Abstract
The proper handling of low-level radioactive waste is crucial to promote the sustainable development of nuclear power. Research into the mechanism for interactions between bacterium and radionuclides is the starting point for achieving successful remediation of radionuclides with microorganisms. Using Sr(II) as a simulation radionuclide and the mixed microorganisms of Saccharomyces cerevisiae and Bacillus subtilis as the biological adsorbent, this study investigates behavior at the interface between Sr(II) and the microorganisms as well as the mechanisms governing that behavior. The results show that the optimal ratio of mixed microorganisms is S. cerevisiae 2.0 g L−1 to B. subtilis 0.05 g L−1, and the optimal pH is about 6.3. Sr(II) biosorption onto the mixed microorganisms is spontaneous and endothermic in nature. The kinetics and the equilibrium isotherm data of the biosorption process can be described with pseudo-second-order equation and the Langmuir isotherm equation, respectively. The key interaction between the biological adsorbent and Sr(II) involves shared electronic pairs arising from chemical reactions via bond complexation or electronic exchange, and spectral and energy spectrum analysis show that functional groups (e.g., hydroxyl, carboxyl, amino, amide) at the interface between the radionuclide and the mixed microorganisms are the main active sites of the interface reactions.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Nuclear energy has already been developed and widely used as an energy source, but treatment and disposal of its by-product, nuclear waste, have become a central problem impeding nuclear energy development. Nuclear waste can be harmful to the biosphere for hundreds or even tens of hundreds of years because of the long half-life of some of the radionuclides it contains (e.g., 90Sr, 137Cs). Radioactive wastewater is the largest component of nuclear waste (Hu et al. 2010). Conventional methods for treating it include chemical precipitation, ion exchange, evaporation, and biochemical and electrochemical techniques. But these methods are not perfected and can easily generate secondary contamination. In addition, their capital and operating costs are often high, especially for treatment of radionuclides in low-level waste. The advance of biotech has included intensive research into the interaction mechanisms between microorganisms and radionuclides, and there has been a growing awareness that the microbial treatment of radioactive wastewater is a very promising method (Lovley et al. 1991; Ahluwalia and Goyal 2007).
Biosorption is one of the treatments that uses the cheap organism to sequester radionuclides and heavy metal ions, and it is particularly suitable for decontaminating of industrial effluents. Due to its high efficiency, low cost, low-energy consumption, environmental friendliness, and easily renewable sorbent, it is widely investigated for treatment and disposal of radioactive wastewater and other wasters containing low concentrations of radionuclide and as a method that makes radionuclide recycling possible (Chojnacka 2010).
A large quantity of organisms have been investigated as biosorbents for the removal of radionuclides, including bacteria (e.g., Bacillus polymyxa) (Shevchuk et al. 2010), fungi (e.g., Rhizopus arrhizus) (Aksu and Balibek 2007), yeast (e.g., Saccharomyces cerevisiae) (Chen and Wang 2008; Wu et al. 2012), and algae (e.g., Cladophora hutchinsiae) (Tuzen and Sari 2010), and the target radionuclides include isotopes of U (Morcillo et al. 2014), Pu (Zotina et al. 2011), Am (Liu et al. 2003), Th (Zhou et al. 2016), and Cs and Sr (Mashkani and Ghazvini 2009). Studies of the removal of radionuclides through bioremediation generally use just one as the biosorbent, and little research has been done involving a mixture of two or more microorganisms, even though a variety of radionuclides and heavy metals is usually present in actual radioactive wastewater. For instance, the wastewater from a nuclear power in Turkey contains 90Sr, 137Cs, 60Co 110mAg, etc. (Osmanlioglu 2006). In such cases, it turns out that one cannot rely on just one microorganism to treat the wastewater effectively. Indeed, experiments concerning the removal of Sr2+, Pb2+, Cs+, and Ag+ from a shared solution by S. cerevisiae have found that the isotopes in question are adsorbed in the order Pb2+ > Ag+ > Sr2+ > Cs+, which means that Sr was not adsorbed preferentially (Chen and Wang 2010). Instead, two or more species of microorganisms with high adsorption efficiency for a particular radionuclide can be combined in order to achieve better adsorption efficiency and complementary advantages. Our previous studies have shown that S. cerevisiae and Bacillus subtilis are potential biosorbents for the treatment of Sr in radioactive wastewater (Liu et al. 2016; Liu et al. 2015). And, since they belong to entirely different biological domains, their outer membrane structures and component of the cell wall are not the same. S. cerevisiae belongs to unicellular fungi, whose cell wall contains D-dextran (main group -OH) and D-mannan (main group -OH) (Mcmurrough and Rose 1967), whereas B. subtilis is a peptidoglycan of gram positive bacteria, whose cell wall contains nearly 90% of peptidoglycan (main group -COOH, -OH, and -NH) and 10% of teichoic acid (main group -PO3H2) (Antelmann et al. 2007; Fang et al. 2011). Due to these differences in cell wall composition, the key functional groups and the adsorption mechanism for Sr removal are not the same for the two microorganisms. Consequently, different functional groups, in a complex system of multiple ions, might complement each other during adsorption, resulting in high adsorption capacity. Pursuing this possibility, the present study focuses on the capacity of a biosorbent composed of a mixture of microorganisms specifically, S. cerevisiae and B. subtilis, to sequester Sr(II) in a simulated solution of the nuclide 90Sr, thereby modeling how one might treat targeted low-level radionuclides in complex wastewater. It covers the ratio of microorganisms, biosorption capacity, biosorption equilibrium, biosorption kinetics, and the parameters that influence biosorption efficiency, such as pH, contact time, coexisting ions, initial concentration of Sr(II), temperature, and so forth, as well as behavior of the microorganisms at the adsorption interface.
Experimental
Preparation of materials
S. cerevisiae was supplied by Hubei Angel Yeast Co. Ltd., China and was grown aerobically in a nutrient broth containing glucose 20.0 g L−1, peptone 5.0 g L−1, and yeast extract 2.0 g L−1 for 48 h at 28 °C. The yeast cells were harvested by centrifuging at 4000 rpm for 15 min, washed with distilled water, vacuum freeze-dried, and then stored in the refrigerator at 4 °C for experimental use.
B. subtilis was supplied by Guangdong Culture Collection Center, China and was grown aerobically in a nutrient broth containing tryptone 10.0 g L−1, meat extract 3.0 g L−1, and NaCl 5.0 g L−1, for 48 h at 37 °C. Microorganism cells were harvested by centrifuging at 4000 rpm for 30 min, washed with distilled water, vacuum freeze-dried, and then stored in the refrigerator at 4 °C for experimental use.
The strontium nitrate, cesium nitrate, copper nitrate, and lead nitrate used were of analytical grade. A 1000 mg L−1 stock solution was prepared for each type of heavy metal ion. Other concentrations were obtained by diluting the stock solution using double-distilled water. The pH was adjusted through 0.1 mol L−1 HNO3 and 0.1 mol L−1 NaOH to the values selected in the experimental design.
Adsorption experiment
S. cerevisiae (S) (0.5, 1.0, 1.5, 2.0 g L−1) and B. subtilis (B) (0.05, 0.10, 0.15 g L−1) were mixed at stated ratios. Batch experiments were conducted by mixing the microorganisms in a solution containing Cu2+, Pb2+, Sr2+, and Cs+ at initial concentrations of 15 to 1000 mg L−1. This mixture was agitated at 150 rpm in a water bath at a controlled temperature. All experiments were conducted in triplicate. The removal efficiency of metal η (%) and the biosorption capacity q (mg g−1) of Sr(II) were calculated according to Eqs. (1) and (2):
where C 0 is the initial mental concentration (mg L−1), C t is the metal concentration (mg L−1) after time t, V is the volume of the solution (L), and M is the mass of the dry biomass dosage (g).
Analytical measurements
The pH was measured using a pH meter (Model PHS-3C+, Fangzhou China), and the metal concentrations using an atomic absorption spectrometer (AA700, PerkinElmer, USA). The organic functional groups on the sorbent structure were indentified using a Fourier transform infrared spectrophotometer (Nicolet 6700, USA) over a range of 400–4000 cm−1. The surface charges of the biomass were measured using a Zeta potential analyzer (ZetaPALS 90Plus, Brookhaven, USA). Field emission scanning electron microscopy (FE-SEM) was done on a Carl Zeiss Irts Libra200 to determine the morphology of the microorganisms before and after biosorption. X-ray photoelectron spectroscopy (XPS) was done using a Kratos XSAM 800 instrument with a magnesium anode at 225 W.
Statistical procedures
The presented data are the mean values obtained from three independent experiments. Experimental errors were estimated and represented with error bars in the figures. To compare the adsorption capacity difference between each microorganism by itself and the mixture of microorganisms, Student’s t test was applied at a 95% confidence level.
Results and discussion
Ratio of mixed microorganisms
According to a preliminary study of the adsorption capacity of each single microorganism, the optimum dosage of S. cerevisiae and B. subtilis at a Sr(II) concentration of c 0 = 15 mg L−1 was 4.0 and 0.3 g L−1, respectively. The dosage for the mixed microorganisms was set at half of the dosage for each microorganism by itself (Liu et al. 2016; Liu et al. 2015). The concentration of pollutants in low-level radioactive waste is usually less than 100 mg L−1, so the goal of the study was to achieve to the optimal adsorption capacity for each single microorganism using the minimum amount of each microorganism in the mixture. The results of different ratios of the microorganisms appear in Tables 1, 2, and 3.
When the mixture consisted of S. cerevisiae 2.0 g L−1 and B. subtilis 0.05 g L−1, (see Table 3), the Sr(II) removal efficiency—96.87 and 84.56% at Sr concentration of c 0 = 15 mg L−1 and c 0 = 100 mg L−1, respectively—was surpassed that for the single microorganism. Thus, peak Sr(II) removal efficiency required only two thirds as much of the microorganisms when they were mixed as it did when they were used separately. When Student’s t test was used to compare the adsorption capacity of the mixed microorganisms to that of the two microorganisms used separately, the value of t for S. cerevisiae versus the mixture and for B. subtilis versus the mixture were 2.10 and 1.61, respectively, both of which are less than t 0.05,2 of 4.30. In other words, the adsorption efficiency of the mixed microorganisms does not differ significantly from that of each microorganism by itself, even though the amount of mixed microorganisms used in the solution is considerably less than each single microorganism. One can say, therefore, that the optimal ratio of the microorganisms when mixed S. cerevisiae at 2.0 g L−1 and B. subtilis at 0.05 g L−1.
Effect of pH on biosorption
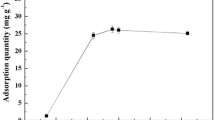
The pH of the solution has been seen as the most important parameter affecting metal biosorption. The results shown in Fig. 1 reveal that little Sr biosorption occurred under highly acidic conditions. The biosorption capacity for mixed biomass sharply increased, from 0.50 to 7.35 mg g−1, over the pH range 2.0 to 6.0. The lower biosorption capacity observed at lower pH could be largely due to mutual exclusion between metal cations and positively charged binding sites on the cell walls of the microorganisms. As the solution pH increases, deprotonation of the cell surfaces proceeds while the density of negatively charged sites on the cells increases, which facilitates metal adsorption by the biosorbent. The pH-dependent changes in the zeta potentials for the mixed microorganisms before and after biosorption presented in Fig. 2 show that the mixed microorganisms’ surface was positively charged when the pH was lower than 1.08, the isoelectric point of the mixed microorganisms. When pH was higher than the isoelectric point, the surface of the mixed microorganisms was negatively charged and the negative zeta potential values increased as the pH values increased under 6.3, which explains why biosorption capacity increases with pH value. At pH 6.3, the peak value of zeta potential was gained. When the pH was higher than 6.3, the negative zeta potential values decreases with pH value. The zeta potential value of the mixed microorganisms before adsorption was more negative than that of after adsorption, which means that electrostatic interaction is one of the adsorption mechanisms. Hence, biosorption studies in this paper were carried out at pH 6.3 (original pH value of Sr(II) solution).
Kinetic modeling of the biosorption process
Figure 3 shows the adsorption efficiency of Sr(II) by mixed microorganisms over time of contact. The data indicate that the adsorption process for the mixed microorganisms is similar to that for each microorganism by itself. Rapid biosorption of Sr(II) (η%, 97.64) at the cell surface was observed at the beginning of the process (i.e., at about 10 min), after which the process involves the transfer of Sr(II) from the cell membrane into the cell interiors (i.e., passive adsorption, which occurs at a slower pace). Therefore, the increase in the removal efficiency for Sr(II) slows, and eventually, it reaches a near-equilibrium maximum value of 98.38%. It was reported (Özacar and Şengil 2003) that the fast kinetics of metal biosorption involving no energy-mediated reactions could be attributed to purely physico-chemical interactions between the biosorbent and metal ions in the biosorption medium.
The pH curve for the aqueous solution during the adsorption process appears in Fig. 4. A downward trend in the pH was observed during the rapid adsorption stage (stage a), and pH increased and then fell during the slow adsorption stage (stage b), where passive adsorption takes over. This indicates that Sr(II) transfer from the cell surface to the cell caused a charge imbalance on the cell surface, which in turn caused part of the H+ to be adsorbed onto the cell surface, thereby increasing the pH of the aqueous solution. But as adsorption proceeded, the pH of the aqueous solution decreased again, presumably because Sr(II) replaced the H+ on the cell surface (stage c), suggesting that ion exchange was one of adsorption mechanisms.
In order to explain these biosorption kinetics, a pseudo-second-order (Günay et al. 2007) equation was applied to the experimental data (see Fig. S1). This figure demonstrates that the kinetics of Sr(II) adsorption by mixed microorganisms is well described by the pseudo-second-order kinetic model. The linearity of the plot shows that the pseudo-second-order kinetic model applies with a regression coefficient of R 2 = 0.9999. According to this pseudo-second-order kinetic model, the equilibrium adsorption amount (q e ) is 7.18 mg g−1 and k 2 is 1.16 g (mg min)−1.
Adsorption isotherms
The Sr(II) biosorption capacity of mixed microorganisms at different temperatures appears in Fig. S2. This figure shows that the biosorption capacity increases with the Sr(II) concentration, and the adsorption equilibrium is achieved at a different temperature, which causes the certain equilibrium of Sr(II) concentration. When the dosage of mixed microorganisms is held constant, an increase in the Sr(II) concentration leads to a lack of enough active adsorption sites, and competition for the sites means that Sr(II) in the solution cannot be completely adsorbed by the limited biomass. Figure S2 also shows that the adsorption capacity increases with temperature at the same concentration of Sr(II). Thus, the adsorption process is an endothermic process (Table S1), which means that higher temperatures are beneficial to adsorption.
This study uses Langmuir and Freundlich adsorption isotherms to examine the interaction between the mixed microorganisms and the metal. The Freundlich model assumes that the sorption surface is heterogeneous and the binding sites have different sorption energies. The Langmuir model assumes a monolayer type of biosorption at the specific homogeneous sites of the biosorbent. The equations for the two models are as follows:
where q e is the equilibrium adsorption amount (mg g−1), and c e is the equilibrium concentration of metal in the solution (mg L−1). In Eq. (3), K L (L mg−1) is the Langmuir adsorption equilibrium constant, q m is the maximum amount of metal biosorption (mg g−1); in Eq. (4), K F is the Freundlich equilibrium binding constant corresponding to the maximum binding energy, and n is a Freundlich constant depicting the sorption intensity (Günay et al. 2007).
The model parameters for the two isotherms along with the R 2 values appear in Table 4. The regression coefficient (R 2) for the Langmuir isotherm equation shows that it is an excellent fit with the experimental data. The calculated maximum Sr(II) sorption capacity using this model is 40.92 mg g−1 at 37 °C, and the maximum K L is 0.1859 L mg−1 at 28 °C, which means that the adsorption rate of mixed microorganisms is highest at 28 °C and their adsorption capacity is highest at 37 °C. Considering the adsorption behavior of each single microorganism and mixed microorganisms, further experiments were conducted at 28 °C.
Effect of coexisting ions on biosorption
Many different ions, such as uranium (Kalin et al. 2005), radium (Warner et al. 2013), thorium (Humelnicu et al. 2011), cesium (Seliman 2012), copper (Zhou et al. 2017), and lead (Yang et al. 2008), are always present in actual low-level radioactive wastewater coming from the nuclear power station or the uranium mine. These coexisting ions will compete with the target ion for the adsorption sites on cell surfaces, thereby reducing the adsorption capacity for the target ion. Given the probable contents of the radioactive wastewater that might be treated using mixed microorganisms, this study focuses on the effects of three coexisting metal ions, namely Pb2+, Cu2+, and Cs+, on the Sr2+ adsorption process and adsorption equilibrium.
Figure 5 shows that, according to the data produced by this study, these three metal ions have little impact on the Sr(II) adsorption process or adsorption equilibrium at low concentrations of metal ions. This is presumably due to the presence of a sufficient number of microorganisms and therefore adsorption sites at those concentrations. Up to a concentration of 40 mg L−1, the selective adsorption order for coexisting metal ions on mixed microorganisms is Sr2+ > Pb2+ > Cu2+ > Cs+. But if the concentration exceeds 40 mg L−1, the order changes to Pb2+ > Sr2+ > Cu2+ > Cs+, which implies that the mixed microorganisms have a strong selective adsorption for Sr(II) at the low metal ion concentrations present in radioactive wastewater and are therefore an effective adsorbent for treating it if the target ion is strontium.
Effects of coexisting ions. a Effect of coexisting ions on biosorption versus time. b Effect of coexisting ions on biosorption versus initial concentration of ions (temperature 28 °C; mixed microorganisms dosage: Saccharomyces cerevisiae 2.0 g L−1 and Bacillus subtilis 0.05 g L−1; initial pH 6.3; initial Sr(II), Cs(I), Pb(II), and Cu(II) concentrations 15 mg L−1)
Interface behavior of biosorption
In order to prove up the mechanism by which the mixed microorganisms adsorbed strontium, this study imaged the microorganisms before and after adsorption, using SEM, EDS, XPS, and in situ IR. These images provide information about the surface functional groups of the microorganisms, the active sites, changes in the microorganisms’ surface, and similarities and differences between single species of microorganisms and the mixed microorganisms.
Figure 6 shows the SEM and EDS images. The mixed microorganisms after adsorption used for characterizing are isolated, washed with distilled water, and vacuum freeze-dried after the adsorption equilibrium reaching. The mixed microorganisms before adsorption are relatively independent of each other, and S. cerevisiae and B. subtilis are evenly distributed across the windows. After adsorption, however, derivatives from the outer cell walls cause the microorganisms to adhere. EDS analysis reveals that there is a strontium ion peak after adsorption in derivatives of the outer cell walls, which means that strontium ions are adsorbed by the mixed microorganisms, and complexes form between the Sr and the cell surfaces. Usually, strontium ions are firstly adsorbed on the surface of the cell membrane, and this process may cause some changes on the cell surface that make it sticky. Subsequently, however, cellular metabolism causes strontium ions adsorbed on the cell surface to exchange with calcium ions, entering calcium storage places within the cell like vacuoles. Eventually, adsorption equilibrium is reached.
Figure 7 shows the results of surface analysis of mixed microorganisms using X-ray photoelectron spectroscopy. It can be seen that the Sr 3d peak (Fig. 7a) is not seen before adsorption, but it is observed at 133.43 eV after absorption (Fig. 7b). This implies that Sr ions are adsorbed by the mixed microorganisms, which the aforementioned EDS results confirm. Then, the fine spectra of Sr peak (Fig. 7c) were calculated by means of XPS peak differentiation imitating analysis. The standard map of Sr 3d has the Sr 3d3/2 and 3d5/2 peaks at 135.5 and 133.7 eV (Young and Otagawa 1985), respectively, with a shift to low electron binding energy at 134.84 and 133.36 eV. It means that the coordination bonds are formed with highly electronegative elements, such as O, N, S, and P, which cause chemical shift of the inner electrons.
Usually, adsorption onto microorganisms happens in two stages. The first involves passive adsorption at the cell surface, which is very rapid and reaches adsorption equilibrium within 10 min, while the second involves the transfer of Sr(II) from the cell membrane into the cell interiors, an active process that necessarily depends on the energy system and metabolism of the cell and is very slow.
Thus, the first stage of adsorption consumes over 90% of the total adsorption capacity, and monitoring the changes in the surface functional groups of microorganisms during passive adsorption suffices to explain their adsorption behavior. Consequently, the in situ infrared spectra for the microorganisms during the first 30 min of adsorption, the fast adsorption stage, were monitored in order to identify the main functional groups in the adsorption process and analyze the differences between the adsorption behavior of the single species of microorganisms and the mixed microorganisms, as Figs. 8 and 9 show. It is clear both that the change in the infrared spectra before and after adsorption is mainly concentrated in the range of 1800–800 cm−1 and that the adsorption behavior of the mixed microorganisms is affected by the concentration of Sr(II). Their adsorption behavior at a low concentration of Sr(II) (15 mg L−1) represents that of B. subtilis, which contributes the smaller share of the two microorganisms, and the adsorption behavior of the mixed microorganisms at a high concentration of Sr(II) (100 mg L−1) represents that of S. cerevisiae, which contributes the larger share of the two microorganisms. The different sizes of the two types of mixed microorganisms, smaller for B. subtilis and larger for S. cerevisiae, lead to different adsorption behaviors. A smaller size means a larger specific surface, more contact sites, and easier bonding between functional groups and Sr. Therefore, as stated above, the adsorption behavior of the mixed microorganisms represents that of B. subtilis at low Sr concentrations, but, since the B. subtilis adsorption sites are saturated first, the adsorption behavior of the mixed microorganisms represents that of S. cerevisiae at high Sr concentrations.
In situ infrared spectra of the microorganisms before and after the first 30 min of biosorption Sr. a Saccharomyces cerevisiae at 100 mg L−1. b Mixed microorganisms at 100 mg L−1. c Amplification of b. d Bacillus subtilis at 15 mg L−1. e Mixed microorganisms at 15 mg L−1. f Amplification of e (temperature 28 °C; mixed microorganisms dosage: S. cerevisiae 2.0 g L−1 and B. subtilis 0.05 g L−1; initial pH 6.3)
According to the scientific literature reported (Fang et al. 2010), the main active groups between 1800 to 800 cm−1 on the surface of S. cerevisiae are hydroxyl, amino, carboxyl, amide, and phosphoryl, and those on the surface of B. subtilis are hydroxyl, amino, carboxyl, phosphoryl, and a sulfur-containing group. The intensity of the characteristic absorption peak corresponding to the main active groups on these two microorganisms increases when they are combined, which provides evidence that the mixed microorganisms synergistically enhance the adsorption of Sr.
As shown in Figs. 8 and 9, the peak at 1638 cm−1 can be attributed to C=O stretching vibration of carboxylate and N-H deformation vibration of the amide I band, which remains unchanged after adsorption at a low Sr(II) concentration and shifts to 1639 cm−1 after adsorption at a high Sr(II) concentration. The peak at 1537 cm−1 can be assigned to N-H bending vibration and C-N stretching vibration, which disappears after adsorption at a low Sr(II) concentration and shifts to 1553 cm−1 after adsorption at a high Sr(II) concentration. The peak at 1403 cm−1 can be ascribed to C-O stretching vibration and C=O bending vibration, which shifts to 1405 cm−1 after adsorption at either low or high Sr concentrations. Similarly, the peak at 1241 cm−1 can be attributed to O-H bending vibration, which shifts to 1243 cm−1. The peak at 1045 cm−1 can be assigned to C-OH stretching vibration in carbohydrate and P-O-C stretching vibration, which shifts to 1048 cm−1 at a low Sr(II) concentration and to 1053 cm−1 at a high Sr(II) concentration. This implies that those organic functional groups involve in adsorption and form coordination bonds of (C=O)-Sr, H-O-Sr, C-N-Sr, and H-N-Sr, which lead to changes in the force constant of their specific chemical bonds, which in turns causes the absorption peaks to drift. This result is the same as for the single types of microorganisms whose functional groups are involved in adsorption.
Conclusions
This study measured the adsorption of Sr(II) onto the mixed microorganisms of S. cerevisiae and B. subtilis after they were vacuum freeze-dried. The dosage of the mixed microorganisms was about 33.3% smaller than that of either of the single microorganism at equal adsorption efficiencies. The negative values of ΔG and the positive values of ΔH show that Sr(II) biosorption by the mixed microorganisms is a spontaneous and endothermic process in nature. The isotherm biosorption process for the adsorption of Sr(II) by the mixed microorganisms is an excellent fit with the Langmuir isotherm equation. The biosorption kinetics follows a pseudo-second-order model (R 2 = 0.9999). The selective order of adsorption by mixed microorganisms for coexisting metal ions is Sr2+ > Pb2+ > Cu2+ > Cs+ under a low concentration of Sr(II), indicating that a mixture of microorganisms is a biologically suitable adsorbent at that concentration. The results from in situ FTIR spectra analysis suggest that C=O, O-H, N-H, and amide may be involved in the biosorption of Sr(II). Synergistic absorption enhancement in the presence of mixed microorganisms is due to association among the functional groups, with complexation between Sr and the highly electronegative atoms on the surface of mixed microorganisms, further promoting adsorption. Preferential adsorption of Sr by mixed microorganisms in the presence of coexisting ions provides an experimental basis for treatment of a target radioactive ion.
References
Ahluwalia SS, Goyal D (2007) Microbial and plant derived biomass for removal of heavy metals from wastewater. Bioresour Technol 98(12):2243–2257
Aksu Z, Balibek E (2007) Chromium(VI) biosorption by dried Rhizopus arrhizus: effect of salt (NaCl) concentration on equilibrium and kinetic parameters. J Hazard Mater 145(1–2):210–220
Antelmann H, Töwe S, Dirk Albrecht A, Hecker M (2007) The phosphorus source phytate changes the composition of the cell wall proteome in Bacillus subtilis. J Proteome Res 6(2):897–903
Chen C, Wang JL (2008) Removal of Pb2+, Ag+, Cs+ and Sr2+ from aqueous solution by brewery’s waste biomass. J Hazard Mater 151(1):65–70
Chen C, Wang JL (2010) Removal of heavy metal ions by waste biomass of Saccharomyces cerevisiae. J Environ Eng 136(1):95–102
Chojnacka K (2010) Biosorption and bioaccumulation—the prospects for practical applications. Environ Int 36(3):299–307
Fang L, Wei X, Cai P, Huang QY, Chen H, Liang W, Rong XM (2011) Role of extracellular polymeric substances in Cu(II) adsorption on Bacillus subtilis and Pseudomonas putida. Bioresour Technol 102(2):1137–1141
Fang LC, Huang QY, Wei X, Liang W, Rong XM, Chen WL, Cai P (2010) Microcalorimetric and potentiometric titration studies on the adsorption of copper by extracellular polymeric substances (EPS), minerals and their composites. Bioresour Technol 101(15):5774–5779
Günay A, Arslankaya E, Tosun I (2007) Lead removal from aqueous solution by natural and pretreated clinoptilolite: adsorption equilibrium and kinetics. J Hazard Mater 146(1):362–371
Hu QH, Weng JQ, Wang JS (2010) Sources of anthropogenic radionuclides in the environment: a review. J Environ Radioactiv 101(6):426–437
Humelnicu D, Dinu MV, Drăgan ES (2011) Adsorption characteristics of UO2 2+ and Th4+ ions from simulated radioactive solutions onto chitosan/clinoptilolite sorbents. J Hazard Mater 185(1):447–455
Kalin M, Wheeler WN, Meinrath G (2005) The removal of uranium from mining waste water using algal/microbial biomass. J Environ Radioactiv 78(2):151–177
Lovley DR, Phillips EJ, Gorby YA, Landa ER (1991) Microbial reduction of uranium. Nature 350(6317):413–416
Liu MX, Dong FQ, Zhang W, Nie XQ, Sun SY, Wei HF, Luo L, Xiang S, Zhang GG (2016) Programmed gradient descent biosorption of strontium ions by Saccaromyces cerevisiae and ashing analysis: a decrement solution for nuclide and heavy metal disposal. J Hazard Mater 314:295–303
Liu N, Liao J, Luo S, Yang YY, Jin JN, Zhang TM, Zhao PJ (2003) Biosorption of 241Am by immobilized Saccharomyces cerevisiae. J Radioanal Nucl Ch 258(1):59–63
Liu X, Hu WY, Huang XJ, Deng HQ (2015) Highly effective biosorption of Sr(II) from low level radioactive wastewater. Water Sci Technol 71(11):1727–1733
Mashkani SG, Ghazvini PTM (2009) Biotechnological potential of Azolla filiculoides for biosorption of Cs and Sr: application of micro-PIXE for measurement of biosorption. Bioresour Technol 100(6):1915–1921
Mcmurrough I, Rose AH (1967) Effect of growth rate and substrate limitation on the composition and structure of the cell wall of Saccharomyces cerevisiae. Biochem J 105(1):189
Morcillo F, Gonzalez-Munoz MT, Reitz T, Romero-Gonzalez ME, Arias JM, Merroun ML (2014) Biosorption and biomineralization of U(VI) by the marine bacterium Idiomarina loihiensis MAH1: effect of background electrolyte and pH. PLoS One 9(3):e91305
Osmanlioglu AE (2006) Treatment of radioactive liquid waste by sorption on natural zeolite in Turkey. J Hazard Mater 137(1):332–335
Özacar M, Şengil IA (2003) Adsorption of reactive dyes on calcined alunite from aqueous solutions. J Hazard Mater 98(1):211–224
Seliman AF (2012) Affinity and removal of radionuclides mixture from low-level liquid waste by synthetic ferrierites. J Radioanal Nucl Ch 292(2):729–738
Shevchuk IA, Klimenko NA, Stavskaya SS (2010) Sorption of ions of U(VI) and strontium by biosorption based on Bacillus polymyxa, IMV 8910 in aqueous systems. J Water Chem Techno 32(3):176–181
Tuzen M, Sari A (2010) Biosorption of selenium from aqueous solution by green algae (Cladophora hutchinsiae) biomass: equilibrium, thermodynamic and kinetic studies. Chem Eng J 158(2):200–206
Warner NR, Christie CA, Jackson RB (2013) Impacts of shale gas wastewater disposal on water quality in western Pennsylvania. Environ Sci Technol 47(20):11849–11857
Wu YH, Wen YJ, Zhou JX, Dai Q, Wu YY (2012) The characteristics of waste Saccharomyces cerevisiae biosorption of arsenic(III). Environ Sci Pollut R 19(8):3371–3379
Yang DJ, Zheng ZF, Liu HW, Zhu HY, Ke XB, Xu Y, Wu D, Sun Y (2008) Layered titanate nanofibers as efficient adsorbents for removal of toxic radioactive and heavy metal ions from water. J Phys Chem C 112(42):16275–16280
Young V, Otagawa T (1985) XPS studies on strontium compounds. Applications of Surface Science 20(3):228–248
Zhou K, Liu YC, Yang ZG, Liu HZ (2017) Biosorption of U(VI) by modified Hottentot fern: kinetics and equilibrium studies. J Environ Radioactiv 167:13–19
Zhou LM, Wang Y, Zou HB, Liang XZ, Zeng K, Liu ZR, Adesina AA (2016) Biosorption characteristics of uranium(VI) and thorium(IV) ions from aqueous solution using CaCl2-modified giant kelp, biomass. J Radioanal Nucl Ch 307(1):635–644
Zotina TA, Kalacheva GS, Bolsunovsky AY (2011) Biochemical fractionation and cellular distribution of americium and plutonium in the biomass of freshwater macrophytes. J Radioanal Nucl Ch 290(2):447–451
Acknowledgements
This study was sponsored by the National Basic Research Program of China (973 Program: 2014CB846003). The author would also like to thank Dr. Patrick Diehl for his help in revising and polishing this paper.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Philippe Garrigues
Electronic supplementary material
ESM 1
(DOCX 113 kb)
Rights and permissions
About this article
Cite this article
Hu, W., Dong, F., Yang, G. et al. Synergistic interface behavior of strontium adsorption using mixed microorganisms. Environ Sci Pollut Res 25, 22368–22377 (2018). https://doi.org/10.1007/s11356-017-9891-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-017-9891-7