Abstract
Acid rain can directly or indirectly affect plant physiological functions, especially photosynthesis. The enzyme ATP synthase is the key in photosynthetic energy conversion, and thus, it affects plant photosynthesis. To clarify the mechanism by which acid rain affects photosynthesis, we studied the effects of acid rain on plant growth, photosynthesis, chloroplast ATP synthase activity and gene expression, chloroplast ultrastructure, intracellular H+ level, and water content of rice seedlings. Acid rain at pH 4.5 remained the chloroplast structure unchanged but increased the expression of six chloroplast ATP synthase subunits, promoted chloroplast ATP synthase activity, and increased photosynthesis and plant growth. Acid rain at pH 4.0 or less decreased leaf water content, destroyed chloroplast structure, inhibited the expression of six chloroplast ATP synthase subunits, decreased chloroplast ATP synthase activity, and reduced photosynthesis and plant growth. In conclusion, acid rain affected the chloroplast ultrastructure, chloroplast ATPase transcription and activity, and P n by changing the acidity in the cells, and thus influencing the plant growth and development. Finally, the effects of simulated acid rain on the test indices were found to be dose-dependent.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Acid rain is a major environmental problem that endangers Earth’s ecosystems and threatens human survival and has thus received extensive attention worldwide (Abbasi et al. 2013). In particular, acid rain is a major problem in Europe, North America, and China (Abbasi et al. 2013). The total area that suffers from acid rain in Europe and North America is estimated to be about 1000 km2 (Lajtha and Jones 2013). Based on nearly 20 years of continuous monitoring, the acidity of European rainwater has increased by 10 %; currently, the pH value of acid rain in Sweden, Denmark, Poland, Germany, Canada, and other European countries is usually between 4.2 and 4.5 (Lajtha and Jones 2013; Menz and Seip 2004). In 15 states of the USA, the pH value of acid rain is currently lower than 4.8 (Lajtha and Jones 2013; Menz and Seip 2004). In China, the area impacted by acid rain has reached two million square kilometers and is gradually increasing (Liu et al. 2011). An estimated 208 cities in China were polluted with acid rain in 2014, accounting for 44.3 % of the total cities in which rainfall was monitored in this year (Department EoCE 2015). Moreover, the cities in which the frequency of acid rain exceeded 25 or 75 % of total rainfall events accounted for 26.6 and 9.1 % of the total monitored cities, respectively (Department EoCE 2015). Moreover, the average pH value of rainfall in 44 of these cities was less than 5.6, and the pH of rainfall in these areas was approximately between 3.8 and 4.5 (Department EoCE 2015). Serious acid rain has had many effects in China (Department EoCE 2015), for example, the continuous acid rain pollution in the south of the Yangtze River has caused lake acidification, a reduction in fish numbers, and forest degradation (Zhao and Hou 2010).
In recent years, there have been many studies into the effects of acid rain on plants, which have shown that acid rain can cause direct or indirect harm to plant leaves (Imran et al. 2014; Macaulay and Enahoro 2015; Ramlall et al. 2015). A study showed that when the pH of acid rain is below 3.0, acid rain harms the plant cuticle, causing blade chlorosis (Macaulay and Enahoro 2015). When the pH of acid rain is below 1.0, acid rain causes necrotic spots on plant tissues, premature defoliation or premature aging, and other visible symptoms (Macaulay and Enahoro 2015). Moreover, many studies have shown that low-acidity acid rain improves seed germination, increases biomass accumulation, and promotes aboveground tissue growth of plants (Imran et al. 2014; Ramlall et al. 2015). With increasing acid rain acidity, the seed germination rate, plant survival rate, and plant biomass are reduced (Ramlall et al. 2015). In particular, highly acidic acid rain significantly inhibits plant growth and development (Ramlall et al. 2015).
It is well known that photosynthesis is the key metabolic basis for plant growth, development, and yield (Bernacchi et al. 2013). Photoreaction is an important component of photosynthesis, associated with light absorption, electron transfer, light phosphorylation, and other processes (Bernacchi et al. 2013). Numerous studies have shown that acid rain can affect plant photosynthesis (Hu et al. 2014; Wang et al. 2014; Wyrwicka and Skłodowska 2006). Wyrwicka and Skłodowska found that acid rain at pH 3.0 and pH 1.8 decreased the chlorophyll content of cucumber (Cucumis sativus) leaves by 25 and 46 %, respectively (Wyrwicka and Skłodowska 2006). Cao et al. showed that acid rain at pH 2.5 caused decreases in the chlorophyll content (30.71 %), net photosynthetic rate (P n, 12.52 %), and transpiration rate (26.89 %) in rape leaves (Cao et al. 2010). Acid rain at pH 4.5 caused 2.17 and 2.41 % increases in the P n and Hill reaction rate, respectively, in rice leaves; however, acid rain at less than pH 4.5 decreased the P n and Hill reaction rate in the leaves (Hu et al. 2014; Wang et al. 2014). In particular, acid rain at pH 2.5 decreased the P n and Hill reaction rate by 26.02 and 24.31 %, respectively (Hu et al. 2014; Wang et al. 2014). Moreover, acid rain at pH 2.5 also changes the chlorophyll fluorescence and damages the PSII reaction center, leading to electron transfer limitation, thereby inhibiting the absorption of light energy by plants (Liu et al. 2015). Furthermore, acid rain can reduce the integrity of chloroplast structures or destroy them entirely, and thus affect some chloroplast functions and photosynthesis (Wen et al. 2011). The aforementioned studies show that effects of acid rain on plant photosynthesis result from the changes in chloroplast composition and structure, and influence various physiological processes, including light reactions and dark reactions.
Chloroplasts are important sites for plant photosynthesis (Bernacchi et al. 2013). The chloroplast coupling factor, adenosine triphosphate (ATP) synthase (CF1–CF0), is the key enzyme of photophosphorylation and an important factor in photosynthetic energy conversion, and thus, it is important for plant photosynthesis (Kartashov et al. 2015). Studies have shown that acid rain can affect the activity and transcription level of plasma membrane H+-ATPase in rice (Liang et al. 2015; Zhang et al. 2015); however, the effects of acid rain on chloroplast ATPase have not been reported. In particular, when acid rain causes increased acidity of the cellular internal environment, the relationship between the integrity of chloroplast structures and the changes of chloroplast ATPase activity is currently unclear. Rice, as an important commercial crop in the agricultural planting structure, provides food for nearly three billion people around the world (Zhang et al. 2013). In China, the rice-growing area overlaps considerably with acid rain-affected areas (Gravois 1998). Therefore, in the present work, effects of simulated acid rain on the growth, P n, activity and gene expression of chloroplast ATP synthase, chloroplast ultrastructure and intracellular H+ level in rice were investigated. On this basis, the possible mechanisms behind the effects of acid rain on rice photosynthesis were analyzed using correlation analysis methods.
Materials and methods
Preparation of rice nutrient solution and simulated acid rain
Modified rice nutrient solution was prepared according to the ionic composition released by the International Rice Research Institute (IRRI) (Zhu et al. 2009). The full-strength modified nutrient solution had the following composition: 1.43 mM NH4NO3, 0.51 mM K2SO4, 1.00 mM CaCl2, 1.64 mM MgSO4, 9.47 μM MnCl2, 0.075 μM (NH4)6Mo7O24, 19.00 μM H3BO4, 0.15 μM ZnSO4, 0.16 μM CuSO4, and 36.00 μM FeCl3. Meanwhile, SiO2 was supplied as 1.67 mM with NaSiO3⋅9H2O in nutrient solution. Then, 0.07 mM C2H4N4 was added as nitrification inhibitor to keep the concentration of NH4 +-N. The nutrient solution pH was adjusted to 5.5 by using a pH meter (PHS-29 A, Shanghai Anting Scientific Instrument Factory, China).
Based on the ionic composition of rainwater sampled in the southeast of China (Xie et al. 2009), the control rain (pH 7.0) containing 0.83 μM Ca2+, 1.32 μM Na+, 0.15 μM K+, 5.34 μM Mg2+, 0.36 μM SO4 2−, 0.64 μM NO3 −, and 0.47 μM Cl− was prepared by adding the appropriate amounts of these ions to deionized water. The ionic composition and content of the control rain was nearly the same as rainwater samples taken in the southeast of China. Simulated acid rain solutions at pH 4.5, 4.0, 3.5, 3.0, and 2.5 were prepared by adjusting the pH value of control rain with the mixture of concentrated H2SO4 and HNO3 (1.1:1, v:v), and the pH value was measured using a pH meter (PHS-29A, Shanghai Anting Scientific Instrument Factory, China) (Kong et al. 2012).
Plant culture and treatments
Rice seeds were surface sterilized in HgCl2 (0.1 %) solution for 10 min and rinsed with deionized water several times. The sterilized seeds were placed in dishes under-laid with three layers of moistened gauze and germinated in the incubator for 2 days. Germinated seeds were sown in a sterilized sand bed. At the two leaf stage, healthy rice plants with similar growth status were transplanted into pots containing rice nutrient solution. Pots were placed in a greenhouse at 25 ± 3 °C, with a light intensity of 1000 μmol m−2 s−1, a day/night cycle of 16/8 h, and a relative humidity of 70–80 %. Modified rice nutrient solution was used to irrigate the plants and to maintain the water content of the substrate at approximately 60 %. When rice seedlings reached the three leaf stage, healthy rice plants with similar growth status were selected to be sprayed with 300 mL simulated acid rain per pot and the control plants were sprayed with control rain every 3 days. The spray amount of simulated acid rain and control rain was calculated based on the precipitation and evaporation in the southeast of China (Kong et al. 2012). Moreover, the modified nutrient solution was supplied every 3 days. All treatments were performed in six replicates, and 1 mM KH2PO4 was sprayed onto the foliage every other day to apply the required inorganic phosphate to the plants. At the tillering stage (35 days after the spraying of acid rain), the fresh leaves from the acid rain and control treatments were sampled for analyses.
Determination of rice growth and P n
Growth indices include leaf area, dry weight of leaves, dry weight of aboveground organs (stems and leaves), and relative growth rate. Leaf area was measured using a handheld laser leaf area meter (CI-203, CID Bio-science, USA). Dry weights of leaves and aboveground organs for each plant were determined using a special-type electronic balance (EL303, Mettler Toledo, Switzerland, accuracy: 0.0001 g) after drying of the material at 80 °C in an oven for 12 h. The relative growth rate (RGR) was calculated based on the dry weight of leaves and stems with the following formula: RGR = (lnW2-lnW1)/35, where W1 (g) and W2(g) represent the dry weight of leaves and stems before and after the acid rain and control treatment, respectively (Kingsbury et al. 1984).
The P n was measured by using a portable photosynthetic system (CIRAS-1, PP Systems International Ltd., UK) at 25 ± 5 °C, 320 μL L−1 CO2, and 80 % relative humidity. The photon flux density was 1000 μmol m−2 s−1, which was provided by the tungsten-halogen lamp on the leaf chamber of CIRAS-1 photosynthesis system.
Determination of Mg2+-ATPase and Ca2+-ATPase activities
Chloroplasts were exacted and isolated, and the Mg2+-ATPase activity was measured based on the previous method (Sun et al. 2012). The chloroplast suspension (0.1 mL) was incubated for 5 min at 20 °C in 0.8 mL reaction solution containing 62.5 mM Tris-HCl (pH 8.0), 25 mM NaCl, 6.25 mM MgCl2, 55 μM PMS, and 2.5 mM C4H10O2S2 under white light (5000 μmol m−2 s−1) and was activated for 5 min under white light (5000 μmol m−2 s−1). Subsequently, 0.1 mL 5 mM ATP was added and incubated for 5 min at 37 °C, and then 0.2 mL 20 % trichloroacetic acid was added to terminate the reaction. The reaction mixture was centrifuged at 2000×g for 10 min. Sulfuric acid molybdate-ferrous sulfate solution (2.5 mL) was added to 0.5 mL of supernatant. After shaking, the OD values at 660 nm of the mixture were measured using a UV/Vis spectrophotometer (S24–752, Shanghai Jinghua Scientific Instrument Factory, China). The Mg2+-ATPase activity was calculated based on the standard curve of inorganic phosphorus.
The Ca2+-ATPase activity was measured as described in the previous method (Sun et al. 2012). Chloroplasts suspension (0.1 mL) was incubated for 10 min at 20 °C in 0.8 mL reaction solution containing 0.25 M Tris-HCl (pH 8.0), 20 mM EDTA, 10 mM ATP, and 2 mg mL−1 trypsin. Subsequently, 0.1 mL of 5 mM ATP was added and incubated for 5 min at 37 °C, and finally, 0.1 mL 10 mg mL−1 bovine serum albumin was added to terminate the reaction. The determination and calculation methods for Ca2+-ATPase activity was the same as Mg2+-ATPase activity.
Transmission electron microscopy observation of chloroplast ultrastructure
Transmission electron microscopy (TEM) observation of chloroplast ultrastructure in rice leaves was performed based on previous method (Helliot et al. 2003). Fresh leaves were sliced into 1.5 × 2 mm sections. After fixation with 3.5 % glutaraldehyde for 24 h, leaf cells were post-fixed with 1 % osmic acid at 4 °C for 4 h and then were dehydrated with ethanol. Next, the samples were embedded in freshly prepared 100 % Epon-812 and polymerized at 80 °C for 24 h. For ultrastructural observations, the samples obtained were cut into ultrathin sections (∼60 nm) on a LKB ultramicrotome. The ultrathin sections were placed upon 250-mesh grids and were post-stained with uranyl acetate and lead citrate. Finally, the TEM images were obtained with a TEM (H-600-A-2, Hitachi Ltd., Japan).
Preparation of total RNA and real-time PCR
Real-time PCR analysis with specific primers for each gene was performed to evaluate the expression of three genes (ɑ subunit, β subunit, and ε subunit) encoding the CF1 of chloroplast ATP synthase and other three genes (I subunit, III subunit, and IV subunit) encoding the CFo of chloroplast ATP synthase. For the normalization of expression of each gene, the actin gene was used as the internal standard. All primers were designed on the basis of sequences published in GenBank by Primer Premier 5.0. Primer sequences of target gene and housekeeping genes are shown in Table 1.
Total RNA of rice leaves were isolated using a column extraction purification kit (SK8661 sangon) according to manufacturer’s instructions and then reverse-transcribed into first-strand cDNA with a high-capacity cDNA reverse transcription kit (AMV first strand cDNA synthesis kit, Shanghai Biological Engineering Ltd., China) according to manufacturer’s instructions. The cDNA was then used as the template for PCR amplification. Each 25 μL reaction system contained 0.5 μL template cDNA, 0.5 μL primer F (10 μM), 0.5 μL primer R (10 μM), 0.5 μL dNTP (10 mM), 2.5 μL Taq Buffer (10×), 2 μL MgCl2 (25 mM), 0.2 μl Taq enzyme (5 U μL−1), and 18.3 μL H2O. The PCR product was observed using 2 % agarose gel electrophoresis. After connection at 4 °C overnight, the PCR products were transformed with the step rapid competent cell preparation kit (SK9307), and then the plasmid DNA were extracted using plasmid extraction kit (SK8191). The OD value of the constructed plasmids was determined using a UV spectrophotometer (TU-1901, Beijing Purkinje General Instrument Ltd., China) at 260 nm and then converted into a copy number.
Water content measurement
Leaf water content was calculated according to the following formula: leaf water content (%) = (W1 − W2)/W1 × 100 %, where W1 is the fresh weight of leaves and W2 is the dry weight of leaves determined after drying at 80 °C for 12 h.
Protoplasts isolation and intracellular H+ level measurement
Rice leaves were cut into 2-mm filaments and put into a test tube to which enzyme solution containing 2 % cellulase, 0.5 % macerase, 0.1 % (w/v) pectolyase (Y-23), 5 mmol L−1 MES, and 0.45 mmol L−1 mannitol was added. Tubes were shaken at 40 rpm and 30 °C in the dark for 3 h. Then, the reaction mixture was filtered through a 400 mesh plug. The filtrate was centrifuged at 1000×g for 5 min. The supernatant was removed by pipette and the precipitate was washed twice with a solution containing 3 % sucrose (w/v) and 0.4 M mannitol. Protoplasts were passed through a 50-μm pore-size nylon mesh filter and then suspended in the sucrose and mannitol solution to maintain 5 × 106 protoplasts ml−1.
Isolated protoplasts were incubated with the pH-sensitive fluorescent dye [2′, 7′-bis (carboxyethyl)-5(6)-carboxy, BCECF] at 37 °C for 60 min. Protoplasts were then washed with HEPES-buffered salt solution (pH 7.4) at 37 °C for 15 min to remove extracellular dye and attain complete de-esterification of the cytosolic dye. Radiometric measurement of BCECF fluorescence was conducted using a workstation (Intracellular Imaging Inc., Cincinnati, OH) consisting of a Nikon TSE 100 Ellipse inverted microscope with epifluorescence attachments. The wavelength of excited light was 500 nm, and the wavelength of emitted light was 530 nm. The fluorescence signal was adjusted to zero background for cells without dying, and then the intracellular H+ level was expressed by the relative BCECF fluorescence (Undem et al. 2012).
Statistical analysis
The data were analyzed by SPSS 17.0 and Origin 8. Student’s t tests were used to determine the significance between different treatments (p < 0.05).
Results
Effects of simulated acid rain on rice growth and P n in rice leaves
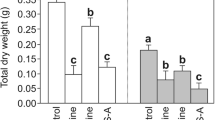
Table 2 shows the effects of simulated acid rain on the growth and photosynthesis in rice seedlings. After treatment with simulated acid rain at pH 4.5, the dry weights of leaves, dry weights of aboveground organs, leaf areas, relative growth rates, and P n were higher than those in the control (pH 7.0). The above indices in other treatment groups decreased, and the magnitude of this decrease increased with decreasing pH of the simulated acid rain.
Effects of simulated acid rain on the activity and transcription level of chloroplast ATPase in rice leaves
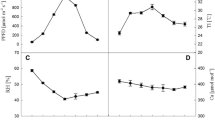
Figure 1 shows that the chloroplast Mg2+-ATPase activity in rice seedlings treated with simulated acid rain at pH 4.5 and pH 4.0 did not differ from that of the control. After rice seedlings were treated with simulated acid rain at pH 3.5, pH 3.0, or pH 2.5, the chloroplast Mg2+-ATPase activity was lower than that of the control, and greater decreases were observed with the pH of the simulated acid rain decreased. In addition, the chloroplast Ca2+-ATPase activity in all treatment groups treated with simulated acid rain decreased relative to the control. In addition, the inhibition effect became clearer as the pH of the simulated acid rain decreased (Fig. 2). Compared with the Mg2+-ATPase activity, the chloroplast Ca2+-ATPase activity was more sensitive to simulated acid rain (Fig. 2).
Table 3 shows that, compared with the control, the expression level of all subunits of chloroplast ATPase initially increased and then decreased as the pH of simulated acid rain decreased. The expression level of each subunit of chloroplast ATPase in plants treated with simulated acid rain decreased in the following order: pH 4.5: β subunit (atp-B) > IV subunit (atp-I) > ε subunit (atp-E) > α subunit (atp-A) > III subunit (atp-H) > I subunit (atp-F). pH 4.0: IV subunit (atp-I) > I subunit (atp-F) > α subunit (atp-A) > III subunit (atp-H) > ε subunit (atp-E) > β subunit (atp-B). pH 3.5: β subunit (atp-B) > ε subunit (atp-E) > IV subunit (atp-I) > I subunit (atp-F) > α subunit (atp-A) > III subunit (atp-H). pH 3.0: ε subunit (atp-E) > III subunit (atp-H) > β subunit (atp-B) > I subunit (atp-F) > α subunit (atp-A) > IV subunit (atp-I). pH 2.5: IV subunit (atp-I) > III subunit (atp-H) > I subunit (atp-F) > ε subunit (atp-E) > α subunit (atp-A) > β subunit (atp-B).
Effects of simulated acid rain on chloroplast ultrastructure in rice leaves
Figure 3 shows that chloroplasts in the control were elliptical and arranged around the plasma membrane. The granum and stroma thylakoids were in an orderly arrangement and the chloroplast membrane was clear and intact. In simulated acid rain at pH 4.5 treatment group, the chloroplast structure remained unchanged. When the pH value of simulated acid rain decreased to 3.5, the thylakoids appeared slightly loose but were arranged in an orderly fashion, and the grana were clearly visible. Some thylakoids disintegrated when the rice was treated with simulated acid rain at pH 2.5.
Effects of simulated acid rain on the intracellular H+ level and water content in rice leaves
The intracellular H+ fluorescence value was lower, which suggests that the intracellular concentration of H+ was higher (Liang et al. 2015; Zhang et al. 2015). Figure 4 shows the water contents of rice leaves treated with simulated acid rain at different pH value. After treatment with simulated acid rain at pH 4.5, pH 4.0, pH 3.5, and pH 3.0, the leaf water content did not change relative to the control. When the pH value of simulated acid rain decreased to 2.5, the leaf water content was significantly lower than the control.
Figure 5 shows the effect of simulated acid rain on the intercellular H+ level of rice leaves. The intracellular H+ fluorescence value in all treatment groups treated with simulated acid rain decreased relative to the control, and the effect became more clear as the pH decreased.
Correlation analysis
Correlation analyses were performed among the different measured growth indices, P n, Mg2+-ATPase activity, Ca2+-ATPase activity, intracellular H+ level, and water content, to determine how simulated acid rain affects the P n and how the change in the P n affects rice seedling growth. Table 4 shows that the growth indices were positively correlated with the P n in rice seedlings. In addition, the P n was positively correlated with chloroplast Mg2+-ATPase activity and Ca2+-ATPase. The chloroplast Mg2+-ATPase and Ca2+-ATPase activities were positively correlated with water content and intracellular H+ level in rice seedlings.
Discussion
Effects of simulated acid rain on rice growth and P n in rice leaves
Acid rain can affect plant physiological functions directly or indirectly and has a particularly strong impact on photosynthesis (Liu et al. 2015). Under the stress of simulated acid rain, the biomass and the relative growth rate of rice first increased and then decreased as the pH value decreased (Table 2), and this finding was consistent with previous studies (Hu et al. 2014; Wang et al. 2014). The correlation results show that the effects of simulated acid rain on rice growth indices were closely associated with the changes in P n (Table 2, Table 4). Low-acidity simulated acid rain improved the P n of leaves and increased the accumulation of assimilation products, laying the material foundations for rice growth (Table 2, Table 4) (Meng et al. 2011). High-acidity simulated acid rain decreased the P n of leaves and reduced the accumulation of assimilation products, impeding rice growth (Table 2, Table 4). In addition, since plant growth is affected by photosynthesis and respiration, we speculate that the effects of weak acid rain on rice growth also occur for respiration processes, and their internal connections will be further investigated (Zhao 2014). Previous research shows that acid rain can influence the chlorophyll content, chlorophyll fluorescence, and Hill reaction activity of plants, which in turn affects the absorption of light by leaves, leading to a change of the P n (Hu et al. 2014; Liu et al. 2015; Wang et al. 2014). In addition to the above factors, the change of the P n might be related to the activity of chloroplast ATPase which determines the formation of photosynthesis assimilation (ATP).
Effects of simulated acid rain on the activity and transcription level of chloroplast ATPase in rice leaves
Chloroplast ATP synthase (CF1–CF0) binds to the surface of thylakoid membranes in chloroplasts. It can combine with Mg2+ or Ca2+ to form Mg2+-ATPase and Ca2+-ATPase, catalyzing the ATP synthesis reaction (Kartashov et al. 2015; Kong et al. 2013; Konno et al. 2012; Walker 2013). The ε subunit is an inhibitor of chloroplast ATP synthase and was dissociated from ATP synthase and product the activity to hydrolysis ATP, leading to a drop in plant endogenous ATP content (del Riego et al. 2006; Feniouk et al. 2006; Schmidt et al. 2007; Shirakihara et al. 2015). Low-acidity-simulated acid rain increased the expression levels of each subunit of chloroplast ATPase; in particular, the increase of β subunit expression level was clearer than that of the ε subunit (Table 3). This effect increased Mg2+-ATPase activity, ATP content, and RuBPase supply, promoting photosynthesis and increasing the P n (Tezara et al. 1999). With the increase in the acidity of simulated acid rain, the increase in the expression level of the ε subunit was clearer than that of the β subunit, and the inhibition of ε subunit caused the observed decrease in the ATP synthase activity (Table 3, Figs. 1 and 2) (Feniouk et al. 2006; Shirakihara et al. 2015). High-acidity simulated acid rain decreased the expression levels and activity of chloroplast ATPase (Table 3). This may have two main causes: first, the inhibition of ε subunit (Feniouk et al. 2006; Shirakihara et al. 2015); and second, the expression levels of the I and IV subunits decreased, and these subunits play key assembly roles in chloroplast ATPase, leading to abnormal assembly of chloroplast ATPase in plants. This abnormal assembly caused the decrease in the synthesis amount of chloroplast ATPase, ATP content, and RuBPase supply, inhibiting photosynthesis and decreasing the P n (Table 2) (Ghulam et al. 2012; Tezara et al. 1999). In addition, the change in acidity of the intracellular environment induced by acid rain stress also causes a change in chloroplast ATPase activity.
Effects of simulated acid rain on chloroplast ultrastructure in rice leaves
The integrity and ordering of grana lamellae and stroma lamellae represent the structural foundation for normal photosynthesis in chloroplast (Zhang et al. 2014). After treatment with low-acidity simulated acid rain, chloroplast lamellae were densely arranged and there were no obvious changes to chloroplast structure (Fig. 3). However, chloroplast ATPase activity and expression levels and P n increased to different degrees (Tables 2 and 3, Figs. 1 and 2). Results indicated that low-acidity simulated acid rain did not change the chloroplast structure but did affect photosynthesis function (Dias et al. 2010; Zhang et al. 2014). High-acidity-simulated acid rain damaged the chloroplast ultrastructure, decreased the quantities of grana and thylakoids (Fig. 3), disorganized grana lamellae, restricted chloroplast ATPase assembly, and reduced the synthesis of chloroplast ATPase. It is also possible that acid rain caused the excessive accumulation of reactive oxygen species (ROS) and induced the peroxidation of chloroplast membrane lipids, leading to the damage to chloroplast ultrastructure (Sena and Chandel 2012; Wen et al. 2011).
Effects of simulated acid rain on the intracellular H+ level and water content in rice leaves
We next consider how simulated acid rain led to the ROS accumulation in mesophyll cells. We found that low-acidity simulated acid rain caused a small number of H+ ions to enter cells, causing slight acidification of the intracellular environment but without significant change in water content (Figs. 4 and 5). Results indicated that low-acidity-simulated acid rain did not affect the metabolic balance of the ROS and did not damage the plasma membrane or chloroplast structure (Chen et al. 2013). A small quantity of H+ entered mesophyll cells, which could increase the expression of the III subunit gene of chloroplast ATPase, which has the ability to transfer H+ (Table 3), and increase the H+ transfer function of chloroplast ATP synthase thus improving chloroplast Mg2+-ATPase activity and P n (Kish Trier et al. 2008). Interestingly, low-acidity-simulated acid rain decreased chloroplast Ca2+-ATPase activity (Fig. 2). We speculate that the increase in intracellular H+ level caused the increase in intracellular Ca2+, and the increased Ca2+ significantly inhibits Ca2+-ATPase activity (Zhu et al. 2013). High-acidity simulated acid rain caused a large number of H+ to enter the cells, leading to serious acidification and loss of intracellular water (Figs. 4 and 5). These effects disrupted metabolism of ROS and increased the ROS accumulation in cell, leading to the observed change in plasma membrane permeability and the damage to chloroplast structure (Fig. 3) (Sena and Chandel 2012; Wen et al. 2011). The gene expressions of all chloroplast ATP synthase subunits were significantly decreased (Table 3), indicating that when considerable H+ entered the cell, there was substantial inhibition of the activities of Ca2+-ATPase and Mg2+-ATPase and a decrease in the P n (Table 4).
Conclusion
Low-acidity-simulated acid rain slightly increased the acidity within the plant leaf cells but did not affect chloroplast ultrastructure, increased the contents of β subunit that play a catalytic role, and increased chloroplast Mg2+-ATPase activity. High-acidity simulated acid rain significantly increased the acidity in the cells, destroyed chloroplast ultrastructure, and increased the inhibition effect of the ε subunit, causing the abnormal assembly of chloroplast ATPase and decreasing Mg2+-ATPase activity. Moreover, unlike chloroplast Mg2+-ATPase activity, acid rain significantly inhibited chloroplast Ca+-ATPase activity. In addition, acid rain affected the chloroplast ultrastructure, chloroplast ATPase transcription and activity, and P n by changing the acidity in the cells and thus influencing the plant growth and development. Finally, the effects of simulated acid rain on the test indices were found to be dose-dependent.
References
Abbasi T, Poornima P, Kannadasan T, Abbasi SA (2013) Acid rain: past, present, and future. Int J Environ Eng 97:229–272
Bernacchi CJ, Bagley JE, Serbin SP, Ruiz Vera UM, Rosenthal DM, Vanloocke A (2013) Modelling C3 photosynthesis from the chloroplast to the ecosystem. Plant Cell Environ 36:1641–1657
Cao CX, Zhou Q, Han LL, Zhang P, Jiang HD (2010) Effects of simulated acid rain on oilseed rape (Brassica napus) physiological characteristics at flowering stage and yield. J Appl Ecol 21:2057–2062
Chen J, Wang WH, Liu TW, Wu FH, Zheng HL (2013) Photosynthetic and antioxidant responses of liquidambar formosana and schima superba seedlings to sulfuric-rich and nitric-rich simulated acid rain. Plant Physiol Biochem 64:41–51
Del Riego G, Casano LM, Martin M, Sabater B (2006) Multiple phosphorylation sites in the beta subunit of thylakoid ATP synthase. Photosynth Res 89:11–18
Department EoCE (2015) China’s environmental bulletin in 2014.37:1
Dias BB, Leite ML, Farago PV, de Oliveira AV, Beruski GC (2010) Sulfur effect by simulated acid rain on morphophysiological parameters of the bean plant. Acta Sci Agron 32:433–439
Feniouk BA, Suzuki T, Yoshida M (2006) The role of subunit epsilon in the catalysis and regulation of FOF1-ATP synthase. Biochim Biophys Acta Bioenerg 1757:326–338
Ghulam MM, Zghidi Abouzid O, Lambert E, Lerbs-Mache S, Merendino L (2012) Transcriptional organization of the large and the small ATP synthase operons, atpI/H/F/a and atpB/E, in Arabidopsis thaliana chloroplasts. Plant Mol Biol Report 79:259–272
Gravois KA (1998) Optimizing selection for rough rice yield, head rice, and total milled rice. Euphytica 101:151–156
Helliot B, Swennen R, Poumay Y, Frison E, Lepoivre P, Panis B (2003) Ultrastructural changes associated with cryopreservation of banana (Musa spp.) highly proliferating meristems. Plant Cell Rep 21:690–698
Hu H, Wang L, Liao C, Fan C, Zhou Q, Huang X (2014) Combined effects of lead and acid rain on photosynthesis in soybean seedlings. Biol Trace Elem Res 161:136–142
Imran MA, Hussain S, Hussain M, Ch MN, Meo AA (2014) Effect of simulated acid rain (SAR) on some morphochemical aspects of mash (Vigna mungo L.). Pak J Bot 46:245–250
Kartashov IM, Opanasenko VK, Malyan AN (2015) Effects of medium viscosity increasing agents on ATP synthesis in chloroplast thylakoids. Biofizika 60:481–486
Kingsbury RW, Epstein E, Pearcy RW (1984) Physiological responses to salinity in selected lines of wheat. Plant Physiol 74:417–423
Kish Trier E, Briere L, Dunn SD, Wilkens S (2008) The stator complex of the a(1)a(0)-ATP synthase-structural characterization of the E and H subunits. J Mol Biol 375:673–685
Kong R, Zheng X, Huang W, Zhou L, Zhang G (2012) Ion composition and its sources of rainwater from shanghai urban and suburban areas. Urban Environ Urban Ecol 25:22–27
Kong M, Wang F, Yang Z, Mi H (2013) ATPG is required for the accumulation and function of chloroplast ATP synthase in Arabidopsis. Chin Sci Bull 58:3224–3232
Konno H, Nakane T, Yoshida M, Ueoka NH, Hara S, Hisabori T (2012) Thiol modulation of the chloroplast ATP synthase is dependent on the energization of thylakoid membranes. Plant Cell Physiol 53:626–634
Lajtha K, Jones J (2013) Trends in cation, nitrogen, sulfate and hydrogen ion concentrations in precipitation in the United States and Europe from 1978 to 2010: a new look at an old problem. Biogeochemistry 116:303–334
Liang CJ, Ge YQ, Su L, Bu JJ (2015) Response of plasma membrane H+-ATPase in rice (Oryza sativa) seedlings to simulated acid rain. Environ Sci Pollut Res 22:535–545
Liu P, Xia F, Pan J, Chen Y, Peng H, Chen S (2011) Discuss on present situation and countermeasures for acid rain prevention and control in China. Environ Sci Manag 21:4–11
Liu MH, Yi LT, Yu SQ, Yu F, Yin XM (2015) Chlorophyll fluorescence characteristics and the growth response of Elaeocarpus glabripetalus to simulated acid rain. Photosynthetica 53:23–28
Macaulay BM, Enahoro GE (2015) Effects of simulated acid rain on the morphology, phenology and dry biomass of a local variety of maize (Suwan-1) in southwestern Nigeria. Environ Monit Assess 187:1–13
Meng H, Dong DM, Wang J, Yang KN, Tian L, Sun W, Fang CS (2011) Effects of simulated acid rain on main nutritional indicators of three leafy vegetables. Chem Res Chin Univ 27:397–401
Menz FC, Seip HM (2004) Acid rain in Europe and the United States: an update. Environ Sci Pol 7:253–265
Ramlall C, Varghese B, Ramdhani S, Pammenter NW, Bhatt A, Berjak P, Sershen (2015) Effects of simulated acid rain on germination, seedling growth and oxidative metabolism of recalcitrant-seeded Trichilia dregeana grown in its natural seed bank. Physiol Plant 153:149–160
Schmidt M, Luff M, Mollwo A, Kaminski M, Mittag M, Kreimer G (2007) Evidence for a specialized localization of the chloroplast ATP-synthase subunits alpha, beta, and gamma in the eyespot apparatus of Chlamydomonas reinhardtii (Chlorophyceae). J Phycol 43:284–294
Sena L, Chandel N (2012) Physiological roles of mitochondrial reactive oxygen species. Mol Cell 48:158–167
Shirakihara Y, Shiratori A, Tanikawa H, Nakasako M, Yoshida M, Suzuki T (2015) Structure of a thermophilic F-1-ATPase inhibited by an epsilon-subunit: deeper insight into the epsilon-inhibition mechanism. FEBS J 282:2895–2913
Sun Z, Wang L, Chen M, Wang L, Liang C, Zhou Q, Huang X (2012) Interactive effects of cadmium and acid rain on photosynthetic light reaction in soybean seedlings. Ecotoxicol Environ Saf 79:62–68
Tezara W, Mitchell VJ, Driscoll SD, Lawlor DW (1999) Water stress inhibits plant photosynthesis by decreasing coupling factor and ATP. Nature 401:914–917
Undem C, Rios EJ, Maylor J, Shimoda LA (2012) Endothelin-1 augments Na+/H+ exchange activity in murine pulmonary arterial smooth muscle cells via rho kinase. PLoS One 7:1602–1603
Walker JE (2013) The ATP synthase: the understood, the uncertain and the unknown. Biochem Soc Trans 41:1–16
Wang LH, Wang W, Zhou Q, Huang XH (2014) Combined effects of lanthanum (III) chloride and acid rain on photosynthetic parameters in rice. Chemosphere 112:355–361
Wen KJ, Liang CJ, Wang LH, Hu G, Zhou Q (2011) Combined effects of lanthanumion and acid rain on growth, photosynthesis and chloroplast ultrastructure in soybean seedlings. Chemosphere 84:601–608
Wyrwicka A, Skłodowska M (2006) Influence of repeated acid rain treatment on antioxidative enzyme activities and on lipid peroxidation in cucumber leaves. Environ Exp Bot 56:198–204
Xie Z, Du Y, Zeng Y, Li Y, Yan M, Jiao S (2009) Effects of precipitation variation on severe acid rain in southern China. J Geogr Sci 19:489–501
Zhang H, Chen TT, Liu LJ, Wang ZQ, Yang JC, Zhang JH (2013) Performance in grain yield and physiological traits of rice in the Yangtze river basin of China during the last 60 yr. Integr Agric 12:57–66
Zhang J, Jiang XD, Li TL, Cao XJ (2014) Photosynthesis and ultrastructure of photosynthetic apparatus in tomato leaves under elevated temperature. Photosynthetica 52:430–436
Zhang YK, Zhu DF, Zhang YP, Chen HZ, Xiang J, Lin XQ (2015) Low pH-induced changes of antioxidant enzyme and ATPase activities in the roots of rice (Oryza sativa L.) seedlings. PLoS One 10:2–6
Zhao Z (2014) Effects of simulated acid rain on photosynthetic physiology characteristics of Ipomoea cairica seedling. Ecotoxicol Environ Sci 19:13–21
Zhao Y, Hou Q (2010) Characteristics of the acid rain variation in China during 1993–2006 and associated causes. Acta Meteorol Sin 24:239–250
Zhu Y, Di T, Xu G, Chen X, Zeng H, Yan F, Shen Q (2009) Adaptation of plasma membrane H+-ATPase of rice roots to low pH as related to ammonium nutrition. Plant Cell Environ 32:1428–1440
Zhu X, Feng Y, Liang G, Liu N, Zhu JK (2013) Aequorin-based luminescence imaging reveals stimulus and tissue-specific Ca2+ dynamics in Arabidopsis plants. Mol Plant 6:444–455
Acknowledgments
The authors are grateful for the financial support of the Natural Science Foundation of China (31170477, 21371100, 21501068), the National Water Pollution Control and Management Technology Major Project (2012ZX07101_013), the Research and Innovation Project for Postgraduate of Higher Education Institutions of Jiangsu Province in 2014 (KYLX_1160), and Ph.D. Programs Foundation of Ministry of Education of China (20130093120006). We thank the LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Responsible editor: Philippe Garrigues
Rights and permissions
About this article
Cite this article
Sun, J., Hu, H., Li, Y. et al. Effects and mechanism of acid rain on plant chloroplast ATP synthase. Environ Sci Pollut Res 23, 18296–18306 (2016). https://doi.org/10.1007/s11356-016-7016-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-016-7016-3