Abstract
Plant–microbe interactions are considered to be important processes determining the efficiency of phytoremediation of heavy metal-contaminated soils. However, relatively little is known about how these interactions are influenced by chromium (Cr) contamination. The effect of Cr stress on metal uptake, root organic acid composition, and rhizosphere bacterial communities was studied using two genotypes of the metallophyte Silene vulgaris, which have shown different tolerance to Cr(VI). The results indicated that root biomass and shoot biomass were not significantly influenced by Cr treatment, but metal uptake in shoots and roots was significantly impacted by the genotype. Principal component analyses (PCA) showed that variation in organic acids oxalic, citric, malic, formic, lactic, acetic, and succinic differed between genotypes. Changes in root organic acid contents in response to Cr revealed a significant increase of oxalic acid in genotype SV-21. The denaturing gradient gel electrophoresis (DGGE) cluster analysis showed that the community structure (determined by PCR-DGGE) was affected by plant genotype and, to a lesser extent, by Cr contamination, the first being the most influential factor shaping the rhizosphere microbiome. Under Cr pollution, a shift in the relative abundance of specific taxa was found and dominant phylotypes were identified as Variovorax in SV-21 and Chitinophaga niastensis, Pontibacter sp., and Ramlibacter sp. in SV-38. These results provided the basis for further studies aimed at the combined use of plants and soil microorganisms in the remediation of Cr-polluted soils.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Chromium pollution is due to a large number of industrial processes such as chrome plating, wood preserving, pigmenting, pulp and paper manufacturing, textile dyeing, tanning, and leather processing (Dhal et al. 2013). Its widespread use has converted Cr into a serious pollutant of air, soil, and water (Zayed and Terry 2003). Chromium is present in the environment in several different forms. The most common oxidation states of chromium are Cr(VI) and Cr(III), although there are various other valence states which are unstable and short-lived in biological systems. Cr(VI) is considered the most toxic form of Cr and is highly soluble and thus mobile and biologically available in the ecosystems. In contrast, Cr(III) is less mobile, less toxic, and displays a high affinity for organics resulting in the formation of complexes that precipitate (Kimbrough et al. 1999). Physicochemical processes such as adsorption and co-precipitation, transformation, ion exchange, and reverse osmosis have been used to treat Cr(VI) contamination, but the environmentally most compatible and cost-effective solution comprises a combination of two or more of these processes. However, the development of new absorbent materials such as CTAB-silica gelatin composite that transform Cr(VI) in Cr (III) without the need to eliminate contaminant sulfate ions represents a highly economic and efficient material (Venditti et al. 2007, 2010). Alternative techniques for the cleanup of polluted soil and water, such as the cost-effective and less disruptive phytoremediation, have gained acceptance in recent years (Pilon-Smits 2005; Vangronsveld et al. 2009). Plant species used for phytoremediation of heavy metal pollution may promote the growth of microorganisms, which in turn have capacity to detoxify metals efficiently by transforming them into insoluble salts or relatively nontoxic oxidation states (Kuiper et al. 2004; Belimov et al. 2005). Improvement of the beneficial associations between microorganisms and plants, particularly in the rhizosphere, is a research area of global interest.
Silene vulgaris (Moench) Garcke is a perennial facultative metallophyte species, phenotypically diverse, and widely distributed across its native range of Eurasia and North Africa (Marsden-Jones and Turrill 1957). It has also been introduced to other regions such as North America, South America, and Australia where it colonizes disturbed habitats such as roadsides, railroad tracks, and agricultural settings (Marsden-Jones and Turrill 1957). The tolerance to a diversity of metals has been described for this species (Paliouris and Hutchinson 1991; Ernst and Nelissen 2000) as well as local adaptation to metal-contaminated soils (Schat et al. 1996). The effectiveness of S. vulgaris in the revegetation of contaminated soils seemed to result in a different reduction of heavy metal toxicity on soil bacteria (Martínez-Iñigo et al. 2009). Recent studies (Pradas del Real et al. 2013) showed that Cr uptake in S. vulgaris increased in the presence of Cr(VI) and a remarkable clonal variability was observed. In addition, specific differences were found in the elemental composition of root exudates in S. vulgaris genotypes due to Cr speciation in the medium (Pradas del Real et al. 2014b). These characteristics along with a wide range of adaptation and high intraspecific diversity (Sloan et al. 2012) make this species of great interest for phytoremediation purposes.
There is a clear evidence that the plant–microorganism interactions in the rhizosphere improve remediation of heavy metals, and this synergism not only facilitates the remediation process by improving phytostabilization (reduction of metal toxicity through metal immobilization in the roots) and phytoextraction (metal accumulation by the parts above the ground as a result of metal mobilization) of metal species but also accelerates the plant growth and development (Kuiper et al. 2004; Abou-Shanab et al. 2008; Bakker et al. 2013). Heavy metals may change soil microbial biomass, activities, and structure (Giller et al. 1998; Kahn and Scullion 2000; Anderson et al. 2009; Wang et al. 2010). In spite of the increasing knowledge of metal–microorganism interactions, few studies have attempted to characterize the bacterial communities in the rhizosphere of metallophytes under heavy metal stress (Wang et al. 2008; Navarro-Noya et al. 2010; Xu et al. 2012; Zhang et al. 2012). In addition, as far we know, no studies have been performed to describe the structure and diversity of rhizobacterial communities under Cr stress.
Plants may actively shape rhizosphere microbial communities due to the release of large amounts of organic carbon by the roots (Bais et al. 2006; Hartmann et al. 2009). Considering that plant root exudates differ among plant species, cultivars, or accessions (López-Bucio et al. 2000), it can be inferred that there are differences in rhizosphere microbiomes of different plant species (Smalla et al. 2001; Costa et al. 2006) and at genotype level within a plant species (Micallef et al. 2009).
Organic acids have been mentioned as playing an important role in the transport, storage, and heavy metal tolerance, including Al, Cd, Cr, Fe, Ni, and Zn (Hall 2002; Zeng et al. 2008). Organic acids can bind heavy metals and may therefore be deployed in response to metal toxicity. Malate and citrate organic acids are involved in root to shoot metal transport through the xylem and in vacuolar sequestration (Rauser 1999). Furthermore, citrate and oxalate have been reported to play an important role in phytoremediation of Cr-contaminated soils by enhancing Cr uptake and increasing translocation to shoots (Davies et al. 2001). Heavy metal stress commonly induces important accumulation of low molecular weight organic acids in various plant organs reinforcing the hypothesis that these molecules are involved in metal-tolerant mechanisms (Mnasri et al. 2015).
Plant–microbe interactions are considered to be important processes determining the efficiency of phytoremediation of heavy metal-contaminated soils. However, relatively little is known about how these interactions are influenced by Cr contamination. Monitoring the changes in some molecular and biochemical parameters involved in Cr stress responses could open potential pathways to enhance plant tolerance to Cr soil contamination. Furthermore, identification of rhizobacteria having potential for heavy metal tolerance is of particular interest for bacteria-assisted phytoremediation. In the present study, two Cr-tolerant genotypes were selected attending to differences in metal uptake, exudation rates, and differences on elemental composition of root exudates in response to Cr under hydroponic conditions (Pradas del Real et al. 2013, 2014b). The purpose of this study was (a) to evaluate the metal-induced response on biomass production, lipid peroxidation (malondialdehyde (MDA)), and root organic acid composition, (b) to assess and compare the effect of Cr contamination on the structure and composition of rhizosphere microbial communities by PCR-DGGE and bacterial 16S rDNA sequencing, and (c) to determine the effect of plant bacterial interactions over Cr uptake.
Materials and methods
Plant material and greenhouse experimental design
S. vulgaris natural populations were collected throughout Madrid Province and, after two growing seasons, rhizome cuttings were used to propagate individual plants. Genetically uniform clones were developed and vegetatively propagated on permanent field plots at the “El Encín” agricultural experiment station (Alcalá de Henares, Spain: 40° 03′ N, 3° 19′ W). Two genotypes from two different populations were selected: genotype SV-21 (Rozas de Puerto Real) and genotype SV-38 (Valdemaqueda). A pot experiment was conducted under greenhouse conditions (humidity 50–65 %, average air temperature 21 °C, and natural light). Plastic pots were filled with 17 kg of agricultural loamy sand soil collected from the top layer (0–30 cm). Soil characteristics were analyzed according to the official soil analysis in Spain (MAPA 1994) and are summarized in Table S1 of Supporting Information. The experiment was performed in a randomized block design with three replicates and a 2 × 3 factorial arrangement with two levels of pollution, (i) no pollution and (ii) K2Cr2O7 to simulate a Cr(VI) spilling, and two plant treatments, (i) S. vulgaris genotype SV-21 and (ii) S. vulgaris genotype SV-38.
Cr pollution was simulated by spiking the soil with K2Cr2O7 solution (1000 mg Cr L−1) to reach a final concentration of total Cr in pots of 100 mg kg−1 soil. The soil was brought to 60 % water holding capacity and maintained by addition of deionized water for 3 months. After soil consolidation, 10 cuttings per genotype were transplanted to each pot and maintained during two growing periods. The total amount of water applied in each irrigation cycle was based on the normal rainfall in Mediterranean conditions.
Plant analysis
At the end of the experimental period, shoots and roots were collected, weighed, and dried in a forced air oven for 48 h at 70 °C. The dry weights were determined, and the samples were digested by adding 0.5 ml of HNO3 (65 % Suprapur®) and 0.5 ml of HClO4 (70 %, Suprapur®). Total Cr concentration was measured by flame atomic absorption spectrophotometer (Varian fast sequential model AA240FS). Bush branches and leaves (DC73348GSV-1) were used as reference material. The translocation factor (TF) was calculated to determine relative translocation of Cr content (mg kg−1) from the root to the aerial part of the plant: TF = C shoot / C root. The ratio of metal concentration in the plant to soil was used to determine the bioconcentration factor (BF): BF = C p / C so, where C p and C so are metal concentrations in the plant (shoot and root) and in the soil, respectively. Tolerance index (TI) was defined as the ratio between plant biomass after Cr treatment and control: TI = W Cr / W control.
Lipid peroxidation of shoots and roots was evaluated as MDA by the method of Reilly and Aust (2001). Absorbance was measure at 532 nm. Absorbance at 600 nm was subtracted to this measure to eliminate the interferences of soluble sugars in the samples. Absorbance values were determined by UV–VIS light spectrophotometer (Thermo Spectronic Helios Alpha).
Organic acids were extracted from fresh root tissues as described by Arnetoli (2008). Samples (1 g) of frozen fresh weight were homogenized in 10 mL of Milli-Q water using a mortar and pestle in liquid N2, then homogenates were centrifuged for 20 min at 10,000 rpm at 4 °C. The supernatant was stored at −20 °C and filtered through 0.20-μm PVC filters before analysis. Organic acids (oxalic, citric, malic, formic, lactic, acetic, and succinic) were measured by ionic chromatography (Dionex DX 500) using a conductivity detector. Chromatographic conditions were as follows: sample loop volume, 25 μL; analytical column, IonPac ICE-AS6; eluent, 0.4 mM heptafluorobutyric acid (flow rate, 1.0 mL min−1); suppressor, MicroMembrane Anion-ICE; regenerant, 5 mM tetrabutylammonium hydroxide (flow rate, 5 mL min−1); and analysis time, 20 min. Organic acids were identified by comparing the retention times of the samples against retention times of the standards. Calibration curves have been performed using Merck reagents from 50 to 280 mg L−1 for oxalic acid; from 1 to 50 mg L−1 for citric, malic, and acetic acids; from 1 to 30 mg L−1 for lactic and succinic acids; and from 0.5 to 10 mg L−1 for formic acid.
Soil DNA extraction, PCR, and DGGE analyses
Rhizosphere soil was collected at flowering time from each of the three replicates. From each pot, three soil samples were bulked into a single sample. After the plants were uprooted and shaken, rhizosphere samples were collected from soil that remained gently adhered to plant roots (Lynch 1990) and collected into sterile Petri dishes and stored at 4 °C until DNA was extracted. Soil DNA was extracted using the UltraClean Soil DNA Isolation Kit (Mo Bio Laboratories, Inc.) according to the manufacturer’s instructions. Bacterial 16S rRNA gene fragments were amplified with the primer set 341F with a GC clamp (40-nucleotide GC-rich sequence, 5′-CCT ACG GGA GGC AGC AG-3′) and 907R (5′ CCG TCA ATT CMT TTG AGT TT-3′) specific for the domain Bacteria (Schäfer and Muyzer 2001). Amplicons were analyzed by DGGE using 6 % (w/v) acrylamide/bisacrylamide (37.5:1) gels containing a 40–60 % linear gradient of formamide and urea (100 % denaturing solution contained 40 % (v/v) formamide and 7 M urea). The wells were loaded with roughly equal amounts of DNA (about 500 ng), and electrophoresis was carried out in 1 × TAE buffer. The electrophoresis was run for 14 h at 100 V and a constant temperature of 60 °C, using the Ingeny Phor-U system (Ingeny International, Goes, The Netherlands). Gels were stained for 90 min in 1 × TAE buffer with SYBR Gold Nucleic Acid Gel Stain (1:10,000; Invitrogen, Breda, The Netherlands). Images of the gels were obtained by the Gel Documentation System GENi (Syngene, Frederick, MS, USA) and analyzed using the software GeneTools v. 4.01. Bands were excised from the gels, eluted in 20 μL of nuclease-free water, and incubated at 37 °C for 60 min. Aliquots (2 μL) were re-amplified by PCR for sequencing using the bacterial primers 341F and 907R without the GC clamp in the forward primer. The PCR products were purified using the NucleoSpin Gel and PCR Clean up Kit (Macherey-Nagel).
Sequencing and phylogenetic analysis
Sequencing was performed on an ABI PRISM 3130XL Genetic Analyzer (Applied Biosystems) using the Big Dye Terminator v3.1 sequencing kit. Analysis of the sequences was done with Chromas (Technelysium Ltd., Australia). Possible chimeras were detected using Chimera Check program from the Ribosome Database Project (Maidak et al. 2001). The partial 16S rRNA gene sequences were compared with sequences in GenBank with the BLASTN nucleotide alignment (Altschul and Lipman 1990) to obtain the nearest phylogenetic neighbors. The 16S rRNA gene partial sequences and their closest relative were analyzed on the Phylogeny.fr web server (Dereeper et al. 2008).
Statistical analysis
The genetic diversity of each of the soil bacterial communities was determined by DGGE profile data. Each detected band was defined as an operational taxonomic unit (OTU), and the number of bands was defined as the species richness (S) of each sample (Bell et al. 2005). To determine the diversity of bacterial communities, the Shannon-Wiener index of diversity (H′) (Shannon and Weaver 1963) and Pielou’s evenness index (E H) (Pielou 1966) were calculated according to the formulas: H′ = −Σpiln(pi) and E H = H′/H′ max = H′/lnS, where pi is the proportion of the bands in the track (it was calculated as follows: pi = ni/N; where ni is the intensity of band i in the densitometric curve, and N is the sum of the intensities of all bands) and S is the number of bands in the track.
The analysis of variance (ANOVA) and principal component analysis (PCA) were performed using the statistical software SPSS version 16.0 (SPSS, Chicago, USA) program. The analysis of one-way or two-way variance (ANOVA) was based on a randomized block design with three replicates. Means were separated by using Duncan’s multiple test (p < 0.05). Principal component analysis was performed to reveal the patterns of organic acids data matrix for genotype differentiation. Eigenvalues, eigenvectors, and principal components were calculated along with 2-D biplot generated between first (PC1) and second principal component (PC2). Similarity among DGGE profiles was estimated using the Dice correlation coefficient to compute the distance matrix. The unweighted pair group with mathematical average method (UPGMA) was used to build the similarity dendrogram from the distance matrix using the NTSYS-pc software (Exeter Software, New York, NY).
Results
Plant growth, Cr uptake, and oxidative stress
The pot experiment showed that Cr treatment led to a decreasing biomass of the two genotypes (Table 1). Compared with the controls, the decrease of root and shoot biomass was not significantly different in both genotypes. The tolerance index values were very similar in SV-21 (0.80) and SV-38 (0.77). According to the two-way analysis of variance, the root biomass and shoot biomass were not significantly influenced by Cr treatment.
Under Cr-treated soil, the total Cr concentration in roots increased from 4.40 to 46.33 mg kg−1 for SV-21 and from 9.90 to 71.39 mg kg−1 for SV-38 and, accordingly, the BF value was higher in SV-38 (0.77) than in SV-21 (0.54). Two-way analysis of variance results indicates that Cr uptake in shoots and roots was significantly impacted by the genotype (p < 0.001). Cr concentration in the roots was much higher than in the aboveground plant parts indicating that the root is the main part for Cr accumulation. Consequently, low TF values were detected in SV-21 (0.18) and SV-38 (0.08) genotypes.
The exposure to Cr stress significantly enhanced the MDA content in roots of genotype SV-38, since significant differences were found between controls and plants exposed to Cr for this genotype. However, there were not significant differences of MDA content in roots of SV-21 and in shoots of both genotypes. According to two-way ANOVA, the effects of both genotype and Cr treatment were significant at p < 0.001 level for root MDA content as well as the interaction effects between them (p < 0.01).
Changes of root organic composition in S. vulgaris genotypes
Root organic acid contents in genotypes SV-21 and SV-38 on Cr-treated and untreated soils are presented in Table 2. Among root organic acids measured in this study, oxalic acid content was much higher than the other six organics acids, followed by citric and malic acid. Different organic acid compositions were observed between both genotypes on control treatment. Thus, significant higher concentrations were found in SV-38 in citric (97 %), malic (236 %), oxalic (48 %), and succinic (34 %) acids and in SV-21 in acetic (21 %), formic (162 %), and lactic (35 %) acids. But also a different response was observed between genotypes under Cr treatment. In this regard, SV-21 shows significantly higher concentrations in citric (60 %), formic (18 %), lactic (168 %), malic (64 %), oxalic (165 %), and succinic (125 %) acids compared to untreated genotype. In contrast, the Cr-treated SV-38 showed an increased content in formic (143 %), lactic (40 %), and succinic (114 %) acids with regard to untreated SV-38.
Organic acid data recorded from three replicates were analyzed with PCA to determine if there was a consistent genotypic and treatment differentiation and to evaluate the contribution of organic acids to the variation. The different response of organic acid accumulation in both genotypes and Cr treatments is successfully summarized as showed in the PCA biplot (Fig. 1). The first principal component (PC1) separates SV-38 and chromium-treated SV-38 from Cr-treated SV-21. On the PC2 axis, genotype SV-21 was the most separated. The first principal component accounted for 49.2 % of the total variation and formic acid (0.96) followed by acetic acid (0.80) that contributed the highest to the component. Malic acid contributed negatively to the first component (−0.73). Second principal component contributed 31.4 % of the total variation. Organic acids that contributed to PC2 include oxalic acid (0.75) and lactic acid (0.72). The third principal component accounted for 19.4 % of the total variation with citric acid (0.70) given the highest contribution, while succinic acid (−0.77) contributed negatively.
Bacterial community structure based on DGGE profiles
Bacterial DGGE profiles generated from the universal bacterial primers (341F and 907R) revealed the structural composition of the communities in the rhizosphere soil samples (Fig. 2). For the analysis, 16S rRNA fragments of S. vulgaris genotypes of triplicates pots were compared by running the reaction products in parallel on the same gel. Repeated DGGE runs of the same PCR product as well as repeated PCR amplification of the same DNA extract followed by DGGE produced similar banding profiles, indicating a high degree of gel reproducibility. Each of the distinguishable bands in the separation pattern represents an individual bacterial species (Luca et al. 2002).
Generally, DGGE patterns of the three replicates for each genotype and treatment were very similar, indicating the reproducibility of the results. The number of prominent DGGE bands ranged from 20 to 21 and 23 to 26 for SV-21 and SV-38 untreated samples, respectively, revealing the presence of a high number of bacterial taxa. Cr-treated samples showed 18 visible bands on genotype SV-21 and from 17 to 21 bands on genotype SV-38 with a decrease of 11.4 and 21.6 %, respectively, compared with untreated samples. This decrease in band number was dependent on the contamination with Cr.
The DGGE patterns exhibited many dissimilarities between both genotypes grown on untreated and Cr-treated soils. In this regard, few prominent bands were unique to genotype SV-21 (6, 13) and several number of bands were specific to genotype SV-38 (4, 5, 9, 10, 11, 12, 14, 17). Conversely, few common intensely stained bands were present in almost all the lanes (1, 2, 3, 7, 8, 15, 16).
Changes in the relative intensity of individual DGGE bands were observed between treatments. Thus, in Cr-treated samples, enhanced intensity of bands 8 and 15 from genotype SV-21 and bands 5, 7, and 17 from genotype SV-38 was observed. The intensity of each band might provide an estimate of the abundance of specific taxa (Nübel et al. 1999), and band intensity might be directly related to the density of the corresponding phylotype in the template mixture, if no bias occurred during the whole extraction-amplification procedure of the microbial genomes (Muyzer et al. 1993).
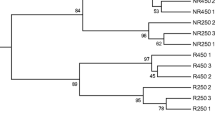
Clustering analysis of the DGGE profiles revealed the differences among the rhizosphere soil samples (Fig. 3). A binary matrix showing the presence or absence of identified bands was made for all gel lanes, and this comparison did not take into account shifts in band intensities. The DGGE profiles were separated into two major clusters with a similarity about 47 %, profiles of samples from genotype SV-21 cluster into one group and profiles from genotype SV-38 into another group, showing that major differences were related to genotype effect. In addition, changes in bacterial communities were observed between Cr-treated soils and controls in both S. vulgaris genotypes. Thus, rhizosphere soil samples from both treatments cluster with a similarity of 70 % in SV-21 and about 65 % in SV-38. Conversely, slight changes in bacterial communities were found among repeats with similarities ranging from 78 to 100 % in both genotypes.
Bacterial diversity analysis
Bacterial diversity of the microbial community was analyzed by DGGE using the Shannon-Wiener index (H′). By computerized image analysis, H′ was calculated on the basis of the bands on the gel tracks and their corresponding relative abundance indicated by band intensity (Boon et al. 2002). A diversity index consist of two components, the total number of species present or species richness (S) and the distribution of the number of individuals among those different species, called species evenness or species equability (E H). The estimate of evenness assumes a value between 0 and 1, with 1 being complete evenness or all relative species densities equal for a population (Boon et al. 2002). The results in Table 3 showed the bacterial diversity indices of all the rhizosphere soils sampled. For the untreated soils, H′ ranged from 1.57 to 1.72 in SV-21 and from 2.10 to 2.78 in SV-38, whereas on chromium-treated soils, H′ ranged from 1.19 to 1.31 in SV-21 and from 1.73 to 2.19 in SV-38. Regarding soil treatments, these results indicate a higher bacterial diversity in rhizosphere of both genotypes on untreated soil samples and, regarding plant genotypes, these results indicate a higher bacterial diversity in rhizosphere of genotype SV-38 in both treatments. Data on species evenness on untreated and Cr-treated soils, respectively, ranged from 0.52 to 0.57 and from 0.41 to 0.46 for genotype SV-21 and from 0.67 to 0.86 and from 0.61 to 0.73 for genotype SV-38. These results reflect differences in species abundance that lead to an unequal representation of species in the bacterial community, being more uneven in genotype SV-21 on both treatments.
Phylogenetic analysis of uncultured bacterial communities
Twenty prominent bands from the gel were excised and sequenced. Bands that were in the same position in the DGGE gel were excised from different lanes and sequenced to confirm that the same position corresponded to the same sequence. The positions of the sequenced bands are shown in Fig. 2. Sequence similarity values compared to previously reported sequences were more than 98 %. The phylogenetic analysis (see Fig. S1 in Supporting Information) showed that the sequences retrieved in this study were assigned to 16 different genera and classified into five major classes, namely Bacilli (Bacillus and Cohnella), Beta-proteobacteria (Acidovorax, Azohydromonas, Massilia, Ramlibacter, and Variovorax), Gamma-proteobacteria (Arenimonas, Dyella, Lysobacter, and Stenotrophomonas), Flavobacteria (Flavobacterium), and Sphingobacteria (Chitinophaga, Pedobacter, Pontibacter, and Terrimonas). All dominant groups were present in the rhizosphere of both plant genotypes, with the exception of class Flavobacteria only present in SV-38. Fifty-three percent of sequences were classified into the beta and gamma subdivisions of Proteobacteria, 23.5 % into the Sphingobacteria class, 17.6 % into the Bacilli class, and 5.8 % into the Flavobacteria class. At the species level, differences in the bacterial community composition were found between the two plant genotypes. In this regard, eight sequences were specific to genotype SV-38 and were related to Flavobacterium sp. (DGGE-4), Terrimonas ferruginea (DGGE-9), Pontibacter sp. (DGGE-5), Cohnella sp. (DGGE-14), Lysobacter sp. (DGGE-12), Stenotrophomonas maltophilia (DGGE-11), Acidovorax ebreus (DGGE-10), and Ramlibacter sp. (DGGE-17). Similarly, two sequences were specific to genotype SV-21 and were related to Massilia timonae (DGGE-6) and Azohydromonas lata (DGGE-13). Furthermore, increasing intensity of bands related to Chitinophaga niastensis (DGGE-7), Pontibacter sp. (DGGE-5), and Ramlibacter sp. (DGGE-17) was observed in Cr-treated genotype SV-38 and Variovorax sp. (DGGE-15) in Cr-treated genotype SV-21. The dominant phylotypes that appear in response to Cr treatment can be regarded as Cr-tolerant bacteria. These data indicated a shift in the relative abundance of these sequences of each Cr-treated genotype.
Discussion
While many plants and bacteria have their own mechanisms for dealing with heavy metal contaminants, the interaction of plants and microorganisms may increase or decrease heavy metal accumulation in plants, depending on the nature of the plant–microbe interaction. Because phytoremediation is a relatively new technology, understanding mechanisms of plant–microbe interactions in removing contaminants from the environment is still not well characterized. Improving knowledge could have practical implications for phytoremediation strategies on Cr-polluted soils.
Differences in heavy metal uptake between both genotypes were observed. The heavy metal toxicity based on tolerance index showed a similar Cr tolerance in both genotypes, but SV-38 showed a higher ability to accumulate Cr on root tissue, based on BF values. However, these genotypic differences in metal uptake are contradictory to those found in previous reports (Pradas del Real et al. 2013; 2014b). Several reasons could account for these contradictory results. The bioavailability of heavy metals will be affected by many interdependent factor such as solid phase sorption/desorption reactions, metal complexation reactions, growth conditions, and, among others, plant–microbe interactions (Morita et al. 2004). Hydroponic studies are useful to screen and to study the response of potential phytoremediation crops in controlled conditions independent of specific soil properties. However, metal concentrations in plant tissues can deviate substantially from soil-grown plants (Wenzel 2009).
Toxicity effects of Cr were observed in roots of both genotypes being more remarkable in SV-38, where lipid peroxidation increased as a root biomass decreased. An increase of the content of MDA reactive compounds is an indicator for physiological stress (Li et al. 2013). According to Pandey et al. (2009), Cr toxicity enhances reactive oxygen species (ROS) generation that causes oxidative stress.
S. vulgaris genotypes had a low extraction efficiency since TF values were lower than 1, although SV-21 showed higher values than SV-38. The poor translocation of Cr to shoots is a major hurdle in using plants and trees for phytoremediation (Hayat et al. 2012). However, organic acids have been reported to play an important role in phytoremediation of Cr-contaminated soils by enhancing Cr uptake and increasing translocation to shoots (Davies et al. 2001). Of the seven internal organic acids analyzed in this study, acetic, citric, malic, and oxalic acids were the most abundant. These organic acids have been reported to be potential metal chelators (Naidu and Harter 1998). Differences in changes in root organic acid contents in response to Cr treatment are evident between genotypes of S. vulgaris. The results confirm the stimulation, by Cr contamination, in six of the seven organic acids in genotype SV-21. The enhancement order with significant increases (p < 0.05) was lactic > oxalic > succinic > malic > citric > formic. Conversely, the genotype (SV-38) showed an increase in four of the seven organic acids and the enhancement order was formic > succinic > lactic > acetic. Differences in changes in root organic acids in response to Cr are also evident among rice genotypes (Zeng et al. 2008). These differences on Cr responses are also reflected in the PCA analysis and suggest that there is variability in the regulation of these responses or different mechanisms are involved in this Cr contamination response, in different genotypes.
The increased concentration of citric and malic found in SV-21 may also be involved in a higher translocation rate of Cr observed in this genotype. Both citric and malic acids have been found to be the major complexants of Cr(III) in the xylem sap of maize (Juneja and Prakasha 2005). Results from previous studies using μ-EXAFS showed that, in the roots of S. vulgaris exposed to 60 μM of Cr(VI), Cr was in a sixfold octahedral coordination which resembles that of the Cr(III)-citrate or Cr(III)-malate standards (Pradas del Real et al. 2014a).
Among organic acids, oxalic was the most abundant in both genotypes, but under Cr stress, a significant increase (165 %) only was observed in genotype SV-21. Several studies of oxalic acid have been reported to play an important role in phytoremediation of Cr-contaminated soils by enhancing uptake and increasing translocation to shoot (Zeng et al. 2008). According to Barlett and James (1988), oxalic acid can convert inorganic Cr to organically bound Cr, making it soluble for a longer period of time and thereby available to plants. However, despite having a high increase in the level of oxalic, SV-21 accumulates less Cr on roots than SV-38, suggesting that plant–bacteria interactions may play a role that influence Cr uptake.
In the adaptation process to polluted environments, bacteria in the rhizosphere of plants might be stimulated by the alterations of root exudates in response to the metal stress (Wang et al. 2008). However, biotic and abiotic factors such as soil type, seasons, heavy metal pollution, plant development stage, root architecture, plant species, cultivars, and genotypes can affect the structure of microbial communities in the rhizosphere (Marschner et al. 2001; Wieland et al. 2001; Berg and Smalla 2009; Buée et al. 2009). The PCR-DGGE cluster analysis of 16S rDNA fragments showed that the community structure was affected by plant genotype and, to a lesser extent, by Cr contamination, the first being the most influential factor shaping the rhizosphere microbiome. The bacterial DGGE profiles showed highest dissimilarity (47 %) between samples from different plant genotypes, being these results in accordance with the different bacterial community composition observed between SV-21 and SV-38, after sequencing DGGE bands. The impact of plant species on rhizobacterial communities has been well documented, but recent studies have shown genotype-dependent influence on the entire rhizobacterial community (Berg and Smalla 2009; Micallef et al. 2009). This phenomenon has been linked to plant genetic constitution (Neal et al. 1973) possibly through differences in composition and concentration of root exudate compounds (Baudoin et al. 2003). Furthermore, due to the allogamous nature of this species (Sloan et al. 2012), a high level of allelic variation among clones is expected.
The microbial community was more diverse in the rhizosphere of both untreated genotypes than in Cr-contaminated samples, based on an inspection of diversity indices. Shannon diversity index (H′), richness (S), and evenness (E H), as calculated from pooled DGGE data, were low in Cr-treated samples of SV-21 and SV-38 suggesting that Cr contamination changed the structure of microbial community to a certain extent. The toxicity of heavy metal on soil bacterial communities depends to the heavy metal involved, with Cr being the most toxic on soil microbial metabolic process (Wang et al. 2010). According to previous reports, heavy metals affect the qualitative and quantitative structure of soil microbial communities, which results in decreasing metabolic activity and diversity (Bååth 1989; Giller et al. 1998). Under heavy metal stress, many microbial species could not cope with Cr pollution, and competitive species can predominate resulting in a lack of diversity, and therefore, the decreases in diversity may reflect changes in the community composition. When comparing diversity index data from the two plant genotypes, a higher diversity was found on genotype SV-38. Bacterial communities characterized by strong species diversity can better face challenging environment perturbations than those of low species diversity (Degens et al. 2001).
Detailed knowledge of the bacterial composition in the rhizosphere of S. vulgaris can be exploited for bacteria-assisted phytoremediation of metal-contaminated sites. The phylogenetic analysis of partial 16S rRNA gene sequences recovered from the DGGE gels shows the distribution of phylogenetic groups in S. vulgaris plants under Cr stress and reveals a different bacterial composition within groups in both genotypes. The bacterial community in the rhizosphere of S. vulgaris was represented by five classes, namely Beta-proteobacteria, Gamma-proteobacteria, Sphingobacteria, Bacilli, and Flavobacteria, which indicate the largest dominance of Gram-negative bacteria. This major group is more associated to the plant than in the surrounding soil (Curl and Truelove 1986). In fact, under conditions of sufficient supply of nutrients, Gram-negative bacteria show high proliferation and activity rate, providing them a competitive advantage in the rhizosphere colonization. In this experiment, the most abundant class was Beta-proteobacteria representing 29.4 % in average of total flora in the rhizospheric area, followed by Gamma-proteobacteria and Sphingobacteria, both with a percentage of 23.5 %. The predominance of phylum Proteobacteria was also observed in the rhizosphere of Pb-, Zn-, and Cu-contaminated soils (Wang et al. 2008; Xu et al. 2012) and Cr-contaminated soils (Sheik et al. 2012). Odum (1985) suggested that r-selected organisms (rapidly reproducing), such as Proteobacteria, are favored after a stressor is applied to an ecosystem, which is perhaps a reason for their dominance.
Band sequencing provided information about the different root-associated bacterial species between SV-21 and SV-38. Most of them have been reported using cultivation-dependent methods in rhizosphere of hyperaccumulating plants growing on heavy metal-contaminated soil (Abou-Shanab et al. 2007; Kuffner et al. 2010). The different bacterial community composition suggests that each clone may have an association to specific bacterial species. Since rhizobacteria can be modulated by exudates, the production of which is, in turn, genetically regulated by plants (Micallef et al. 2009), and it can be expected that different plant genotypes would select for specific microbial consortia based on the assumption of their unique exudate profile. Previous studies on root exudates of Cr-treated genotypes SV-21 and SV-38 (Pradas del Real et al. 2014b) showed a higher root exudation release in SV-21 than in SV-38, suggesting that both genotypes could present different mechanisms in the rhizosphere to promote beneficial rhizobacteria. Furthermore, band intensity can be related to ribosome content and hence related to the activity of the microbial populations (Smalla et al. 2001).
In this context, it may be noted that under Cr contamination, an increasing intensity in specific DGGE bands was observed and related to genera Chitinophaga, Pontibacter, and Ramlibacter in SV-38 and Variovorax in SV-21. These dominant bacterial genera were reported in rhizosphere soils as in Cr-contaminated soils and groundwater sediments (Abou-Shanab et al. 2007; Brodie et al. 2011). Interestingly, Variovorax was described as highly tolerant to Cr in minimum inhibitory concentration (MIC) determinations (Abou-Shanab et al. 2007). Bacteria have developed different strategies of chromate tolerance including chromate efflux and chromate reduction based on the biotransformation of Cr(VI) to Cr(III) (Cheung and Gu 2007). Furthermore, after an organic C application, a chromium-reducing activity was observed on bacterial communities that included, among others, Variovorax genera (Brodie et al. 2011). In that same study, stimulation in growth was not detected in Ramlibacter and Chitinophaga genera. On the basis of these studies, we may speculate that exudates with large amounts of C-rich compounds can stimulate a chromium-reducing activity in Variovorax. Previous research on these plant genotypes found an increment of the exudation rate in the genotype SV-21 under Cr(VI) exposure as well as a great increase in C concentration (Pradas et al. 2014b). In this plant–bacteria interaction, Variovorax could protect its host plant from the phytotoxicity of Cr changing the speciation from bioavailable to the less bioavailable form and then leading, with regard to SV-38, to a decrease of Cr uptake.
The rhizosphere is a rich source of bacteria exhibiting enormous degree of plant growth-promoting (PGP) activities. PGP bacteria have the ability to increase the phytoavailability of nutrients and metals in soils through organic acids and siderophore production or phosphorus solubilization. Many studies have shown that bacteria could solubilize metals and nutrients in soils and improve the metal uptake by the plant (Sessitsch et al. 2013). Braud et al. (2009) observed that siderophore-producing bacteria facilitate soil metal mobilization and enhanced Cr uptake in maize after soil inoculation with Ralstonia metallidurans. In this context, it is not unreasonable to assume that dominant bacteria with PGP activities on the rhizosphere of SV-38 may enhance Cr uptake in this genotype. However, further studies are needed to assess PGP activities and chromate-reducing capacity in these bacterial taxa.
In conclusion, Cr treatment contributes to differentiate the abundance of particular organic acids as well as dominant specific rhizobacteria. The mobility and bioavailability of heavy metals in the soils, particularly at the rhizosphere where root uptake or exclusion takes place, are critical factors that affect phytoremediation processes. Based on the discussion above, specific bacterial taxa showing good PGP activities and/or chromium-stress evasion will be useful for improving current bacterial-assisted phytoremediation on Cr-polluted soils.
References
Abou-Shanab RAI, van Berkum P, Angle JS (2007) Heavy metal resistance and genotypic analysis of metal resistance genes in gram-positive and gram-negative bacteria present in Ni-rich serpentine soil and in the rhizosphere of Alyssum murale. Chemosphere 68:360–367
Abou-Shanab RA, Ghanem K, Ghanem N, Al-Kolaibe A (2008) The role of bacteria on heavy metal extraction and uptake by plants growing on multi-metal contaminated soils. World J Microbiol Biotechnol 24:253–62
Altschul SF, Lipman DJ (1990) Protein database searches for multiple alignments. Proc Natl Acad Sci U S A 87:5509–5513
Anderson J, Hooper M, Zak J, Cox S (2009) Molecular and functional assessment of bacterial community convergence in metal-amended soils. Microb Ecol 58:10–22
Arnetoli M, Montegrossi G, Buccianti A, Gonnelli C (2008) Determination of organic acids in plants of Silene paradoxa L. by HPLC. J Agric Food Chem 56:789–795
Bååth E (1989) Effects of heavy metals in soil on microbial processes and populations: a review. Water Air Soil Poll 47:335–379
Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM (2006) The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol 57:233–266
Bakker PAHM, Berendsen RL, Doornbos RF, Wintermans PCA, Pieterse CMJ (2013) The rhizosphere revisited: root microbiomics. Front Plant Sci 4:1–7
Bartlett RJ, James BR (1988) Mobility and bioavailability of chromium in soils. In: Nriagu JO and Nieboer (ed) Chromium in the natural and human environments. John Wiley & Sons, New York, pp 267–306
Baudoin E, Benizri E, Guckert A (2003) Impact of artificial root exudates on the bacterial community structure in bulk soil and maize rhizosphere. Soil Biol Biochem 35:1183–1192
Belimov AA, Hontzeas N, Safronova VI, Demchinskaya SV, Piluzza G, Bullita S, Glick BR (2005) Cadmium-tolerant plant growth-promoting bacteria associated with the roots of Indian mustard (Brassica juncea L. Czern.). Soil Biol Biochem 37:241–250
Bell T, Ager D, Song JI, Newman JA, Thompson IP, Lilley AK (2005) The contribution of species richness and composition to bacterial services. Nature 436:1157–1160
Berg G, Smalla K (2009) Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol Ecol 68:1–13
Boon N, De Windt W, Verstraete W, Top EM (2002) Evaluation of nested PCR-DGGE (denaturing gradient gel electrophoresis) with group-specific 16S rRNA primers for the analysis of bacterial communities from different wastewater treatment plants. FEMS Microbiol Ecol 39:101–112
Braud A, Jézéquel K, Bazot S, Lebeau T (2009) Enhanced phytoextraction of an agricultural Cr- and Pb-contaminated soil by bioaugmentation with siderophore-producing bacteria. Chemosphere 74:280–286
Brodie EL, Joyner DC, Faybishenko B, Conrad ME, Rios-Velazquez C, Malave J, Martinez R, Mork B, Willett A, Koenigsberg S, Herman DJ, Firestone MK, Hazen TC (2011) Microbial community response to addition of polylactate compounds to stimulate hexavalent chromium reduction in groundwater. Chemosphere 85:660–665
Buée M, de Boer W, Martin F, van Overbeek L, Jurkevitch E (2009) The rhizosphere zoo: an overview of plant-associated communities of microorganisms, including phages, bacteria, archaea, and fungi, and of some of their structuring factors. Plant Soil 321:189–212
Cheung KH, Gu JD (2007) Mechanism of hexavalent chromium detoxification by microorganisms and bioremediation potential: a review. Int Biodeter Biodegr 59:8–15
Costa R, Gotz M, Mrotzek N, Lottmann J, Berg G, Smalla K (2006) Effects of site and plant species on rhizosphere community structure as revealed by molecular analysis of microbial guilds. FEMS Microbiol Ecol 56:236–249
Curl EA, Truelove B (1986) The rhizosphere. Springer, Berlin-Heidelberg
Davies FT, Puryear JD, Newton RJ, Egilla JN, Grossi JAS (2001) Mycorrhizal fungi enhance accumulation and tolerance of chromium in sunflower (Helianthus annuus). J Plant Physiol 158:777–786
MAPA (Ministerio de Agricultura) (1994) Métodos Oficiales de Análisis. Ed: Secretaría General Técnica. Vol III, Spain
Degens BP, Schipper LA, Sparling GP, Duncan LC (2001) Is the microbial community in a soil with reduced catabolic diversity less resistant to stress or disturbance? Soil Biol Biochem 33:1143–1153
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465–469
Dhal B, Thatoi HN, Das NN, Pandey BD (2013) Chemical and microbial remediation of hexavalent chromium from contaminated soil and mining/metallurgical solid waste: a review. J Hazard Mater 250–251:272–291
Ernst WHO, Nelissen HJM (2000) Life-cycle phases of a zinc and cadmium resistant ecotype of Silene vulgaris in risk assessment of polymetallic soils. Environ Pollut 107:329–338
Giller KE, Witter E, McGrath SP (1998) Toxicity of heavy metals to microorganisms and microbial processes in agricultural soils: a review. Soil Biol Biochem 30:1389–1414
Hall JL (2002) Cellular mechanisms for heavy metal detoxification and tolerance. J Exp Bot 53:1–11
Hartmann A, Schmid M, Van Tuinen D, Berg G (2009) Plant-driven selection of microbes. Plant Soil 321:235–257
Hayat S, Khalique G, Irfan M, Wani AS, Tripathi BN, Ahmad A (2012) Physiological changes induced by chromium stress in plants: an overview. Protoplasma 249:599–611
Juneja S, Prakasha S (2005) The chemical form of trivalent chromium in xylem sap of maize (Zea mays L.). Chem Spec Bioavailab 17:161–169
Kahn M, Scullion J (2000) Effect of soil on microbial responses to metal contamination. Environ Pollut 110:115–125
Kimbrough DE, Cohen Y, Winer AM, Creelman L, Mabuni C (1999) A critical assessment of chromium in the environment. Crit Rev Env Sci Tec 29:1–46
Kuffner M, De Maria S, Puschenreiter M, Fallmann K, Wieshammer G, Gorfer M, Strauss J, Rivelli AR, Sessitsch A (2010) Culturable bacteria from Zn- and Cd-accumulating Salix caprea with differential effects on plant growth and heavy metal availability. J Appl Microbiol 108:1471–1484
Kuiper I, Lagendijk EL, Bloemberg GV, Lugtenberg BJJ (2004) Rhizoremediation: a beneficial plant-microbe interaction. Mol Plant Microbe Interact 17:6–15
Li Y, Zhang S, Jiang W, Liu D (2013) Cadmium accumulation, activities of antioxidant enzymes, and malondialdehyde (MDA) content in Pistia stratoides L. Environ Sci Pollut Res 20:1117–1123
López-Bucio J, Nieto-Jacobo MF, Ramírez-Rodríguez V, Herrera-Estrella L (2000) Organic acid metabolism in plants: from adaptive physiology to transgenic varieties for cultivation in extreme soils. Plant Sci 160:1–13
Luca C, Daniele A, Marisa M, Carlo C, Giuseppe C (2002) An application of PCR-DGGE analysis to profile the yeast population in raw milk. Int Dairy J 12:407–411
Lynch JM (1990) Soil rhizosphere. John Wiley and Sons, New York
Maidak BL, Cole JR, Lilburn TG, Parker CT Jr, Saxman PR, Farris RJ, Garrity GM, Olsen GJ, Schmidt TM, Tiedje JM (2001) The RDP-II (Ribosomal Database Project). Nucleic Acids Res 29:173–174
Marschner P, Yang CH, Lieberei R, Crowley DE (2001) Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biol Biochem 33:1437–1445
Marsden-Jones EM, Turrill WB (1957) The bladder campions (Silene maritima and S. vulgaris). The Ray Society, London
Martínez-Iñigo MJ, Pérez-Sanz A, Ortiz I, Alonso J, Alarcón R, García P, Lobo MC (2009) Bulk soil and rhizosphere bacterial community PCR-DGGE profiles and β-galactosidase activity as indicators of biological quality is soils contaminated by heavy metals and cultivated with Silene vulgaris (Moench) Garcke. Chemosphere 75:1376–1381
Micallef SA, Shiaris MP, Colon-Carmona A (2009) Influence of Arabidopsis thaliana accessions on rhizobacterial communities and natural variation in root exudates. J Exp Bot 60:1729–1742
Mnasri M, Ghabriche R, Fourati E, Zaier H, Sabally K, Barrington S, Lutts S, Abdelly C, Ghnaya T (2015) Cd and Ni transport and accumulation in the halophyte Sesuvium portulacastrum: implication of organic acids in these processes. Front Plant Sci 6:156
Morita A, Horie H, Fujii Y, Takatsu S, Watanabe N, Yagi A, Yokota H (2004) Chemical forms of aluminum in xylem sap of tea plants. Phytochemistry 65:2775–2780
Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Naidu R, Harter RD (1998) Effect of different organic ligands on cadmium sorption by and extractability from soils. Soil Sci Soc Am J 62:644–650
Navarro-Noya YE, Jan-Roblero J, González-Chávez MC, Hernández-Gama R, Hernández-Rodríguez C (2010) Bacterial communities associated with the rhizosphere of pioneer plants (Bahia xylopoda and Viguiera linearis) growing on heavy metals-contaminated soils. Antonie Van Leeuwenhoek 97:335–349
Neal J, Ruby J, Atkinson T, Larson R (1973) Changes in rhizosphere populations of selected physiological groups of bacteria related to substitution of specific pairs of chromosomes in spring wheat. Plant Soil 39:209–212
Nübel U, Garcia-Pichel F, Kühl M, Muyzer G (1999) Quantifying microbial diversity: morphotypes 16S rRNA genes and carotenoids of oxygenic phototrophs in microbial mats. Appl Environ Microbiol 65:422–430
Odum EP (1985) Trends expected in stressed ecosystems. Bioscience 35:419–422
Paliouris G, Hutchinson TC (1991) Arsenic, cobalt and nickel tolerances in two populations of Silene vulgaris (Moench) Garcke from Ontario, Canada. New Phytol 117:449–459
Pandey V, Dixit V, Shyam R (2009) Chromium effect on ROS generation and detoxification in pea (Pisum sativum) leaf chloroplasts. Protoplasma 236:85–95
Pielou EC (1966) The measurement of diversity in different types of biological collections. J Theor Biol 13:131–144
Pilon-Smits E (2005) Phytoremediation. Annu Rev Plant Biol 56:15–39
Pradas del Real AE, García-Gonzalo P, Alarcón R, González-Rodríguez A, Lobo MC, Pérez-Sanz A (2013) Effect of genotype, Cr(III) and Cr(VI) on plant growth and micronutrient status in Silene vulgaris (Moench.). Span J Agric Res 11:685–694
Pradas del Real AE, Lobo MC, Pérez-Sanz A, McNear DH (2014a) The chromium detoxification pathway in the multimetal accumulator Silene vulgaris. Environ Sci Technol 48:11479–11486
Pradas del Real AE, García-Gonzalo P, Lobo MC, Pérez-Sanz A (2014b) Chromium speciation modifies root exudation in two genotypes of Silene vulgaris. Environ Exp Bot 107:1–6
Rauser WE (1999) Structure and function of metal chelators produced by plants. The case for organic acids, amino acids, phytin and metallothioneins. Cell Biochem Biophys 31:19–48
Reilly CA, Aust SD (2001) Measurement of lipid peroxidation. Curr Protoc Toxi-col http://dx.doi.org/10.1002/0471140856.tx0204s00
Schäfer H, Muyzer GS (2001) Denaturing gradient gel electrophoresis in marine microbial ecology. In: Paul JH (ed) Methods in microbiology. Academic, London, pp 425–468
Schat H, Vooijs R, Kuiper E (1996) Identical major gene loci for heavy metal tolerances that have independently evolved in different local populations and subspecies of Silene vulgaris. Evolution 50:1888–1895
Sessitsch A, Kuffner M, Kidd P, Vangronsveld J, Wenzel WW, Fallmann K, Puschenreiter M (2013) The role of plant-associated bacteria in the mobilization and phytoextraction of trace elements in contaminated soils. Soil Biol Biochem 60:182–194
Shannon CE, Weaver W (1963) The mathematical theory of communication. University of Illinois Press, Urbana
Sheik CS, Mitchell TW, Rizvi FZ, Rehman Y, Faisal M, Hasnain S, McInerney MJ, Krumholz LR (2012) Exposure of soil microbial communities to chromium and arsenic alters their diversity and structure. PLoS One 7:40059
Sloan DB, Keller SR, Berardi AE, Sanderson BJ, Karpovich JF, Taylor DR (2012) De novo transcriptome assembly and polymorphism detection in the flowering plant Silene vulgaris (Caryophyllaceae). Mol Ecol Resour 12:333–343
Smalla K, Wieland G, Buchner A, Zock A, Parzy J, Kaiser S, Roskot N, Heuer H, Berg G (2001) Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl Environ Microbiol 67:4742–4751
Vangronsveld J, Herzig R, Weyens N, Boulet J, Adriaensen K, Ruttens A, Thewys T, Vassilev A, Meers E, Nehnevajova E, van der Lelie D, Mench M (2009) Phytoremediation of contaminated soils and groundwater: lessons from the field. Environ Sci Pollut Res 16:765–794
Venditti F, Ceglie A, Palazzo G, Colafemmina G, Francesco Lopez F (2007) Removal of chromate from water by a new CTAB–silica gelatin composite. J Colloid Interface Sci 310:353–361
Venditti F, Cuomo F, Ceglie A, Ambrosone L, Lopez F (2010) Effects of sulfate ions and slightly acidic pH conditions on Cr(VI) adsorption onto silica gelatin composite. J Hazard Mater 173:552–557
Wang Y, Li Q, Shi J, Lin Q, Chen X, Wu W, Chen Y (2008) Assessment of microbial activity and bacterial community composition in the rhizosphere of a copper accumulator and a non-accumulator. Soil Biol Biochem 40:1167–1177
Wang F, Yao J, Si Y, Chen H, Russel M, Chen K, Qian Y, Zaray G, Bramanti E (2010) Short-time effect of heavy metals upon microbial community activity. J Hazard Mater 173:510–516
Wenzel WW (2009) Rhizosphere processes and management in plant-assisted bioremediation (phytoremediation) of soils. Plant Soil 321:385–408
Wieland G, Neumann R, Backhaus H (2001) Variation of microbial communities in soil, rhizosphere, and rhizoplane in response to crop species, soil type, and crop development. Appl Environ Microbiol 67:5849–5854
Xu ZY, Tang M, Chen H, Ban YH, Zhang HH (2012) Microbial community structure in the rhizosphere of Sophora viciifolia grown at a lead and zinc mine of northwest China. Sci Total Environ 435–436:453–464
Zayed AM, Terry N (2003) Chromium in environment: factors affecting biological remediation. Plant Soil 249:139–156
Zeng F, Chen S, Miao Y, Wu F, Zhang G (2008) Changes of organic acid exudation and rhizosphere pH in rice plants under chromium stress. Environ Pollut 155:284–289
Zhang W, Huang Z, He L, Sheng X (2012) Assessment of bacterial communities and characterization of lead-resistant bacteria in the rhizosphere soils of metal-tolerant Chenopodium ambrosioides grown on lead-zinc mine tailings. Chemosphere 87:1171–1178
Acknowledgments
The authors thank the financial supports provided by EIADES (project S2009/AMB-1478, Comunidad de Madrid) and INIA (project RTA-000150-00-00).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Yi-Ping Chen
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 145 kb)
Rights and permissions
About this article
Cite this article
García-Gonzalo, P., del Real, A.E.P., Lobo, M.C. et al. Different genotypes of Silene vulgaris (Moench) Garcke grown on chromium-contaminated soils influence root organic acid composition and rhizosphere bacterial communities. Environ Sci Pollut Res 24, 25713–25724 (2017). https://doi.org/10.1007/s11356-016-6667-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-016-6667-4