification of “green capping agents” extracted from Nasturtium officinale (Brassicaceae) leaves for the synthesis of MoO3 nanoparticles. J Sep Sci 1-10
Shaheen I, Ahmad KS, Jaffri SB, Zahra T, Azhar S (2016) Evaluating the adsorption and desorption behavior of triasulfuron as a function of soil physico-chemical characteristics. Soil Environ 35:99
Sharma V, Kaushik S, Pandit P, Dhull D, Yadav JP, Kaushik S (2019) Green synthesis of silver nanoparticles from medicinal plants and evaluation of their antiviral potential against chikungunya virus. Appl Microb Biotech 103(2):881
Shelar A, Sangshetti J, Chakraborti S, Singh AV, Patil R, Gosavi S (2019) Helminthicidal and larvicidal potentials of biogenic silver nanoparticles synthesized from medicinal plant Momordica charantia. Med Chem 15(7):781–789
Smanalieva J, Iskakova J, Oskonbaeva Z, Wichern F, Darr D (2019) Determination of physicochemical parameters, phenolic content, and antioxidant capacity of wild cherry plum (Prunus divaricata Ledeb.) from the walnut-fruit forests of Kyrgyzstan. Euro Food Res Tech 245:2293
Stierlin E, Azoulay S, Massi L, Fernandez X, Michel T (2019) Osmetic potentials of Prunus domestica L. leaves. J Sci Food Agric 98:726
Thatikayala D, Jayarambabu N, Banothu V, Ballipalli CB, Park J, Rao KV (2019) Biogenic synthesis of silver nanoparticles mediated by Theobroma cacao extract: enhanced antibacterial and photocatalytic activities. J Mat Sci: Mat Elect 30:17303
Varshney R, Bhadauria S, Gaur MS (2010) Biogenic synthesis of silver nanocubes and nanorods using sundried Stevia rebaudiana leaves. Adv Mater Lett 1(3):237
Zahra T, Ahmad KS, Shaheen I, Azhar S, Jaffri SB (2017) Determining the adsorption and desorption behavior of thiabendazole fungicide for five different agricultural soils. Soil Envir 36:13
Zhou X, Zhang J, Pan Z, Li D (2019) Review of methods for the detection and determination of malachite green and leuco-malachite green in aquaculture. Crit Rev Ana Chem 49(1):1
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Current research has not employed any experimentation on animals and humans.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Responsible editor: Philippe Garrigues
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Jaffri, S.B., Ahmad, K.S. Biomimetic detoxifier Prunus cerasifera Ehrh. silver nanoparticles: innate green bullets for morbific pathogens and persistent pollutants. Environ Sci Pollut Res 27, 9669–9685 (2020). https://doi.org/10.1007/s11356-020-07626-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-020-07626-6