Abstract
A culture supernatant from Micrococcus luteus containing resuscitation-promoting factor (SRpf) was used to enhance the biological nutrient removal of potentially functional bacteria. The obtained results suggest that SRpf accelerated the start-up process and significantly enhanced the biological nutrient removal in sequencing batch reactor (SBR). PO4 3−-P removal efficiency increased by over 12 % and total nitrogen removal efficiency increased by over 8 % in treatment reactor acclimated by SRpf compared with those without SRpf addition. The Illumina high-throughput sequencing analysis showed that SRpf played an essential role in shifts in the composition and diversity of bacterial community. The phyla of Proteobacteria and Actinobacteria, which were closely related to biological nutrient removal, were greatly abundant after SRpf addition. This study demonstrates that SRpf acclimation or addition might hold great potential as an efficient and cost-effective alternative for wastewater treatment plants (WWTPs) to meet more stringent operation conditions and legislations.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
In recent years, phosphorus and nitrogen are nutrients of primary concern about surface water eutrophication. Since most wastewater treatment plants (WWTPs) are facing increasingly stringent effluent limitations, removing nutrients from wastewater is becoming a more and more acute problem (Wang et al. 2015; Winkler et al. 2011). Therefore, there is an urgent need for technique solutions to enhance nutrient removal. Conventional activated sludge systems are widespread in Chinese WWTPs, which will face enormous challenges for modifying treatment systems due to the limitation of land and capital, creating an acute demand for more cost-efficient and less land-occupied technology (Chen et al. 2015). Activated sludge is the essential functional element of biological wastewater treatment. It is well known that prokaryotic microorganisms in activated sludge catalyze the most important transformations in WWTPs (Juretschko et al. 2002), and the efficient and stable operation of activated sludge process relies upon bacterial communities. Thus, improving the biological activity of functional bacterial communities in activated sludge could be a promising way to enhance nutrient removal.
Several studies showed that traditional model functional organisms composed only a small fraction of total bacteria cells in the activated sludge (Van Loosdrecht et al. 1997; Wagner et al. 1994). This bias arised mainly because of the low fraction of culturable organisms present in the sludge (Wagner et al. 1994; Wagner and Loy 2002). Most bacteria of the biosphere that cannot be cultivated, referred to as the “viable but non-culturable” (VBNC) or uncultured bacteria with their typically low levels of metabolic activity, can also play an important role in the function of the natural environment (Epstein 2013; Su et al. 2013a). It was reported that only 1–15 % of bacteria in activated sludge were culturable (Amann et al. 1995). In other words, a large portion of the functional microbial communities, including bacteria responsible for nitrogen removal or catalyzing phosphorus removal, might fail to grow on routinely employed media with their typically low levels of metabolic activity (Su et al. 2013a). As the biodiversity of microbial communities in activated sludge were revealed at astonishing speed with 16S rRNA sequences-based culture-independent methods and modern cultivation procedures, a great amount of VBNC or uncultured but key potential nutrient removal bacteria were discovered during the past decades (Fujitani et al. 2014; Kong et al. 2005; Wan et al. 2014), such as Rhodocyclus-related bacteria (also known as Candidatus Accumulibacter), Acinetobacter, Microlunatus sp., Tetrasphaera-related species, Lampropedia sp., Malikia sp., and several gram-positive Actinobacteria for phosphorus removal and Nitrosomonas, Dechloromonas sp., Thauera sp., Proteobacterium, Hyphomicrobium, Azoarcu, and Nitrospira for nitrification or denitrification process. Since abundant VBNC or uncultured bacteria are potential bacterial resource, how to stimulate and resuscitate VBNC or uncultured bacteria in activated sludge might be of great significance in domestic wastewater treatment process.
The discovery of a bacterial cytokine called the resuscitation-promoting factor (Rpf), which is secreted by Micrococcus luteus, was a very exciting development in resuscitating and promoting VBNC or uncultured bacteria (Kaprelyants and Kell 1993; Mukamolova et al. 1998). To this time, there are some possible Rpf action mechanisms which had been reported. Some studies indicated that the mechanism about Rpf resuscitating VBNC bacteria or uncultured bacteria is that the Rpfs are peptidoglycan hydrolases, which are involved in the complex process of cell wall digestion in order to allow cell division to occur (Hett and Rubin 2008; Kana and Mizrahi 2010; Wyckoff et al. 2012). Moreover, Reissbrodt et al. (2002) indicated that Rpf was a heat-stable “autoinducer of growth,” and it was secreted by a variety of gram-positive and gram-negative bacterial species (Senoh et al. 2012). In addition, Mukamolova et al. (2006) demonstrated that Rpf possessed muralytic activity that was probably responsible for its observed influence, which would have major implications for the resuscitation process. However, the mechanism of Rpf activity, both in resuscitating VBNC bacteria and in stimulating growth when bacteria are in a state of low activity under adverse environmental conditions, remains to be uncovered. Up to date, it is realized that Rpf can stimulate a large number of gram-positive bacteria, including the phylum Actinobacteria, such as the genera Mycobacterium, Rhodococcus, Arthrobacter, Leifsonia, and Kitasatospora (Nikitushkin et al. 2011). Ding and Yokota (2010) discovered that Rpf also stimulated the growth of some other gram-negative organisms, like several Proteobacteria. Although it remains to be uncovered about the mechanism of Rpf activity in either resuscitating VBNC or uncultured bacteria or stimulating growth when bacteria are in a state of low activity, several studies have been done with Rpf exploring the potential environmental functions of VBNC or uncultured bacteria (Su et al. 2013a, b). In addition, a previous study showed that neither pure Rpf protein from M. luteus culture supernatant nor recombinant Rpf could maintain its activity for over 1 week at 4 °C (Su et al. 2013b). What is more, several other functionally equivalent proteins were found in supernatant from M. luteus (Mukamolova et al. 2006). Therefore, it is more cost-effective and advantageous to resuscitate and stimulate VBNC or uncultured bacteria using the culture supernatant from M. luteus containing Rpf, also referred to as SRpf, than that using purified Rpf protein.
In this study, the nutrient removal efficiencies of three different sequencing batch reactors (SBRs; control reactor R1 with inactivated SRpf addition, treatment reactor R2 with SRpf addition, and blank reactor R3 without SRpf) were compared and the bacteria community compositions in these sludge samples were assessed. It was reported that 10–15 % (v v−1) was the effective dosage of SRpf when it was applied to promote biphenyl-capability of bacterial community from PCB-contaminated soil (Su et al. 2013b). However, since SRpf itself contained highly concentrated organic matter and nutrients (COD 2000–5000 mg L−1, TN 457–766 mg L−1, PO4 3−-P 150–166 mg L−1), it would cause serious disturbances of influent if such amount of SRpf dosage was applied, which might inhibit removal efficiency and interfere experimental results. Therefore, we proposed to introduce a small amount of SRpf to nutrient removal process by acclimating part of the condensed activated sludge periodically.
Our goal was to determine whether SRpf could be a potential effective additive for wastewater treatment and characterize the bacterial community composition which is responsible for enhancing nutrient removal efficiency. We hypothesized that SRpf enhanced the biological phosphorus and nitrogen removal capability of potential bacteria in activated sludge, which may open up a new avenue for wastewater treatment to meet more stringent operation conditions and legislations in a more cost-effective and less land-occupied way. This is, to our knowledge, the first study to resuscitation and stimulation of the potential phosphorus and nitrogen removal bacterial community in activated sludge by using SRpf.
Materials and methods
M. luteus Rpf (SRpf)
M. luteus (AJ409096.1) was inoculated on a rotary shaker (30 °C, 160 rpm) for 36 h in lactate minimal medium (LMM), which was described by the previous study (Kaprelyants and Kell 1992). SRpf was obtained according to the methodology described by the previous study (Su et al. 2013b), then the SRpf was stored at −20 °C for further experiment.
Lab-scale sequencing batch reactors
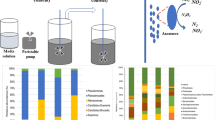
Activated sludge collected from the secondary sedimentation tank in Qi Ge domestic wastewater treatment plant (Hangzhou, China) was used as inoculum for SBRs. The inoculums were completely mixed and equally divided into three portions before the reactors were seeded. The sludge was elutriated and filtered with distilled water in order to remove both inorganic and organic components and suspended solids. The mixed liquor suspended solids (MLSS) in every SBR was maintained at 4.5–5.5 g L−1.
Three parallel SBRs (2.5 L working volume each) were used in this work, which comprised 3-L glass cylindrical containers, an aerator, a peristaltic pump, three intake pumps, and three electric stirrers. Every bioreactor had an 8-h cycle with the following sequence: 10 min of feed of 1.25 L influent synthetic wastewater, 50 min of anaerobic phase, 3.5 h of aerobic phase, 2.5 h of anoxic phase, 35 min of sedimentation, and 25 min for extraction of 1.25 L of supernatant. Dissolved oxygen (DO) was controlled between 3.5 and 5 mg L−1 during the aerobic phase. The sludge retention time (SRT) was kept at 20 days by periodic wasting sludge mixed liquor at the end of anoxic stage. The primary components of synthetic wastewater used in the test are shown in Table 1. The wastewater had a chemical oxygen demand (COD) of about 620 mg L−1, ammonia nitrogen (NH4 +-N) of 48 mg L−1, total nitrogen (TN) of 54 mg L−1, total phosphorous (TP) of 16 mg L−1, and pH of 6.9–7.3. The SBRs were operated under constant temperature of 25 ± 0.5 °C.
Acclimation of activated sludge using SRpf
SRpf was fed into R2, also known as the treatment reactor, from the beginning of start-up phase: during the sedimentation phase in every cycle, certain amount (120 mL) of the condensed sludge was removed from R2 for acclimation using SRpf (10 %, v v−1), then same amount of sludge which has already been removed and acclimated for 1 h in a rotary shaker (25 °C, 150 rpm) was returned to the reactor before the next cycle started. The inactivated SRpf (sterilized at 121 °C for 15 min) was added to the sludge removed from R1 (control reactor), and synthetic wastewater which contains the same COD and nutrient concentrations as that SRpf has was added to the sludge removed from R3 (blank reactor), then same amount of sludge after same treatment in rotary shaker were returned to R1 and R3, respectively. The COD, NH4 +-N, NO3 −-N, NO2 −-N, and PO4 3−-P concentrations in the influent and effluent, MLSS, and sludge volume index (SVI) of R1, R2, and R3 were monitored every 2 days.
Chemical analysis
COD, BOD, NH4 +-N, TN, and TP in the influent and effluent were regularly measured to monitor the performance and stability of the SBRs, according to the APHA standard methods (APHA 2005).
Dissolved oxygen (DO) and temperature were measured by a DO sensor (Mettler Toledo, Switzerland), and pH levels were measured by a pH monitor (Mettler Toledo, Switzerland).
Specific oxygen uptake rate (SOUR) was measured in triplicate using batch extent respirometric assays described elsewhere (Hu et al. 2002; Wang et al. 2015). Briefly, biomass aliquots (150 mL) were collected from each reactor. The assay was performed at final pH of 7.0 ± 0.05 in replicate 290-mL water-jacketed glass vessels, maintained at 25 °C. Biomass suspensions were aerated with pure oxygen before substrate (acetate, NH4 +-N, or NO2 −-N) was injected. Initial concentration of 100 mg L−1 COD, 5 mg L−1 NH4 +-N, and 10 mg L−1 NO2 −-N were determined to yield substrate-independent specific oxygen uptake rates. A decrease in the DO level due to substrate oxidation was measured by a DO monitor (YSI model 5100, USA). SOUR were calculated using the equation SOUR (mg O2/g MLSS min) = −G/X (Zhang and Zhao 2013), where G is the slope of the linear of the DO decline curve in milligrams per liter per minute, and X is the MLSS of biomass suspensions in grams per liter.
Microbiological analysis
DNA extraction
Twelve activated sludge samples were collected from R1 (R1-1, R1-2, R1-3, and R1-4), R2 (R2-1, R2-2, R2-3, and R2-4), and R3 (R3-1, R3-2, R3-3, and R3-4) on four dates shown (Fig. 1) according to different stages in the long-term operation. Each sample was condensed and dispended into a 50-mL tube. The supernatant was decanted, and the samples were stored at −20 °C prior to analysis.
Research timeline showing time points for all sampling investigations in this study. Twelve activated sludge samples were collected from R1 (R1-1, R1-2, R1-3, and R1-4), R2 (R2-1, R2-2, R2-3, and R2-4), and R3 (R3-1, R3-2, R3-3, and R3-4) on four dates (DNA 1, DNA 2, DNA 3, and DNA 4) according to different stages in the long-term operation
The activated sludge samples were washed three times by centrifugation using sterile high-purity water for 5 min at 10,000×g. DNA extraction was then performed using a Fast DNA SPIN Kit for Soil (Bio101 Inc., USA), according to the manufacturer’s protocol. The DNA extracts were stored at −20 °C for further analysis.
Illumina high-throughput sequencing
To determine the diversity and composition of the bacterial communities in those 12 activated sludge samples, 16S rRNA gene sequencing was performed according to the protocol described by Caporaso et al. (2010). About 500 ng of DNA of each sample was mixed and sent to a commercial company (Guheinfo, Hangzhou, China) for Illumina MiSeq sequencing. PCR amplifications were conducted with the 319F/806R primer set that amplifies the V3–V4 region of the 16S rRNA gene. The primer set selected yielded relatively accurate phylogenetic and taxonomic information because of the few biases. Sequencing was conducted on an Illumina MiSeq platform. All raw reads have been deposited into the NCBI Sequence Read Archive (Accession Number SRR1948176).
According to the unique barcode of each sample, sequencing reads were assigned to each sample. Sequences were analyzed with the Quantitative Insights Into Microbial Ecology (QIIME) software package, in addition to custom Perl scripts to analyze alpha and beta diversity (Wang et al. 2007). Sequences were assigned to operational taxonomic units (OTUs) at 97 % similarity.
Statistical analysis
Data Processing System (DPS) software (version 11.0) was used for data analysis. Data represented the means and standard deviations (SD). The least significant difference (LSD) test was used to determine the statistical significance of the SRpf acclimation effects. The level of statistical significance was set at a p < 0.05 for all statistical analyses.
Chao1 metric estimates the species richness, the observed species metric is simply the count of unique OTUs found in the sample, and the Shannon index was calculated to assess bacterial diversity.
Results and discussion
Effects of SRpf on long-term performance of R1, R2, and R3
The objectives of the continuous operation were to test whether R2 process could enhance the nutrient removal more than R1 and R3 could in the long-term.
Figure 2 shows the continuous operating results of nutrient removal in R1, R2, and R3. During the start-up phase, the effluent qualities of three SBRs were very close in the first 3 days, then R2 gradually performed better than R1 and R3. The effluent PO4 3−-P concentration in R2 was the only one reduced to less than 3 mg L−1 after 1-week operation among those three SBRs, and it reduced to 0.72 mg L−1 after another week, while PO4 3−-P concentrations in R1 and R3 were 2.78 and 2.92 mg L−1 at the same time. Similarly, effluent TN concentration in R2 was below 7 mg L−1 after 1-week acclimation and kept decreasing until 15 days, while effluent TN concentrations in R1 and R3 were 7.36 and 8.23 mg L−1, respectively, after 1-week operation and could not obtain significant lower effluent since 10 days. Those indicated that SRpf accelerated the SBR start-up process, which was probably due to the stimulating ability of SRpf towards VBNC or uncultured bacteria cells under adverse conditions, such as influent loads shock. Steady states of all three SBRs were reached after 15-day operation. The average effluent PO4 3−-P concentration in R1 and R3 were 2.69 ± 0.27 and 2.20 ± 0.35 mg L−1, respectively, and effluent TN concentrations of R1 and R3 was between 7.0 and 7.7 mg L−1. The average TN removal efficiency of R1 and R3 were 69.94 ± 1.13 and 67.32 ± 1.43 %, respectively, and the average PO4 3−-P removal rate were 85.88 ± 2.24 and 82.75 ± 1.72 %, respectively. The reactors did not show significant difference in the effluent TN and PO4 3−-P between R1 and R3. Compared to R3, the fragile advantage of R1 in nutrient removal is possibly attributed to the various components in inactivated SRpf, such as polysaccharide and trace element, which could affect the functional bacteria in activated sludge. Meanwhile, effluent PO4 3−-P and TN of R2 were 0.67 ± 0.27 and 5.63 ± 0.32 mg L−1 with the removal efficiency as 95.71 ± 1.71 and 76.03 ± 1.38 %, respectively, indicating that PO4 3−-P removal efficiency increased by over 12 %, and TN removal efficiency increased by over 8 % in treatment reactor under the same operating conditions compared with those of control or blank reactor.
To further verify the impact of SRpf on the bioreactor, SRpf addition in the reactor was stopped for 90 days. It was worth noting that removal efficiency of both PO4 3−-P and TN in R2 were gradually decreased. After 10 days, the effluent PO4 3−-P and TN even showed no significant difference between R2 and R3, and they maintained this level for a week (Fig. 2). Then, at the 107th day, we restarted adding SRpf. Interestingly, the removal efficiency of R2 almost regained its previous level just after 1 week of SRpf acclimation. Therefore, SRpf did play an essential part in facilitating the nutrient removal during long-term operation. Moreover, it was indicated that the promoting effects could last for 10 days even without SRpf constantly addition, and it could quickly regain previous high level of nutrient removal after restarting SRpf acclimation. Therefore, SRpf might be regarded as a promising way to not only enhance removal capability but also improve the activity of activated sludge from deterioration.
Effects of SRpf on a typical cycle of R1, R2, and R3
Phosphorus removal
Each of the three phosphorous (P) cycling was consistent with current EBPR theory (Smolders et al. 1994), and comparatively better P removal was achieved in R2 (Fig. 3a; Table 2). During anaerobic phase, the amount of P release in R2 was 11.32 mg L−1, while the amount of P release in R1 and R3 was only 6.74 and 7.20 mg L−1, respectively. The P release rate was 3.72 mg gVSS−1 h−1, while P release rate in R1 and R3 were only 2.23 and 2.35 mg gVSS−1 h−1, respectively. Thus, SRpf significantly enhance (p < 0.05) the P release in anaerobic phase. According to previous studies, the P release rate could reach to 5–30 mg gVSS−1 h−1 (Monti et al. 2007). Although even R2 did not reach that level, it did show dominance in P release. Correspondingly, the biomass in R2 assimilated significantly more phosphorus than (p < 0.05) that in R1 and R3. The amount of P uptake in R2 was 23.21 mg L−1. Meanwhile, the amount of P uptake in R1 and R3 were only 18.36 and 17.95 mg L−1, which were significantly less (p < 0.05) than that in R2. In terms of the anoxic phase, P uptake was also observed in all three SBRs. Previous study demonstrated that secondary P release could be avoided during the post anoxic phase as long as nitrate/nitrite remained available (Carvalho et al. 2007; Winkler et al. 2011), which was consistent with the herein results in our study. Notably, the biomass of R2 had the most P uptake amount (9.77 mg gVSS−1 h−1) among those three SBRs in the equal times, suggesting that SRpf might accelerate the process of denitrifying phosphorus removal by promoting specific functional bacteria.
Nitrogen removal
Figure 3b shows the typical profiles of different nitrogen species (NH4 +-N, NO3 −-N, and NO2 −-N) in three reactors at a steady state. All three SBRs showed typical nitrogen removal via nitrification and denitrification during the aerobic and anoxic phases, respectively. Table 3 summarizes the process performances in aerobic phase of R1, R2, and R3. Nitrification, the two-step process by which ammonia is oxidized to nitrate via nitrite, plays a key role in the biological removal of nitrogen in aerobic phase in wastewater treatment systems. The process involves two groups of obligately chemolithotrophic bacteria. Ammonia-oxidizing bacteria (AOB) first oxidize ammonia to nitrite, and nitrite was then oxidized by nitrite-oxidizing bacteria (NOB) (Fudala-Ksiazek et al. 2014; Limpiyakorn et al. 2005). Ammonia oxidation process, driven by AOB, is a rate-limiting step of nitrogen removal because of their slow growth rate, their high sensitivity to many environmental factors, and their inability to outcompete heterotrophs (Wagner et al. 1995). As shown in Table 3, 91.8 and 90.1 % of NH4 +-N were transformed at the end of the aerobic phase in R1 and R3, respectively, while 94.2 % of NH4 +-N was transformed in R2. The specific nitrification rate in R2 was found to be 1.09 mg gVSS−1 h−1, which is significantly higher (p < 0.05) than that in R1 as 0.74 mg gVSS−1 h−1 and that in R3 as 0.85 mg gVSS−1 h−1. During the aerobic phase, most ammonia in all three SBRs were oxidized into N-oxides (nitrate and nitrite). In comparison with that of R1 and R3, significantly larger (p < 0.05) production of nitrate occurred in R2 from the beginning to the end of aerobic phase. The nitrite concentration in three SBRs increased at first and decreased after 2 h aeration, and this phenomenon has been reported by Winkler et al. (2011). The explanation for the variation of nitrite during aerobic phase, involving many aspects, such as aeration rate, ammonia concentration, and competition of related bacteria species, is very complicated and remains unclear according to current research (Nielsen et al. 2010). Noticeably, R2 produced the most amount of nitrite in the first 2 h but ended up with the least amount of nitrite. Those results indicate that R2, acclimated by SRpf, enhanced NH4 +-N removal and oxidation efficiency.
Obviously, carbon source in three SBRs had already been completely consumed before the end of the aerobic phase (Fig. 3a). Meanwhile, N-oxides in all SBRs were observed to be still gradually decreased during the anoxic phase, demonstrating that post-denitrification, using intracellular glycogen as potential electron donor, occurred in all three reactors, and this is consistent with similar studies (Coats et al. 2011; Vocks et al. 2005). Recognizing that the denitrification rate (DNR) in R2 was significantly better (p < 0.05) than that in R1 and R3 (Table 4) thus indicates that SRpf probably induced better utilization of intracellular glycogen and other positive biochemical reactions of functional bacteria that could drive denitrification.
Effects of SRpf on SOUR
As oxygen is required to remove both nutrients and carbon source, SOUR represents an important process in wastewater treatment. SOUR is influenced by MLSS concentration, mixing characteristics, substrate availability, nutrient availability, and toxin presence during an activated sludge process. Thus, SOUR reflects strength of microbial metabolism and hydraulic loading effects on substrate utilization rate (Zhang and Zhao 2013).
The slope of linear of DO decline curve of three-substrate (acetate, ammonia, and nitrite) oxidation process in the biomass from R1, R2, and R3 were calculated to demonstrate SOUR. The specific oxygen uptake rate of ammonia oxidation was calculated from the difference between the measured specific oxidation rate with ammonium as substrate (also known as the specific oxygen uptake rate of autotrophic bacteria, SOURA) and the measured specific oxidation rate with nitrite as substrate nitrite oxidation rate (SOURNO2): SOURNH4 = SOURA − SOURNO2.
Figure 4 further depicts the difference in SOUR of activated sludge from R1, R2, and R3. The average SOURCOD, also considered as the specific oxygen uptake rate of heterotrophic bacteria, were 0.38 ± 0.02 mg O2/(g MLSS min), 0.66 ± 0.04 mg O2/(g MLSS min), and 0.33 ± 0.02 mg O2/(g MLSS min) from the activated sludge in R1, R2, and R3, respectively. Through statistical analysis, we could tell that SOURCOD of R2 was significantly more (p < 0.05) than that of R1 and R3, indicating that SRpf significantly enhanced the metabolism for organic matter removal. Moreover, significant enhancements (p < 0.05) were also observed in the ammoxidation process [0.19 ± 0.02 mg O2/(g MLSS min) for R1, 0.26 ± 0.02 mg O2/(g MLSS min) for R2, and 0.16 ± 0.01 mg O2/(g MLSS min) for R3] and nitrite oxidation process [0.13 ± 0.01 mg O2/(g MLSS min) for R1, 0.18 ± 0.01 mg O2/(g MLSS min) for R2, and 0.10 ± 0.01 mg O2/(g MLSS min) for R3] from the activated sludge acclimated by SRpf (Fig. 4). These results suggest that SRpf successfully enhanced the strength of microbial metabolism on substrate utilization in activated sludge, which to some extent, explained the promotion of the better efficiency on COD and nutrient removal in R2.
Shifts in the bacterial community structure
The key to reveal the effects of SRpf addition on nutrient removal ability is to identify the main bacteria involved in the process and to elucidate how their activities may be enhanced.
As shown in Table 5, after filtering the low-quality reads, there were 19,536–48,684 effective reads for 12 activated sludge samples. Operational taxonomic units (OTUs) were adopted to estimate bacterial species richness among those samples at a distance level of 3 %. The average OTUs of the samples in R2 under SRpf acclimation was 1192; however, the average OTUs of the samples in R1 and R3 were 918 and 808, respectively. Notably, the OTUs of the sample in R2 after stopping SRpf addition was only 968 and went back to 1167 after reusing SRpf. It is clear that SRpf acclimation significantly increase the amount of OTUs in activated sludge. With SRpf addition, the samples (R2-1, R2-3, and R2-4) showed the highest Shannon diversity, and the samples without SRpf or inactivated SRpf addition showed the lowest Shannon diversity, and the promotion effect of SRpf (averaging 6.47) was significantly better (p < 0.05) than inactivated SRpf (averaging 5.77). Overall, the order of bacterial diversity in long-term operation with SRpf was R2 > R1 > R3. SRpf acclimation obviously caused a boost in the bacteria diversity in activated sludge.
The distributions of sequences at the phyla level and class level in Proteobacteria for each sample are shown in Fig. 5. Proteobacteria was the predominant phylum in all those samples constituting 33.9–56.3 % of total effective bacteria sequences. The abundance of Proteobacteria in samples from R2-1 (56.3 %), R2-3 (50.8 %), and R2-4 (54.6 %) were higher (p < 0.05) than that in either R1 (R1-1 38.0 %, R1-3 44.3 %, R1-4 43.4 %), or R3 (R3-1 33.9 %, R3-3 42.1 %, R3-4 37.7 %). And the order of average abundance for Proteobacteria in operation with SRpf was R2 > R1 > R3. Notably, there was no significant difference among the abundance of Proteobacteria in R1-2 (40.0 %), R2-2 (39.8 %), and R3-2 (41.1 %), which were collected from R2 during operation without SRpf. The abundance of Proteobacteria in R2, corresponding to effluent quality change, decreased from 56.3 to 39.8 % within 10 days since SRpf was no longer added, then increased to 54.6 % after reusing SRpf for 2 weeks. It was obvious that long-term acclimation of SRpf have increased the abundance of Proteobacteria. With respect to the Actinobacteria phylum, which was the subdominant group (20.6–30.1 %), the order of average abundance under SRpf acclimation was also R2 (29.0 %) > R1 (23.8 %) > R3 (22.8 %). Compared with the control reactor with inactivated SRpf and the blank reactor, R2 had a higher abundance of Proteobacteria and Actinobacteria, which could be attributed to the function of some proteins in SRpf (Su et al. 2013b). It was reported that Rpf protein from M. luteus promoted the resuscitation and growth of high G+C gram-positive organisms, and several other proteins in SRpf also had the same muralytic activity as Rpf protein (Su et al. 2014). Those primary bacteria in R2 contained a good deal of polyphosphate accumulating organisms (PAOs), nitrifiers, and denitrifiers (Accumulibacter, Nitrosomonas, Dechloromonas, Pseudomonas, Bacillus, etc.).
Within Proteobacteria, β-Proteobacteria was the most dominant class in all the activated sludge samples, and the abundance of α-Proteobacteria and γ-Proteobacteria composed the second and third largest proportion, respectively. However, the abundance of β-Proteobacteria (averaging 42.9 %) and γ-Proteobacteria (averaging 4.5 %) in the sludge samples of R2 is significantly higher (p < 0.05) than that in R1 (averaging 33.3 % for β-Proteobacteria, averaging 2.3 % for γ-Proteobacteria) and R3 (averaging 29.0 % for β-Proteobacteria, averaging 2.2 % for γ-Proteobacteria) under SRpf acclimation, while no significant difference was observed in terms of the abundance of γ-Proteobacteria among those samples. δ-Proteobacteria (0.9–2.0 %) and ε-Proteobacteria (0–0.3 %) composed a very low proportion in 12 activated sludge samples. In conclusion, SRpf increased the abundance of β-Proteobacteria and γ-Proteobacteria and finally induced the increase of the abundance of Proteobacteria in R2.
To further explore the changes in bacterial community with SRpf addition, the sequence assignment results at the genus level in all samples were investigated using heat map. The top 15 most abundant genera in each sample were selected (a total of 66 genera for all 12 samples), and their abundances were compared with those in other samples (Fig. 6).
SRpf influenced the abundance of nitrifying bacteria, denitrifying bacteria, and PAOs. In the activated sludge samples of R2, Nitrosomonas and Nitrospira were two typical genera of AOB and NOB, which were mainly responsible for the oxidation of ammonia to nitrate. The average abundance of Nitrosomonas accounted for 0.08 % of total biomass in R2 under SRpf acclimation. In contrast, the abundance of Nitrosomonas in sludge samples in R1 and R3 were negligibly small (0–0.01 %). The average abundance of Nitrospira (0.30 %) in the activated sludge of R1 using inactivated SRpf was similar to that in R3 (0.25 %), which were both significantly less (p < 0.05) than that in R2 (0.62 %). Obviously, SRpf increase the abundance of Nitrospira and Nitrosomonas, and it might explain the promotion of the ammonia oxidation process in the presence of SRpf. In addition, the abundance of Dechloromonas and Pseudomonas were observed to be increased in the activated sludge samples of R2, and all these genera could drive denitrification process, which might be the main reason for its better denitrifying capability. In fact, species in the genus Pseudomonas were observed to be denitrifying phosphate-accumulating organisms (DNPAOs), which were known to perform both denitrification processes and polyphosphate accumulation (Jørgensen and Pauli 1995). With regard to carrying out wastewater phosphorus removal, Acinetobacter and Pseudomonas were the main genera in activated sludge. The results indicate that the average abundance of Acinetobacter and Pseudomonas in R2, acclimated by SRpf, accounted for 1.37 % of total biomass, which was much more than that in R1 (0.55 %) and R3 (0.12 %). Moreover, the abundance Candidatus Accumulibacter and Rhodocyclus, which were two functional but uncultured genera previously discovered in EBPR, were also observed to be much more than those in R1 and R3. In conclusion, SRpf resuscitated and promoted several functional bacteria communities in activated sludge, and this might be the main reason for the better capability of nutrient removal of R2.
Conclusions
The obtained results suggest that SRpf played a key role in significantly accelerating the start-up process, enhancing the biological nutrient removal in SBR and in shifts in the composition and diversity of bacterial community. SRpf could also improve the activity of activated sludge from deterioration. The phyla of Proteobacteria and Actinobacteria, which increased significantly after SRpf addition, were mainly responsible for the enhanced nutrient removal capability. We can speculate that SRpf or Rpf addition might be a promising and cost-effective alternative for WWTP to meet more stringent operation conditions and legislations. Further studies are needed to investigate the effects of SRpf on highly concentrated nutrients of wastewater and to estimate the optimal dosage of SRpf addition for activated sludge system.
References
Amann RI, Ludwig W, Schleifer K-H (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59(1):143–169
APHA W (2005) Standard methods for the examination of water and wastewater. American Public Health Association, Washington, DC
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335–336
Carvalho G, Lemos PC, Oehmen A, Reis MAM (2007) Denitrifying phosphorus removal: linking the process performance with the microbial community structure. Water Res 41(19):4383–4396
Chen HB, Wang DB, Li XM, Yang Q, Zeng GM (2015) Enhancement of post-anoxic denitrification for biological nutrient removal: effect of different carbon sources. Environ Sci Pollut Res 22(8):5887–5894
Coats ER, Mockos A, Loge FJ (2011) Post-anoxic denitrification driven by PHA and glycogen within enhanced biological phosphorus removal. Bioresour Technol 102(2):1019–1027
Ding L, Yokota A (2010) Curvibacter fontana sp. nov., a microaerobic bacteria isolated from well water. J Gen Appl Microbiol 56(3):267–271
Epstein S (2013) The phenomenon of microbial uncultivability. Curr Opin Microbiol 16(5):636–642
Fudala-Ksiazek S, Luczkiewicz A, Fitobor K, Olanczuk-Neyman K (2014) Nitrogen removal via the nitrite pathway during wastewater co-treatment with ammonia-rich landfill leachates in a sequencing batch reactor. Environ Sci Pollut Res 21(12):7307–7318
Fujitani H, Ushiki N, Tsuneda S, Aoi Y (2014) Isolation of sublineage I Nitrospira by a novel cultivation strategy. Environ Microbiol 16(10):3030–3040
Hett EC, Rubin EJ (2008) Bacterial growth and cell division: a mycobacterial perspective. Microbiol Mol Biol Rev 72(1):126–156
Hu Z, Chandran K, Grasso D, Smets BF (2002) Effect of nickel and cadmium speciation on nitrification inhibition. Environ Sci Technol 36(14):3074–3078
Jørgensen KS, Pauli AS-L (1995) Polyphosphate accumulation among denitrifying bacteria in activated sludge. Anaerobe 1(3):161–168
Juretschko S, Loy A, Lehner A, Wagner M (2002) The microbial community composition of a nitrifying-denitrifying activated sludge from an industrial sewage treatment plant analyzed by the full-cycle rRNA approach. Syst Appl Microbiol 25(1):84–99
Kana BD, Mizrahi V (2010) Resuscitation‐promoting factors as lytic enzymes for bacterial growth and signaling. FEMS Immunol Med Microbiol 58(1):39–50
Kaprelyants A, Kell D (1992) Rapid assessment of bacterial viability and vitality by rhodamine 123 and flow cytometry. J Appl Microbiol 72(5):410–422
Kaprelyants AS, Kell DB (1993) Dormancy in stationary-phase cultures of Micrococcus luteus: flow cytometric analysis of starvation and resuscitation. Appl Environ Microbiol 59(10):3187–3196
Kong YH, Nielsen JL, Nielsen PH (2005) Identity and ecophysiology of uncultured actinobacterial polyphosphate-accumulating organisms in full-scale enhanced biological phosphorus removal plants. Appl Environ Microbiol 71(7):4076–4085
Limpiyakorn T, Shinohara Y, Kurisu F, Yagi O (2005) Communities of ammonia-oxidizing bacteria in activated sludge of various sewage treatment plants in Tokyo. FEMS Microbiol Ecol 54(2):205–217
Monti A, Hall ER, van Loosdrecht MM (2007) Kinetics of phosphorus release and uptake in a membrane-assisted biological phosphorus removal process. J Environ Eng 133(9):899–908
Mukamolova GV, Kaprelyants AS, Young DI, Young M, Kell DB (1998) A bacterial cytokine. Proc Natl Acad Sci 95(15):8916–8921
Mukamolova GV, Murzin AG, Salina EG, Demina GR, Kell DB, Kaprelyants AS, Young M (2006) Muralytic activity of Micrococcus luteus Rpf and its relationship to physiological activity in promoting bacterial growth and resuscitation. Mol Microbiol 59(1):84–98
Nielsen PH, Mielczarek AT, Kragelund C, Nielsen JL, Saunders AM, Kong Y, Hansen AA, Vollertsen J (2010) A conceptual ecosystem model of microbial communities in enhanced biological phosphorus removal plants. Water Res 44(17):5070–5088
Nikitushkin V, Demina G, Kaprelyants A (2011) Effect of secreted Rpf protein on intracellular contacts in Micrococcus luteus and Mycobacterium smegmatis cultures. Microbiology 80(2):143–149
Reissbrodt R, Rienaecker I, Romanova J, Freestone P, Haigh R, Lyte M, Tschäpe H, Williams P (2002) Resuscitation of Salmonella enterica serovar Typhimurium and enterohemorrhagic Escherichia coli from the viable but nonculturable state by heat-stable enterobacterial autoinducer. Appl Environ Microbiol 68(10):4788–4794
Senoh M, Ghosh‐Banerjee J, Ramamurthy T, Colwell RR, Miyoshi SI, Nair GB, Takeda Y (2012) Conversion of viable but nonculturable enteric bacteria to culturable by co‐culture with eukaryotic cells. Microbiol Immunol 56(5):342–345
Smolders G, Van der Meij J, Van Loosdrecht M, Heijnen J (1994) Stoichiometric model of the aerobic metabolism of the biological phosphorus removal process. Biotechnol Bioeng 44(7):837–848
Su X, Chen X, Hu J, Shen C, Ding L (2013a) Exploring the potential environmental functions of viable but non-culturable bacteria. World J Microbiol Biotechnol 29(12):2213–2218
Su X, Shen H, Yao X, Ding L, Yu C, Shen C (2013b) A novel approach to stimulate the biphenyl-degrading potential of bacterial community from PCBs-contaminated soil of e-waste recycling sites. Bioresour Technol 146:27–34
Su X, Zhang Q, Hu J, Hashmi M Z, Ding L, Shen C (2015). Enhanced degradation of biphenyl from PCB-contaminated sediments: the impact of extracellular organic matter from Micrococcus luteus. Appl Microbiol Biotechnol 99(4):1989–2000
Van Loosdrecht M, Hooijmans C, Brdjanovic D, Heijnen J (1997) Biological phosphate removal processes. Appl Microbiol Biotechnol 48(3):289–296
Vocks M, Adam C, Lesjean B, Gnirss R, Kraume M (2005) Enhanced post-denitrification without addition of an external carbon source in membrane bioreactors. Water Res 39(14):3360–3368
Wagner M, Loy A (2002) Bacterial community composition and function in sewage treatment systems. Curr Opin Biotechnol 13(3):218–227
Wagner M, Erhart R, Manz W, Amann R, Lemmer H, Wedi D, Schleifer K (1994) Development of an rRNA-targeted oligonucleotide probe specific for the genus Acinetobacter and its application for in situ monitoring in activated sludge. Appl Environ Microbiol 60(3):792–800
Wagner M, Rath G, Amann R, Koops H-P, Schleifer K-H (1995) In situ identification of ammonia-oxidizing bacteria. Syst Appl Microbiol 18(2):251–264
Wan CL, Yang X, Lee DJ, Sun SP, Liu X, Zhang P (2014) Influence of hydraulic retention time on partial nitrification of continuous-flow aerobic granular-sludge reactor. Environ Technol 35(14):1760–1765
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73(16):5261–5267
Wang ZC, Gao MC, She ZL, Jin CJ, Zhao YG, Yang SY, Guo L, Wang S (2015) Effects of hexavalent chromium on performance and microbial community of an aerobic granular sequencing batch reactor. Environ Sci Pollut Res 22(6):4575–4586
Winkler M, Coats ER, Brinkman CK (2011) Advancing post-anoxic denitrification for biological nutrient removal. Water Res 45(18):6119–6130
Wyckoff TJ, Taylor JA, Salama NR (2012) Beyond growth: novel functions for bacterial cell wall hydrolases. Trends Microbiol 20(11):540–547
Zhang Y, Zhao X (2013) The effects of powdered activated carbon or ferric chloride on sludge characteristics and microorganisms in a membrane bioreactor. Desalination and Water Treatment (ahead-of-print) 1–10
Acknowledgments
We gratefully acknowledge the financial support provided by the Fundamental Research Funds for the Central Universities (2014QNA6012).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Gerald Thouand
Rights and permissions
About this article
Cite this article
Liu, Y., Su, X., Lu, L. et al. A novel approach to enhance biological nutrient removal using a culture supernatant from Micrococcus luteus containing resuscitation-promoting factor (Rpf) in SBR process. Environ Sci Pollut Res 23, 4498–4508 (2016). https://doi.org/10.1007/s11356-015-5603-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-015-5603-3