Abstract
Chromium pollution is a problem that affects different areas worldwide and, therefore, must be solved. Bioremediation is a promising alternative to treat environmental contamination, but finding bacterial strains able to tolerate and remove different contaminants is a major challenge, since most co-polluted sites contain mixtures of organic and inorganic substances. In the present work, Bacillus sp. SFC 500-1E, isolated from the bacterial consortium SFC 500-1 native to tannery sediments, showed tolerance to various concentrations of different phenolic compounds and heavy metals, such as Cr(VI). This strain was able to efficiently remove Cr(VI), even in the presence of phenol. The detection of the chrA gene suggested that Cr(VI) extrusion could be a mechanism that allowed this strain to tolerate the heavy metal. However, reduction through cytosolic NADH-dependent chromate reductases may be the main mechanism involved in the remediation. The information provided in this study about the mechanisms through which Bacillus sp. SFC 500-1E removes Cr(VI) should be taken into account for the future application of this strain as a possible candidate to remediate contaminated environments.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Chromium is one of the most widely distributed heavy metals in nature. It is mainly discharged in industrial effluents from metal smelting, electroplating, leather tanning, and stainless steel manufacturing (Saranraj and Sujitha 2013). It occurs in various oxidation states, but Cr(VI) is the most soluble, oxidative, and toxic form, and for the past 15 years, it has been in the top 20 on the list of priority hazardous substances (Liu et al. 2015).
Due to its hazardousness, international organizations for environmental protection have established very low tolerance limits of Cr(VI) in surface water, and the industries concerned must minimize Cr(VI) concentrations in their effluents (Dhal et al. 2013). Therefore, it is crucial to find an effective method to treat these effluents before they are discharged into natural water bodies. Treatment should also cope with co-pollutants present in the effluent, such as other heavy metals and organic compounds like phenols and derivatives. This constitutes the greatest difficulty for remediation (Abha and Singh 2012). Bacterial bioremediation is one of the most interesting methods for cleaning polluted environments. The metabolic versatility of some bacteria allows them to diminish the toxicity and bioavailability of heavy metals even in the presence of co-pollutants (Megharaj et al. 2011). In particular, several microorganisms have been reported to reduce Cr(VI) to the less toxic form Cr(III) and to carry out its precipitation at near neutral pH. Cr(VI) efflux, exclusion, biosorption, and bioaccumulation are other mechanisms commonly displayed by tolerant bacteria (Thatoi et al. 2014). Both reduction and immobilization in the biomass constitute cost-effective green technologies for the treatment of wastes contaminated with Cr(VI), in contrast to the traditional physicochemical methods (Ahemad 2014).
It is well known that chromate resistance and reduction are not necessarily interrelated, and not all Cr(VI)-resistant bacteria are able to reduce Cr(VI) to Cr(III). The understanding and characterization of tolerance/removal mechanisms and the identification of their genetic determinants could be essential to devise an efficient strategy for Cr(VI) remediation (Viti et al. 2014). However, most studies in this area focus on the removal performance of microorganisms, but they do not go deep into the molecular and physiological basis of the process (Huang et al. 2017). In this work, Bacillus sp. SFC 500-1E., which belongs to the consortium SFC 500-1 isolated from tannery sludges, was studied in regard to its capabilities for removing Cr(VI) and tolerating other organic and inorganic contaminants. The mechanisms involved in Cr(VI) tolerance and bioremediation were studied through genetic and enzymatic analysis.
The results obtained may help to elucidate the involvement of this strain in the capacity of consortium SFC 500-1 to simultaneously remove Cr(VI) and phenol (Ontañon et al. 2015a, 2017), since Acinetobacter sp. SFC 500-1A, the other component of the system, has already been characterized (Ontañon et al. 2015b; Fernández et al. 2017).
Materials and methods
Identification of the bacterial strain
A gram positive bacterial strain, isolated from a consortium with high capability for simultaneously removing Cr(VI) and phenol (Ontañon et al. 2015a), was used in this work. DNA extraction and 16S rRNA gene amplification and sequencing were carried out by the “MacroGen” company (Korea) in order to identify the strain. The sequence for the 16S rRNA gene of Bacillus sp. SFC 500-1E was deposited in GenBank under accession number JQ701739.
A phylogenetic tree was constructed with MEGA7 software (Kumar et al. 2016) based on the alignment with other 16S rRNA gene sequences deposited in GenBank.
Moreover, two commercially available kits for bacterial biochemical analysis were employed (API 20 NE and API 20 E system, BioMerieux® SA). Additional biochemical tests and matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF), coupled to a mass spectrometer MALDI-TOF-TOF Autoflex III (BrukerDaltonics GmbH, Leipzig, Germany), were used for identification at species level. For MALDI-TOF analysis, a score of ≥ 2 indicated species level identification, a score between 1.7 and 1.99 indicated identification at the genus level, and a score below 1.7 was interpreted as no identification.
Assays of tolerance to heavy metals and phenolic compounds
Tolerance was evaluated through the ability of the bacterial strain to grow in TY medium plates containing the following (expressed in g L−1): 5.0 tryptone, 3.0 yeast extract, 0.5 CaCl2, and 13 agar (Beringer 1974), supplemented with increasing metal or metaloid concentrations (expressed in mg L−1): K2Cr2O7 [Cr(VI) 50; 100; 150]; NaAsO2 [As(III) 150, 375]; AsHNa2O4 [As(V) 1500, 3000]; ZnSO4 (Zn 150, 300); HgCl2 (Hg 2, 10, 20); CdCl2 (Cd 10, 30, 60), or increasing phenolic compound concentrations (also expressed in mg L−1): phenol (1000, 1500, 2000); 2,4 dichlorophenol (50, 100, 200); pentachlorophenol (1, 10, 20); and guaiacol (500, 1000, 2000). Plates were spread with different dilutions of a bacterial culture grown overnight in TY medium, at 150 rpm and 28 ± 2 °C. Cell viability was determined by the microdroplet method following Somasegaran and Hoben (1994), and the results were expressed as total colony forming units CFU mL−1 in the maximum tolerated concentration (MTC).
Removal assays
Cr(VI) removal assays were performed in Erlenmeyer flasks containing 20 mL of TY medium (pH 7) supplemented with different Cr(VI) concentrations (10, 25, 50 mg L−1) for 72 h. Bacterial efficiency in removing Cr(VI) in the presence of phenol (300 mg L−1) was also evaluated. All flasks were inoculated with a bacterial culture grown overnight in TY medium, in order to achieve an initial absorbance of 0.1 at 600 nm. Then, they were incubated at 28 ± 2 °C and 150 rpm. Abiotic controls were performed with the same concentrations of Cr(VI) and phenol but without inoculation. At predetermined time intervals, aliquots were withdrawn for evaluation of bacterial growth and quantification of residual contaminants (sections “Chromium determination” and “Phenol determination”).
Amplification of genes related to Cr(VI) removal
ChRF and ChR primers designed by Patra et al. (2010) were used for amplifying the coding gene of chromate reductases described for other strains. PCR amplification was carried out following the original protocol (Patra et al. 2010).
Moreover, the pair of primers CHRBAC-F (5′GTY GCT CAT GCH ATA HGD GGA ATG GC 3′) and CHRBAC-R (5′ GGH ARH ACR ACG TGH CCN CCD CC 3′) was designed to monitor the presence of the gene chr A, which encodes a chromate efflux pump. Known sequences of such gene in different strains of Bacillus sp. were obtained from the NCBI database using the BLAST program (www.ncbi.nlm.nih.gov/blast/BLAST.cgi), and multiple alignments were done using Clustal W (www.ebi.ac.uk/Tools/clustalw). Highly conserved portions in the alignment were selected for designing degenerate primers.
For chr A amplification, PCR conditions were as follows: an initial denaturation step at 95 °C for 5 min, then 30 cycles of 95 °C for 45 s, 60 °C for 45 s, and 72 °C for 1 min, with a final extension of 72 °C for 5 min. Negative controls included deionized water.
The PCR products obtained were sequenced and analyzed with the Bioedit program. Homology searches were performed using BLAST.
Detection and characterization of chromate reductase activity
Preparation of cell-free extracts
Bacterial cultures grown overnight in 50 mL of TY broth with or without contaminants were harvested by centrifugation (10,000 rpm, 15 min, 4 °C), washed, and resuspended in 2 mL of 50 mM phosphate buffer, pH 7. Cells were then disrupted by ultrasonication in ice at 60 W (Sonics VC 500, USA) using 6 cycles of 0.5 min each, with a gap of 0.5 min between cycles (Paisio et al. 2013). The cell lysates were then centrifuged at 15,000 rpm for 10 min at 4 °C. Filtered supernatants were employed as total cell-free extracts (CFEs) for a chromate reductase assay.
CFEs were obtained from three different culture conditions: TY broth without contaminants [CFE-Cr(VI)], TY broth supplemented with 25 mg L−1 Cr(VI) [CFE + Cr(VI)], and with 25 mg L−1 Cr(VI) plus 300 mg L−1 phenol [CFE + Ph + Cr(VI)].
Protein concentration in all CFE was calculated following the Bradford method (Bradford 1976).
Chromate reductase activity
The ability of the CFE to reduce Cr(VI) was evaluated as described by McLean and Beveridge (2001), with some modifications. The reaction mixture (3 mL) contained CFE (1 mg mL−1 of protein), 10 mg L−1 Cr(VI), and 50 mM phosphate buffer at pH 7, with and without 6.5 mg L−1 NADH. The mixtures were incubated in agitation at 30 °C for 60 min, and residual Cr(VI) was quantified every 15 min with diphenylcarbazide (DPC) reagent. One unit (U) of chromate reductase activity was defined as the amount of enzyme that reduced 1 nmol of Cr(VI) per min under the conditions assayed. Specific activity (SA) was defined as units of chromate reductase activity per mg of protein.
Two controls were performed to evaluate non-enzymatic Cr(VI) reduction: mixtures of 10 mg L−1 Cr(VI) and 6.5 mg L−1 NADH prepared in 50 mM phosphate buffer without CFE and with CFE, pre-heated at 100 °C for 5 min.
Chromate reductase localization
Subcellular fractioning was carried out using the technique outlined by Murugavelh and Mohanty (2012), with some modifications. An aliquot of CFE was centrifuged for 60 min at 30,000 rpm and 4 °C, and the filtered supernatant was used as the cytoplasmic fraction. The pellet obtained was used as the membrane fraction.
Cytoplasmic and membrane fractions were assayed for chromate reductase activity as described above, and their specific activities were compared.
Transmission electron microscopy
Ultrastructural changes of cells grown in the presence of Cr(VI) for 16 h were studied using TEM (JEOL JEM 12EXII, Japan). The bacterial culture was centrifuged and the pellets were fixed using 2.5% glutaraldehyde in s-collidine buffer solution (0.2 M, pH 7.4) for 3 h, at 4 °C. Then, the cells were washed with s-collidine buffer solution and fixed with 1% osmium tetra oxide. Fixed samples were dehydrated with acetone, the pre-inclusion was made with 812 1:1 Embedepoxi resin in 100% acetone, and the inclusion was made with EMbed 812 at 56 °C. Semi-thin sections were cut using a manual ultramicrotome (Sorvall MT-1A, DuPont), then stained with toluidine blue, and observed under an optical microscope. Ultrathin sections (20–60 nm) were made with a diamond blade (PelcoR) and placed on copper grid sand contrasted with uranyl acetate and lead citrate (Bencosme and TsutSumi 1970; Ackerley et al. 2006).
Chromium determination
Cr(VI)
Residual Cr(VI) was determined through reaction with DPC in acid solution at 540 nm (APHA-AWWA 1989). The reaction mixture contained 500 μL of the sample, 500 μL of H2SO4 0.2 N, and 200 μL of DPC (5 mg L−1) in a total volume of 5 mL. The absorbance data were converted to Cr(VI) concentrations using a calibration curve.
Cr(III)
In the supernatants, total Cr concentration was determined by atomic absorption spectrometry (AAS) at the end of the assay (APHA-AWWA 1989). Cr(III) concentration was calculated as the difference between total Cr and Cr(VI).
To determine the presence of Cr(III) in the biomass, Erlenmeyer flasks containing TY medium plus Cr(VI) (10, 25, and 50 mg L−1) were inoculated with bacterial cells as described in section “Removal assays” and incubated for 72 h. Then, cells were washed three times with saline solution (0.9% NaCl), harvested by centrifugation (15,000 rpm–15 min), dried, and weighed. Samples were exposed to a cationic resin, and Cr(III) associated to biomass was determined following EPA SW 846 CAP 7000-EAA in a specialized laboratory using a Perkin Elmer Analyst.
Phenol determination
Phenol was determined spectrophotometrically following a standard method described by Wagner and Nicell (2002). Previously centrifuged 100-μL aliquots of each sample were mixed with 700 μL sodium bicarbonate (0.25 M, pH 8.4), 100 μL 4-aminoantipyrine (20.8 mM), and 100 μL potassium ferricyanide (83.4 mM). After 5 min, the absorbance of the colored compound formed was determined at 510 nm. Phenol concentrations were calculated by comparison with a calibration curve.
Statistical analysis
All the experiments were done at least three times by triplicate. Data were analyzed using ANOVA, followed by Tukey test (p < 0.05), with the Infostat (Version 2012 E) software.
Results and discussion
Identification of the strain
A phylogenetic tree was constructed based on 16S rRNA gene sequences of Bacillus sp. SFC 500-1E and other Bacillus strains. As can be seen in Fig. 1, the phylogenetic analysis showed a close relationship between SFC 500-1E and strains belonging to the B. cereus, B. toyonensis, and B. thuringiensis species, which are members of the Bacillus cereus group. For identification at species level, we used the MALDI-TOF MS technique, given that biochemical tests did not yield conclusive information to differentiate between the cereus and thuringiensis species (online resource). Regarding the mass spectrometry pattern, the strain SFC 500-1E could be classified as B. cereus. However, the score obtained (1.96) was below the confidence limit defined for this analysis (Saffert et al. 2011). These findings demonstrated that Bacillus sp. SFC 500-1E belongs to the Bacillus cereus group of bacteria, but were not reliable enough to classify it as a particular species.
Phylogenetic tree based on 16S rDNA sequences of strain SFC 500-1E and related species. The evolutionary history was inferred using the neighbor-joining method (Saitou and Nei 1987). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches (Felsenstein 1985). The evolutionary distances were computed using the Kimura 2-parameter method (Kimura 1980) and are in the units of the number of base substitutions per site
Tolerance to heavy metals and phenolic compounds
Industrial effluents usually contain complex mixtures of contaminants (Tripathi et al. 2011). Thus, the search for microorganisms that can treat them must include an analysis of their resistance to various toxic compounds that may be present in those environments. Such a strategy was followed in the present study, in which we evaluated the growth of Bacillus sp. SFC 500-1E at different Cr(VI) and phenol concentrations, as well as with other heavy metals and aromatic compounds. As can be seen in Table 1, this bacterium displayed suitable tolerance to Cr(VI), As(V), Zn, and Cd, but it was not able to grow in the presence of As(III) and Hg, probably because of the high toxicity of the latter two metals at relative low concentration (Gadd 1992).
Additionally, the strain tolerated high concentrations of phenol and its substituted derivatives guaiacol and dichlorophenol. However, bacterial growth was inhibited by pentachlorophenol, a highly toxic compound used as a biocide in the leather tanning process. This performance is common among phenol-tolerant microorganisms, whose tolerance varies with the number and position of substituents in the phenolic ring. As the number of substituents increases, these compounds become more toxic and less degradable (Al-Khalid and El-Naas 2012).
Different Bacillus strains exhibit multiple resistances to heavy metals and/or organic compounds like phenols. Such resistance has been directly related to the presence of contaminants in the isolation site, since the microorganisms have developed mechanisms to overcome toxicity as adaptation strategies. Biosorption, accumulation, and biotransformation to less toxic compounds have been described among these tolerance mechanisms (Oves et al. 2013; Banerjee and Ghoshal 2016).
Phenol and Cr(VI) removal assays
In some cases, tolerance and bioremediation potential may be unrelated. For instance, while some poorly tolerant strains are efficient for bioremediation, other strains selected on the basis of their tolerance are unsuccessful (Huang et al. 2017).
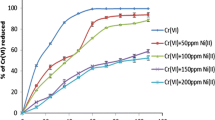
In agreement with this data, Bacillus sp. SFC 500-1E was unable to remove phenol, even when low concentrations were tested throughout long time periods (data not shown). This behavior suggests the absence of a metabolic pathway that allows the strain to degrade phenol, despite being a hypertolerant bacterium. On the contrary, Cr(VI) removal was almost complete for 10 mg L−1 and was above 80% for 25 mg L−1 and 43% for 50 mg L−1 (Fig. 2). The addition of phenol caused a significant decrease (p < 0.05) in the removal of 25 and 50 mg L−1 Cr(VI), reaching values of 70 and 24%, respectively. The maximum Cr(VI) removal registered after 72 h was around 22 mg L−1 for media without phenol, and 17 mg L−1 in the presence of both contaminants.
These results indicate that this strain is suitable to remove the moderate concentrations of Cr(VI) usually found in many industrial effluents worldwide, such as those from leather tanning and dying, wood treatment, paint, pigment, and textile industries. In general, most of the traditional remediation methods are unviable at metal concentrations below 100 mg L−1 and, therefore, those biological systems capable of dealing with these concentrations are very attractive (Narayani and Shetty 2013; Verma and Singh 2013).
The ability to remove Cr(VI) in the presence of phenol is an additional advantage of this strain, considering that many microorganisms display appropriate efficiency for heavy metal removal but are not able to tolerate organic co-pollutants (Sandrin and Maier 2003). Previous publications by our group showed that Bacillus sp. SFC 500-1E remains part of the consortium SFC 500-1 even in the simultaneous presence of high concentrations of phenol and Cr (VI) (Ontañon et al. 2015a). This could be related to the high tolerance of the strain to phenol, although it probably participates only in Cr(VI) removal.
Molecular characterization of Cr(VI) metabolism
Metal efflux and biotransformation are among the main molecular mechanisms involved in bacterial tolerance to Cr(VI) (Gutiérrez-Corona et al. 2016).
Efflux of chromate ions, carried out by the chromate ion transporter protein (ChrA), is a widespread mechanism of resistance among microorganisms, and the chrA gene has been a useful marker for the selection of Cr(VI)-tolerant bacteria from polluted sources (Huang et al. 2017). In the present work, a 314 bp fragment from the genomic DNA of Bacillus sp. SFC 500-1E was amplified employing primers designed from known sequences of the chrA gene in other strains belonging to Bacillus sp. (Fig. 3a) (GenBank accession: KY656903). It exhibited 100% homology with the chrA gene from B. toyonensis BCT-7112 (GenBank accession number CP006863, region 2324187..2324500), a relatively novel species from the B. cereus group (Jiménez et al. 2013). This result would suggest the presence of some metal efflux mechanisms in this strain, which is in agreement with previous reports that describe the presence of a plasmid-encoded chrA gene flanked by transposase and resolvase sequences in various Cr(VI)-tolerant Bacillus strains (He et al. 2010).
Although chromate efflux is an important mechanism that allows bacteria to tolerate this pollutant, such ability does not reduce its toxic effect for other organisms. On the contrary, Cr(VI)-reducing bacteria are able to detoxify Cr(VI) by transforming it into the less toxic Cr(III). It is well known that those mechanisms are in general encoded independently, but if a bacterial strain has molecular determinants for both activities, it will probably survive under chromate stress and could be an interesting candidate for Cr(VI) detoxification (Thatoi et al. 2014). For instance, (Baaziz et al. 2017) suggested that a Cr(VI) efflux pump improved S. oneidensis MR1 survival by lowering intracellular Cr(VI) concentration and consequently Cr-induced damages inside the cells, which allowed the cells to grow and reduce Cr(VI) over an extended period of time.
Several authors have tried to find universal markers for identifying Cr(VI)-reducing bacteria, but it has not been an easy task, considering the structural variability among chromate reductases. With this aim, Patra et al. (2010) suggested a pair of primers designed from the chrR gene of E. coli K12, which were useful to amplify the chrR gene from other Cr(VI) reducing strains belonging to the Bacillus and Arthrobacter genera. The 250 bp fragment amplified from the genome of Bacillus sp. SFC 500-1E using these primers presented 100% identity with the chromate reductase gene from E. coli (GenBank accession number KY656902) (Fig. 3b). Its translated amino acid sequence also presented high homology with the conserved family of bacterial NAD(P)H-dependent FMN reductases, previously characterized by Eswaramoorthy et al. (2012).
These results suggest the presence of genetic determinants for chromate reductases in Bacillus sp. SFC 500-1E.
Characterization of chromate reductase activity
Chromate reductase activity
Cell-free extracts (CFE) obtained from Bacillus sp. SFC 500-1E grown in the presence and in the absence of contaminants were able to remove Cr(VI) in a short reaction time (Fig. 4a). Since almost negligible removal was detected in controls without CFE and heated CFE, such results may be mainly related to the activity of chromate reductases present in the extracts.
a Cr(VI) removal and b specific chromate reductase activity (SRA) in CFE of Bacillus sp. SFC 500-1E obtained from cells growing in the absence [-Cr(VI)] or in the presence of Cr(VI) [+Cr(VI)], or Cr(VI) plus phenol [+Cr(VI) + Ph]. The effect of adding NADH to CFE is also plotted. Dotted lines represent Cr(VI) removed by reaction mixtures without CFE (control) and heated CFE [CFE(H)]. All reaction mixtures contained 10 mg L−1 Cr(VI). In b, different letters indicate SSD between treatments (p < 0.05)
As can be seen in Fig. 4b, the chromate reductase activity of CFE from both Cr(VI)-induced [+Cr(VI)] and uninduced cells [-Cr(VI)] were similar. In this regard, chromate reductases are a group of diverse enzymes with primary metabolic functions different from Cr(VI) reduction, such as nitroreductase, quinonereductase, and flavinreductase (Ackerley et al. 2004; Gonzalez et al. 2005). Thus, the detection of this constitutive activity is not surprising.
On the other hand, the addition of NADH to the reaction mixture caused an increase of above 100% in the activity detected in both CFE-Cr(VI) and CFE + Cr(VI). These results provide additional evidence of the enzymatic nature of Cr(VI) reduction in Bacillus sp. SFC 500-1E, as most of the described reductases are activated by NADH, which serves as a source of reducing power (Thatoi et al. 2014).
It was demonstrated that certain compounds, usually found as co-pollutants in Cr(VI)-contaminated wastewaters, decreased the reductase activity of bacterial CFE (Pal et al. 2005). In this sense, CFE obtained from Bacillus sp. SFC 500-1E growing in the presence of Cr(VI) plus phenol [CFE + Cr(VI) + Ph + NADH] showed 42% less activity compared to those obtained from cells growing only with Cr(VI) [CFE + Cr(VI) + NADH]. This result is in agreement with the removal assays, which showed a diminishment in Cr(VI) removal caused by the addition of phenol.
Chromate reductase localization
As reviewed by Thatoi et al. (2014), bacterial Cr(VI) reductases can be localized in cytosolic or membrane fractions and, in some cases, they are also secreted into the culture medium. In the present study, higher chromate reductase activity was detected in the cytoplasm (3.7 U mg−1 protein), compared to the membrane fraction (2 U mg−1 protein). However, no extracellular activity was measured (Fig. 5).
In agreement with our results, some microorganisms have shown to have combined reductase activity in both fractions, such as certain Actinobacteria strains (Camargo et al. 2004; Polti et al. 2010) and different strains of the Bacillus genus (Iftikhar et al. 2007; Mary Mangaiyarkarasi et al. 2011). Such behavior could be due to the existence of a single chromate reductase with more than one location, or several isozymes with different intracellular locations acting in a synergistic manner (Shen and Wang 1993; Sau et al. 2010).
Chromium destination after removal
The capability of Bacillus sp. SFC 500-1E to reduce Cr(VI) to Cr(III) was confirmed, given that Cr(III) was found associated to biomass after 72 h (Fig. 6a). This interaction between Cr(III) and biomass could be due to either surface sorption or accumulation within cells (Polti et al. 2011). However, as chromate reductase activity was intracellular in this strain, it is likely that Cr(III) had been complexed to intracellular particles rather than adsorbed into the cellular surface. It has been reported that Cr(III) binds to ionic sites of nucleic acids and proteins and later acts as a template for heterogeneous nucleation and crystal growth (Pei et al. 2009). Around 33% of 25 mg L−1 Cr(VI) was accumulated in the reduced and less toxic trivalent form, but an additional 19% was associated to biomass as Cr(VI) (Fig. 6b). Some researchers have highlighted the proper potential of some Bacillus strains to bioadsorb Cr(VI) due to their cellular envelope with plenty of interaction sites (Quintelas et al. 2008; Oves et al. 2013). However, heat-inactivated cells were unable to remove Cr(VI) from the culture medium (data not shown), which suggests that this strain needs living biomass to achieve bioremediation of Cr(VI).
Furthermore, 32% Cr(III) was found to be soluble in the culture medium (Fig. 6b). This suggests the existence of some mechanism of Cr(III) efflux or Cr(III) release by cellular lysis. In line with this assumption, some studies have characterized soluble organo-Cr(III) products, such as NAD+-Cr(III), thiol-Cr(III), and amino-Cr(III), released into culture supernatants during Cr(VI) bioremediation (Puzon et al. 2005; Dong et al. 2013).
TEM photographs of Bacillus sp. SFC 500-1E growing in TY medium supplemented with Cr(VI) showed the presence of electron-dense bodies within the cell, on the bacterial surface, and dispersed in the culture medium (Fig. 7). The intracellular aggregates, organo-Cr(III) complexes, and insoluble Cr(III) precipitates formed after Cr(VI) reduction have been observed as electron-opaque bodies through electronic microscopy (Polti et al. 2011; Chen et al. 2012; Dong et al. 2013; Pan et al. 2014). Coupling TEM to EDX/FTIR analysis would be useful to characterize these bodies and confirm the presence of Cr.
Conclusions
Bacillus sp. SFC 500-1E, which was isolated from the consortium SFC 500-1 and belongs to the B. cereus group of bacteria, tolerated various concentrations of Cr(VI) and other heavy metals, and was hyper-tolerant to phenol. Such tolerance was not necessarily related to the bioremediation potential of the strain, since it was unable to degrade phenol. However, it was able to remove Cr(VI) in co-contaminated synthetic media, probably by the activity of intracellular constitutive NADH-dependent chromate reductases. Such Cr(VI) reduction to the less toxic Cr(III) and its subsequent immobilization into the biomass constitutes a useful strategy for bioremediation purposes. In addition, the strain presented an encoding gene for the protein ChrA suggesting Cr(VI) extrusion; however, this activity should be verified.
The ability to reduce Cr(VI) and grow in the presence of other contaminants makes this strain interesting for future application in bioremediation, either isolated or as part of the consortium SFC 500-1. The information provided in the present work may also be useful for a further optimization of the bioremediation process carried out by this microorganism.
References
Abha S, Singh CS (2012) Hydrocarbon pollution: effects on living organisms, remediation of contaminated environments, and effects of heavy metals co-contamination on bioremediation. Introduction to Enhanced Oil Recovery (EOR) processes and bioremediation of oil-contaminated sites 185–206
Ackerley DF, Gonzalez CF, Park CH et al (2004) Chromate-reducing properties of soluble Flavoproteins from pseudomonas putida and Escherichia coli. Society 70:873–882
Ackerley DF, Barak Y, Lynch SV, Curtin J, Matin A (2006) Effect of chromate stress on Escherichia coli K-12. J Bacteriol 188:3371–3381
Ahemad M (2014) Bacterial mechanisms for Cr(VI) resistance and reduction: an overview and recent advances. Folia Microbiol 59:321–332
Al-Khalid T, El-Naas MH (2012) Aerobic biodegradation of phenols: a comprehensive review. Crit Rev Environ Sci Technol 42:1631–1690
APHA-AWWA (1989) Standard methods for the examination of water and wastewater, 17th edn
Baaziz H, Gambari C, Boyeldieu A et al (2017) ChrASO, the chromate efflux pump of Shewanella oneidensis, improves chromate survival and reduction. PLoS One 12:1–15
Banerjee A, Ghoshal AK (2016) Biodegradation of real petroleum wastewater by immobilized hyper phenol-tolerant strains of Bacillus cereus in a fluidized bed bioreactor. 3 Biotech 6:1–4
Bencosme SA, TsutSumi V (1970) Fast method for processing biologic material for electron microscopy. Lab Investig 23:447–450
Beringer JE (1974) R factor transfer in Rhizobiurn Zegurninosarum. J Gen Microbiol 84:188–198
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 254:248–254
Camargo FAO, Bento FM, Okeke BC, Frankenberger WT (2004) Hexavalent chromium reduction by an actinomycete, Arthrobacter crystallopoietes ES 32. Biol Trace Elem Res 97:183–194
Chen Z, Huang Z, Cheng Y, Pan D, Pan X, Yu M, Pan Z, Lin Z, Guan X, Wu Z (2012) Cr(VI) uptake mechanism of Bacillus cereus. Chemosphere 87:211–216
Dhal B, Thatoi HN, Das NN, Pandey BD (2013) Chemical and microbial remediation of hexavalent chromium from contaminated soil and mining/metallurgical solid waste: a review. J Hazard Mater 250–251:272–291
Dong G, Wang Y, Gong L, Wang M, Wang H, He N, Zheng Y, Li Q (2013) Formation of soluble Cr(III) end-products and nanoparticles during Cr(VI) reduction by Bacillus cereus strain XMCr-6. Biochem Eng J 70:166–172
Eswaramoorthy S, Poulain S, Hienerwadel R et al (2012) Crystal structure of ChrR-A quinone reductase with the capacity to reduce chromate. PLoS One 7:1–7
Fernández M, Morales GM, Agostini E, González PS (2017) An approach to study ultrastructural changes and adaptive strategies displayed by Acinetobacter guillouiae SFC 500-1A under simultaneous Cr(VI) and phenol treatment. Environ Sci Pollut Res 24:20390–20400
Gadd GM (1992) Metals and microorganisms: a problem of definition. FEMS Microbiol Lett 100:197–203
Gonzalez CF, Ackerley DF, Lynch SV, Matin A (2005) ChrR, a soluble quinone reductase of Pseudomonas putida that defends against H2O2. J Biol Chem 280:22590–22595
Gutiérrez-Corona JF, Romo-Rodríguez P, Santos-Escobar F, Espino-Saldaña AE, Hernández-Escoto H (2016) Microbial interactions with chromium: basic biological processes and applications in environmental biotechnology. World J Microbiol Biotechnol 32:191
He M, Li X, Guo L et al (2010) Characterization and genomic analysis of chromate resistant and reducing Bacillus cereus. BMC Microbiol 10:221
Huang Y, Feng H, Lu H, Zeng Y (2017) A thorough survey for Cr-resistant and/or -reducing bacteria identified comprehensive and pivotal taxa. Int Biodeterior Biodegrad 117:22–30
Iftikhar S, Faisal M, Hasnain S (2007) Cytosolic reduction of toxic Cr (VI) by indigenous microorganism. Res J Environ Sci 1:77–81
Jiménez G, Urdiain M, Cifuentes A, López-López A, Blanch AR, Tamames J, Kämpfer P, Kolstø AB, Ramón D, Martínez JF, Codoñer FM, Rosselló-Móra R (2013) Description of Bacillus toyonensis sp. nov., a novel species of the Bacillus cereus group, and pairwise genome comparisons of the species of the group by means of ANI calculations. Syst Appl Microbiol 36:383–391
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Liu Y, Mou H, Chen L, Mirza ZA, Liu L (2015) Cr(VI)-contaminated groundwater remediation with simulated permeable reactive barrier (PRB) filled with natural pyrite as reactive material: environmental factors and effectiveness. J Hazard Mater 298:83–90
Mary Mangaiyarkarasi MS, Vincent S, Janarthanan S, Subba Rao T, Tata BVR (2011) Bioreduction of Cr(VI) by alkaliphilic Bacillus subtilis and interaction of the membrane groups. Saudi J Biol Sci 18:157–167
McLean J, Beveridge TJ (2001) Chromate reduction by a pseudomonad isolated from a site contaminated with chromated copper arsenate. Appl Environ Microbiol 67:1076–1084
Megharaj M, Ramakrishnan B, Venkateswarlu K, Sethunathan N, Naidu R (2011) Bioremediation approaches for organic pollutants: a critical perspective. Environ Int 37:1362–1375
Murugavelh S, Mohanty K (2012) Bioreduction of hexavalent chromium by free cells and cell free extracts of Halomonas sp. Elsevier B.V
Narayani M, Shetty KV (2013) Chromium-resistant bacteria and their environmental condition for hexavalent chromium removal: a review. Crit Rev Environ Sci Technol 43:955–1009
Ontañon OM, González PS, Agostini E (2015a) Optimization of simultaneous removal of Cr (VI) and phenol by a native bacterial consortium: its use for bioaugmentation of co-polluted effluents. J Appl Microbiol 119:1011–1022
Ontañon OM, González PS, Agostini E (2015b) Biochemical and molecular mechanisms involved in simultaneous phenol and Cr(VI) removal by Acinetobacter guillouiae SFC 500-1A. Environ Sci Pollut Res 22(17):13014–13023
Ontañon OM, González PS, Barros GG, Agostini E (2017) Improvement of simultaneous Cr(VI) and phenol removal by an immobilised bacterial consortium and characterisation of biodegradation products. New Biotechnol 37:172–179
Oves M, Khan MS, Zaidi A (2013) Biosorption of heavy metals by Bacillus thuringiensis strain OSM29 originating from industrial effluent contaminated north Indian soil. Saudi J Biol Sci 20:121–129
Paisio CE, Talano MA, González PS et al (2013) Characterization of a phenol-degrading bacterium isolated from an industrial effluent and its potential application for bioremediation. Environ Technol (UK) 34:485–493
Pal A, Dutta S, Paul a K (2005) Reduction of hexavalent chromium by cell-free extract of Bacillus sphaericus AND 303 isolated from serpentine soil. Curr Microbiol 51:327–330
Pan X, Liu Z, Chen Z, Cheng Y, Pan D, Shao J, Lin Z, Guan X (2014) Investigation of Cr(VI) reduction and Cr(III) immobilization mechanism by planktonic cells and biofilms of Bacillus subtilis ATCC-6633. Water Res 55:21–29
Patra RC, Malik S, Beer M, Megharaj M, Naidu R (2010) Soil biology & biochemistry molecular characterization of chromium (VI) reducing potential in gram positive bacteria isolated from contaminated sites. Soil Biol Biochem 42:1857–1863
Pei QH, Shahir S, Santhana Raj AS, Zakaria ZA, Ahmad WA (2009) Chromium(VI) resistance and removal by Acinetobacter haemolyticus. World J Microbiol Biotechnol 25:1085–1093
Polti MA, Amoroso MJ, Abate CM (2010) Full paper chromate reductase activity in Streptomyces sp. MC1. Cultures 18:11–18
Polti MA, Amoroso MJ, Abate CM (2011) Intracellular chromium accumulation by Streptomyces sp. MC1. Water Air Soil Pollut 214:49–57
Puzon GJ, Roberts AG, Kramer DM, Xun L (2005) Formation of soluble organo-chromium(III) complexes after chromate reduction in the presence of cellular organics. Environ Sci Technol 39:2811–2817
Quintelas C, Fernandes B, Castro J, Figueiredo H, Tavares T (2008) Biosorption of Cr(VI) by a Bacillus coagulans biofilm supported on granular activated carbon (GAC). Chem Eng J 136:195–203
Saffert RT, Cunningham SA, Ihde SM, Monson Jobe KE, Mandrekar J, Patel R (2011) Comparison of Bruker Biotyper matrix-assisted laser desorption ionization-time of flight mass spectrometer to BD phoenix automated microbiology system for identification of gram-negative bacilli. J Clin Microbiol 49:887–892
Sandrin TR, Maier RM (2003) Impact of metals on the biodegradation of organic pollutants. Environ Health Perspect 111:1093–1101
Saranraj P, Sujitha D (2013) Microbial bioremediation of chromium in tannery effluent: a review. International journal of. Microbiol Res 4:305–320
Sau GB, Chatterjee S, Mukherjee SK (2010) Chromate reduction by cell-free extract of Bacillus firmus KUCr1. Pol J Microbiol 59:185–190
Shen H, Wang YT (1993) Characterization of enzymatic reduction of hexavalent chromium by Escherichia coli ATCC 33456. Appl Environ Microbiol 59:3771–3777
Somasegaran P, Hoben HJ (1994) Methods in legume-rhizobium technology
Thatoi H, Das S, Mishra J, Prasad B (2014) Bacterial chromate reductase , a potential enzyme for bioremediation of hexavalent chromium : a review. J Environ Manag 146:383–399
Tripathi M, Vikram S, Jain RK, Garg SK (2011) Isolation and growth characteristics of chromium(VI) and pentachlorophenol tolerant bacterial isolate from treated tannery effluent for its possible use in simultaneous bioremediation. Indian J Microbiol 51:61–69
Verma T, Singh N (2013) Isolation and process parameter optimization of Brevibacterium casei for simultaneous bioremediation of hexavalent chromium and pentachlorophenol. J Basic Microbiol 53:277–290
Viti C, Marchi E, Decorosi F, Giovannetti L (2014) Molecular mechanisms of Cr(VI) resistance in bacteria and fungi. FEMS Microbiol Rev 38:633–659
Wagner M, Nicell JA (2002) Detoxification of phenolic solutions with horseradish peroxidase and hydrogen peroxide. Water Res 36:4041–4052
Acknowledgements
Paola S. González and Elizabeth Agostini are researchers from Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) (Argentina). Ornella M. Ontañon and Marilina Fernandez are on a fellowship from CONICET. This work was supported by grants from PPI (SECyT-UNRC), CONICET, MinCyT Córdoba, and PICT (FONCyT).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Responsible editor: Philippe Garrigues
Electronic supplementary material
ESM 1
(DOCX 12 kb)
Rights and permissions
About this article
Cite this article
Ontañon, O.M., Fernandez, M., Agostini, E. et al. Identification of the main mechanisms involved in the tolerance and bioremediation of Cr(VI) by Bacillus sp. SFC 500-1E. Environ Sci Pollut Res 25, 16111–16120 (2018). https://doi.org/10.1007/s11356-018-1764-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-018-1764-1