Abstract
Histology and immunohistochemistry of thin tissue sections have been the standard diagnostic procedure in many diseases for decades. This method is highly specific for particular tissue regions or cells, but mechanical sectioning of the specimens is required, which destroys the sample in the process and can lead to non-uniform tissue deformations. In addition, regions of interest cannot be located beforehand and the analysis is intrinsically two-dimensional. Micro X-ray computed tomography (μCT) on the other hand can provide 3D images at high resolution and allows for quantification of tissue structures, as well as the localization of small regions of interest. These advantages advocate the use of μCT for virtual histology tool with or without subsequent classical histology. This review summarizes the most recent examples of virtual histology and provides currently known possibilities of improving contrast and resolution of μCT. Following a background in μCT imaging, ex vivo staining procedures for contrast enhancement are presented as well as label-free virtual histology approaches and the technologies, which could rapidly advance it, such as phase-contrast CT. Novel approaches such as zoom tomography and nanoparticulate contrast agents will also be considered. The current evidence suggests that virtual histology may present a valuable addition to the workflow of histological analysis, potentially reducing the workload in pathology, refining tissue classification, and supporting the detection of small malignancies.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Background
Histological analysis continues to be one of the most common techniques in pathological assessment of tissue samples at cellular level in both preclinical research and clinical routine. In combination with the many existing specific staining protocols or by applying immunohistochemistry techniques, this method can provide high specificity for instance in localizing tumor cells. However, mechanical sectioning of specimens is required, which destroys the sample in the process and can lead to non-uniform deformations. In addition, the analysis cannot be targeted to sites of interest, because the sectioning is performed without any knowledge of their location. To compensate for this, serial sectioning is often applied, which is very labor intensive. Moreover, analyzing three dimensional (3D) features such as volumes of structures as for example metastases, 3D textures, and fiber orientations is limited by the 2D nature of histology and by the deformation which occurs during the mechanical sectioning process.
Therefore, virtual sectioning of 3D data sets at cellular level would be a great tool to complement existing histological techniques. Several so-called virtual histology (VH) approaches have recently emerged, each with their own limitations:
(I) Confocal microscopy allows focused imaging on a single plane within tissue sections of up to about 4 mm3 [1] and can provide 3D representations of these specimens. (II) Light sheet microscopy uses tissue samples which are optically cleared, focussing the light into deeper locations; however, only fluorescence dyes can be imaged [2]. (III) Micro magnetic resonance imaging (μMRI) can reach resolutions in the range of ~ 25 μm and allows a depiction of larger samples, due to its high soft-tissue contrast and its 3D nature [3]. However, this comes at the expense of acquisition time (~ hours to days) and is therefore not feasible on a large scale and not very cost-effective. Moreover, MRI is only possible with hydrated tissue, excluding the use of routine paraffin-embedded histological samples.
X-ray-Based Virtual Histology
In contrast to the imaging techniques mentioned above, X-ray imaging can supply high-resolution data sets of larger volumes. Classical X-ray imaging is based on the absorption of X-rays by the sample. In comparison with classical microscopy, X-rays can penetrate larger samples, due to their high energies. Especially using synchrotron light sources, an incident X-ray beam with an extremely high flux in the range of 1010 photons/s/mm2 can be generated, which is about 106 times brighter than a normal X-ray tube [4]. This ensures a high signal-to-noise ratio (SNR) and allows extremely high spatial resolutions below 1 μm. Moreover, by using a tomography approach, 3D images can be generated—called computed tomography (CT). μCT thus provides ideal prerequisites for X-ray-based virtual histology (xVH).
Hard tissue analysis has always been a domain of high-resolution X-ray imaging ranging from planar imaging of bone sections [5] to μCT evaluation of the 3D structure of bone tissue [6]. Due to the strong difference between the X-ray attenuation of bone and soft-tissue or void spaces, bone is especially suited for this type of analysis. In combination with a phantom containing different known concentrations of hydroxyapatite—a main component of bone—the X-ray attenuation values, which represent electron density, can be translated to bone mineral density (BMD) [7]. In addition, due to the 3D nature of μCT, typical morphometric parameters of bone such as trabecular thickness (Tb.Th), trabecular separation (Tb.S), and the ratio of bone to tissue volume (BV/TV) can reliably be assessed [8]. Due to the extremely high resolution of xVH not only the trabecular structure, but also the canaliculi network [9], the osteon-structure [10] and finally even the single mineralized collagen fibers can be imaged [11]. Along with bone, xVH is used regularly in dental studies, especially the formation of root canals is of interest [12]. In contrast to xVH, the mechanical sectioning of teeth and bones for classical histology is very labor intensive. Moreover, a reliable analysis of a 3D structure such as the root canal cannot easily be performed, because a major part of the sample material is lost during the sectioning process. xVH is therefore a clear improvement compared to classical methods and is already used routinely for these applications.
Similar to the density differences between bone and soft tissue, the air soft-tissue differences in the lung provide strong intrinsic contrast for imaging this organ with CT. In addition, these interfaces generate strong scattering and phase shifts, which makes lung imaging especially suited for the use of phase-contrast or darkfield imaging techniques [13, 14]. μCT of excised lungs, when dose is not a concern, can offer 3D insight with even higher resolution. xVH of the lung thereby reaches a similarly high resolution as microscopic approaches on tissue sections, but with the additional advantage that it allows for analysis of 3D-imaging-derived biomarkers [15]. Because xVH can cover parts of or even the entire lung, it provides regional information, and sampling errors as with classical 2D histology are avoided. Because of the power of (micro-) CT-derived readouts, we foresee an enormous impact of μCT as an efficient technology for routine use in lung disease research and diagnostics because of the significant added value it creates to classical histological approaches.
In contrast to hard-tissue samples, which have an intrinsically high absorption of X-rays, soft-tissue samples are low absorbing, but can still be investigated by xVH. A simple method to accomplish sufficient X-ray absorption, the old technique of corrosion casting can be used. This method utilizes a contrast enhancing liquid to fill void spaces such as the airways or vessels. After the solution has solidified, the surrounding biological tissue is dissolved, leaving a negative replica of the bronchi or vessel tree. Since the replica is now surrounded by air, it has a high contrast for CT imaging, which can be further improved if the liquid is mixed with an X-ray contrast agent containing, for example, iodine or barium [16]. xVH then provides a 3D data set, which can be quantified, similarly to hard-tissue applications. To successfully perform corrosion casting, the pressure and viscosity of the filling material must be carefully controlled; otherwise, it will either not reach small capillaries or they will rupture. In addition, because the biological tissue is destroyed within the process, no additional histological analysis can be performed. Figure 1 shows a μCT dataset of a mouse kidney after applying corrosion casting. The vessel tree can be depicted down to its finest structures. The technique of vascular filling has been optimized to allow imaging within the surrounding soft tissue, preserving the organ architecture with the additional advantage that this technique is compatible with subsequent standard histological sectioning and staining for 2D analysis [18, 19].
a Reconstructed scan of an entire mouse kidney corrosion cast. A finely textured layer of capillaries was seen lining the entire cortical surface of the kidney. These were densely packed and probably lie just beneath the connective tissue capsule of the kidney. Res. 7.5 μm voxel pitch. b, c Gradual elimination of the smaller vessels from the model by thresholding revealed the larger vessels that comprised the vascular tree of the entire kidney. Res. 7.5 μm voxel pitch. Figure reproduced from Wagner et al. [17].
To directly apply xVH for soft-tissue analysis, the X-ray attenuation of these tissues needs to be increased. For this purpose, ex vivo staining procedures using solutions that contain heavy ions such as tungsten, iodine, silver, or barium have been developed [20]. The staining is mainly related to differences in the diffusion speed between different tissue types and can therefore be applied to various types of samples and applications. In recent years, this approach has been used in several experiments ranging from phenotyping of mouse embryos [21] to the analysis of specific organs like the heart [22] or kidneys [23]. In addition, staining protocols developed for electron microscopy usually based on osmium also work well for xVH and allow for a combination of those two methods [24]. The development of new sample preparation procedures is an active area of research that might greatly improve soft tissue contrast in the future [14].
Although many studies describe a successful combination of xVH and heavy ion staining protocols, a main disadvantage of this approach is the low diffusion speed of heavy ions, which in some applications results in long staining times of several days and which effectively limits this technique to smaller specimens. In addition, the diffusion process of such staining protocols creates artificial contrast gradients at tissue interfaces, which limit the spatial resolution of this approach as discussed by Saccomano et al. [25]. Thus, label-free xVH of soft-tissue samples would be of significant benefit. All prior discussed applications have in common that the image content is based on differences in X-ray attenuation between adjacent tissues or structures. This attenuation of the incident X-ray beam is predominately related to absorption of energy by the sample. However, this is not the only interaction the X-ray waves undergo with the specimen. Also, phase shifts and scattering occur, which can be exploited to raise the contrast of low absorbing structures similar to techniques in classical microscopy [26]. Since phase shifts do not alter the intensity of the wave, they can only be measured indirectly, but the interference effects caused by the phase shift can be recorded. However, this requires a stable correlation between the waves within the X-ray beam, the so-called coherence criterium. Common X-ray sources like normal light bulbs do not generate coherent light, which explains why these phase effects have not been exploited in conventional X-ray imaging so far. Alternative X-ray sources exist, which achieve a sufficient degree of spatial coherence in the X-ray beam and thus allow the use of phase-sensitive imaging techniques. These include synchrotron light sources, micro or nano-focus X-ray tubes commonly employed in high-resolution μCTs or a combination with specific optical elements such as gratings. In recent years, it has been demonstrated that the application of X-ray phase-sensitive techniques allows for a strong increase in soft-tissue applications especially at high spatial resolutions [13, 27], thereby lowering the applied X-ray dose. Thus, label-free xVH at dedicated μCT setups becomes feasible. Setups available at synchrotron light sources are particularly efficient, because of the high brilliance of their source and their intense X-ray fluxes.
In contrast to classical histology, where specific stainings can be multiplexed by using different color labels, X-ray detectors can only measure intensities, thereby generating greyscale pictures. This drawback in xVH may be solved in the future by applying a novel generation of energy discriminating X-ray detectors [28]. These detectors permit the definition of several energy bands for detection that could be tailored to the specific absorption edges in metals used for contrast agents such as iodine, barium, gold etc. This would allow multiplexing contrast agents within a single specimen and could thus increase the sensitivity for the detection of these elements, similar to k-edge subtraction imaging performed with monochromatic X-rays [29], but without the need of multiple acquisitions at different energies.
Application of xVH
Assessment of 3D Features
A main advantage of xVH is the possibility to quantify 3D features of tissue. In many applications, alterations in the 3D structure of tissues is of crucial interest for monitoring disease progression or therapy response. Due to the intrinsic high contrast of bone, the analysis of bone structure by xVH is well established. In addition, the structural assessment of the bone in osteological diseases in rat or mice models requires higher resolution than in human applications, due to their small size. xVH is the only 3D imaging method that can provide this necessary spatial resolution for bones and thus enables the investigation of sub- or even ultrastructures. Resolutions in micron or submicron range can be especially reached, if synchrotron light sources are utilized. The combination of a high resolution with 3D image processing software facilitates pore analysis to quantify the structural changes in mouse bones, as demonstrated by Tavella et al. [6] (Fig. 2). They used this approach to discriminate between mice kept in a deep-sea vivarium, at sea level, as well as in the international space station (ISS) under micro gravity conditions. The trabecular structure of the femura, as well as the lumbar spine, was assessed by xVH and quantified applying pore analysis (Pore3D, [30]). Not only did they find a structural difference between the groups housed at different altitudes, but also variations in the efficacy of different treatments (Fig. 2). New applications of 3D xVH are also emerging in the field of neurology, mostly using phase-contrast approaches. They include the assessment of neurodegenerative diseases, both in human autopsy samples [31] and in mice models [32,33,34,35], as well as the study of human [36, 37] and mouse [38, 39] neuroanatomy, whole rodent brain vasculature [40, 41], uncut mouse nerves [42], and unlabelled brain tumors [43,44,45]. An interesting approach for simultaneously analyzing adipocyte distribution and bone morphology was presented by Kerckhofs et al. [46]. They used hafnium-POM (polyoxometalate) for staining of soft tissue, thereby generating a strongly negative contrast for the adipocytes and allowing for their visualization even in the presence of bone.
Bone trabecular thickness distribution. Wt2 (a), PTN-Tg1 (b), and PTN-Tg2 (c) color map of bone trabecular thickness distribution in the femur of a representative vivarium, the ground and the flight mice. Wt2 (d), PTN-Tg1 (e), and PTN-Tg2 (f) color map of bone trabecular thickness distribution in the seventh lumbar vertebra of representative vivarium, the ground and the flight mice. Figure reproduced from Tavella et al. [6].
Computer-Aided Diagnosis
The possibility to measure 3D features will improve the pathological classification of tissues, which could lead to the design of a computer-aided diagnosis (CAD) systems. CAD systems have already been successfully introduced for mammographic breast cancer screening [47]. The advantage of CAD systems, which are usually based on artificial neuronal networks, is the possibility of training them using annotated data sets without an extensive knowledge of specific features. In case the training process is successful, the CAD system generalizes the task and can be applied to data that have not been part of the training process. Given the recent progress in computer science and hardware development, very complex artificial networks can be realized and trained with a tremendous amount of data. CAD has already outperformed radiologists in mammography screening [48]. It can therefore be expected that CAD will also be an extremely valuable tool in pathology, by identifying regions of interest and providing a second opinion for a medical indication [49]. However, the success of CAD is based on the information contained in the feature space; thus, xVH will play an important role in this regard.
Guided Sectioning
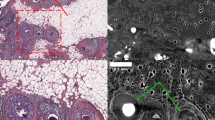
As outlined above, one of the main limitations in classical histology is the need for mechanical sectioning. To facilitate the cutting process, tissue samples need to be dehydrated and embedded (or frozen, for cryosectioning), making it difficult to target sectioning to a specific position. xVH allows scanning the prepared histology samples and identifying regions of interest in relation to the surface of the embedding material. This information can be used to guide the sectioning process, making labour intensive serial sectioning of the whole block unnecessary [50,51,52,53]. Soft-tissue histology usually requires an additional staining step with a solution containing heavy ions to provide a sufficient contrast for xVH. Saccomano et al. demonstrate the application of three different staining protocols for μCT imaging of the lung and heart tissue based on iodine, tungsten, and chromium [25]. Such an approach can be integrated into the standard workflow of histological analysis and does not interfere with subsequent histological analysis as shown by Dullin et al. They performed classical histology, as well as immunohistochemistry, on mouse heart tissues which were stained with phosphotungstic acid (PTA) beforehand [22]. However, due to the slow penetration of the staining solutions, pre-processing time of larger tissue samples will increase significantly. Martin et al. report staining periods of about 6 days for mouse brain, and Dullin et al. about 12 days for mouse heart tissue [22, 54]. Figure 3 shows a PTA-stained mouse lung in comparison with histological sections. The sample was embedded in resin and cut with a laser microtome to avoid sectioning artifacts. Figure 3(C) shows a near perfect overlay of the histology and μCT data.
Comparison and overlay of the μCT scan and histological image of a resin-embedded and laser cut mouse lung sample: (A) Shows an exemplary slice of the reconstructed μCT data set of a resin-embedded PTA-stained mouse lung. In addition to alveolar lung tissue, a lung metastasis (m), the esophagus (o), larger bronchi (b), and blood vessels (v) are displayed. (B) Shows the microscope image of the Sanderson Rapid Stain (SRS)-stained and van Gieson (VG)-counterstained cut performed with the laser microtome at the predefined position. No cutting artifacts are visible. (C) Depicts the overlay of both data sets. The histological data is shown in pseudo SRS/VG colors (the data set was colorized for better vizualisation), while the CT data is shown in green. The overlay of the CT data with the histology image reveals a precise match of both data sets. Figure adapted from Albers et al. [55].
Scott et al. demonstrated that unstained formalin-fixed and paraffin-embedded human lung tissue samples can successfully be imaged by μCT [51]. Such a label-free xVH approach would be of great interest as it could be applied on conserved tissue and would not interfere with subsequent histological analysis. However, the scans that were presented in this publication took 12 h. Therefore, a routine application of this technique seems not feasible for now. Label-free xVH using phase-contrast imaging techniques enables the examination of soft-tissue samples without an additional staining and potentially more rapidly than absorption-based approaches [39]. Phase-contrast label-free xVH could therefore be integrated into the workflow of clinical pathology. Figure 4 shows μCT images acquired with a laboratory-based setup. The images were pseudo-colored to resemble histological slices. Single cell nuclei can be visualized. Since a certain degree of spatial coherence of the incident X-ray beam, or the use of more complex systems like grating-interferometers are needed, label-free xVH was so far limited to synchrotron applications. Recent studies by Bidola et al. [56] show that commercially available X-ray microscopes can provide sufficient coherence and allow performing label-free xVH outside of synchrotron, grating-based or edge illumination setups. These systems, however, are quite costly and require long scanning times. Since the combination of xVH and guiding of the sectioning process sounds so intriguing, the development of combined μCT-microtome systems seems to be the next logical step.
Volume rendering down to the cellular level of a mouse cerebellar vermis. a Transverse slice through the reconstructed volume, showing the molecular layer (ML), granular layer (GL), white matter (WM), and Purkinje cell layer (PCL) of the cerebellar vermis at cellular resolution. Prior to the tomographic reconstruction, the individual projections were resampled by a factor 2. b A longitudinal slice through the volume shows sufficient contrast to identify axon bundles within the white matter. c Automatic volume rendering of the sample with a cut within the volume, indicating the position of the cellular segmentation shown in (d). d Cellular segmentation of a small part of the sample. Scalebars 200 μm. Figure reproduced from Töpperwien et al. [39].
Specific Staining Nanoparticles
The multiple advantages of xVH discussed so far come with one major flaw: there are no CT staining or contrasting techniques, which can specifically highlight certain cell types or tissues. In contrast, immunohistochemistry uses labeled antibodies, antibody fragments, or other targeting compounds and can therefore provide the detection of tumor cells etc. This weakness of xVH might be compensated in the future by applying nanoparticles with a metal core made for example of gold. Gold nanoparticles have already been successfully applied for preclinical X-ray imaging and are commercially available [57]. Their surface can also be modified and labeled with antibodies. Such conjugates have successfully been used in electron microscopy (EM) [58]. Since xVH is less sensitive than EM, these compounds need to be either larger or more concentrated at the site of interest. Both would significantly reduce tissue penetration, which would limit the analysis to tissue slices. Therefore, only a few studies using antibodies in combination with μCT have been reported. Metscher et al. showed an interesting general approach using enzyme-mediated silver deposition in combination with antibodies and demonstrated successful μCT imaging of entire chick embryos [59]. Nevertheless, in applications in which nanoparticles are actively transported within a living organism by for instance macrophages [60,61,62] or tumor cells [63, 64] or penetrate tumor tissue due to the EPR effect [65, 66], subsequent xVH can be used to track these particles on a cellular or even subcellular level in much larger samples compared to EM. Moreover, xVH might be an interesting tool to study the biodistribution of nanoparticles in studies of employment protection or environmental hazard [67].
Zoom in X-ray Imaging Approach
Due to the high energy of X-ray photons, large biological specimens can easily be penetrated and scanned. In contrast to this, high-resolution imaging can usually only be applied on small fields of view (FOV) for two reasons. In classical cone-beam μCT systems, this is mainly because the increased resolution is generated by geometric magnification, requiring the sample to be positioned close to the source, which in turn prevents scanning larger specimens. Secondly, for more practical reasons, because a large object scanned at very high resolution generates an extremely memory consuming data set that could not be easily processed. The 3D reconstruction of the acquired data, especially when using the classical filtered back projection algorithm, shows artifacts if the scanned object exceeds the FOV. For this reason, local area reconstruction algorithms were developed that suppress such artifacts and produce reliable outcomes at least in the center of the reconstructed data [68]. These local area scans enable “zoom tomography”, which consists of a low-resolution scan with a large FOV or of the entire object that facilitates the subsequent positioning of a high-resolution scan. This was demonstrated by Krenkel et al. in imaging of in situ mouse lungs [60] (Fig. 5). Figure 5c shows a fusion of a high-resolution local area scan with a low-resolution overview scan. Thus, xVH in combination with such a zoom approach is one of very few techniques that can achieve cellular resolution within relatively large objects without the need of mechanical sectioning.
a Orthoslice perpendicular to the rotation axis of the “large FOV” tomographic reconstruction. The inset shows a zoom into the lung area marked by the dashed rectangle with adjusted contrast. b 3D rendering of the “large FOV” volume showing the thorax, containing automatically segmented bones (gray), the heart (red), and lung tissue (pink). The dashed area denotes the region shown in the inset in (a). c 3D rendering of the “zoom-move” dataset (see Fig. 2c, d), obtained by tomographic reconstruction of the phase-retrieved projections. Three orthoslices together with a volume rendering of the soft-tissue structure are shown. Scale bars denote 2 mm in (a, b) and 500 μm in (c). Figure reproduced from Krenkel et al. [60].
Rapid 3D Assessment
The extremely high flux of X-rays that can be produced at a synchrotron light source (by a factor of 1 × 106 higher than in a standard X-ray tube) allows for high-resolution imaging with a sufficient contrast-to-noise ratio at extremely short exposure times. If modern X-ray detectors with high frame rates are applied and automated sample positioning systems are used, high-throughput 3D imaging can be realized. Van de Kamp et al. demonstrated that wasps embedded in amber could be scanned at the ANKA synchrotron light source with a resolution below 4 μm in 100 s per tomogram [69]. Recent publications show that by utilizing free propagation phase-contrast imaging (PBI), a more than 1000 fold increase in contrast-to-noise ratio (CNR) can be achieved [13]. This gain in CNR can be traded for a dramatic reduction of detector exposure time and would further speed-up the imaging process. Currently, the speed and precision of the mechanical components, i.e., mainly the rotational stage and the bandwidth of the data transfer, are the limiting factor for rapid 3D imaging, not the performance of the detector. Dullin et al. demonstrated that a local area scan of an in situ mouse lung at a resolution of 25 μm can be achieved in less than 10 s and with a dose below 70 mGy by utilizing the novel detector MÖNCH [70]. Given the vast technical developments, it can be expected that high-resolution xVH can soon be performed within larger specimens with a comparable radiation dose and at short acquisition times. This would support the further development of this approach into a virtual biopsy technique for instance in the lungs.
Virtopsy
Using CT for the 3D assessment of pathological tissue is an approach with a long and successful history. To provide 3D data sets of corpses prior to the invasive approach of autopsy, CT in combination with the infusion of ex vivo contrast agents has been introduced. This so-called virtopsy approach has been successfully used particularly in forensic medicine [71]. It results in a digitalization of the entire body that allows for a subsequent and repeatable analysis independent of the operating pathologist. The application of CT on these large specimens prevents the use of high-resolution imaging, but to further aid this approach, xVH of excised tissue specimen may be used to provide additional information on a different scale.
Conclusion
xVH is an emerging tool, which facilitates the 3D assessment of hard-tissue, as well as in soft-tissue samples, when combined with staining procedures. If novel imaging approaches, such as phase-contrast techniques, can be exploited, xVH could even be performed label free on soft-tissue samples. xVH has a great potential to supplement classical histology and immunohistochemistry by the 3D nature of its data. It can be used for guidance of sectioning techniques and can be integrated into the standard workflow of pathological assessments of tissue samples. It has therefore the potential to reduce the workload in pathology and refine tissue classification. Since it allows for a 3D digitalization of the sample, it could also be used to ensure that the original shape and structure of a specimen is secured before sectioning. Given its rapid and extensive technological development xVH will certainly play an important role in biomedical research and clinical pathology in the future.
References
Sands GB, Gerneke DA, Hooks DA, Green CR, Smaill BH, Legrice IJ (2005) Automated imaging of extended tissue volumes using confocal microscopy. Microsc Res Tech 67:227–239
Silvestri L, Bria A, Sacconi L, Iannello G, Pavone FS (2012) Confocal light sheet microscopy: micron-scale neuroanatomy of the entire mouse brain. Opt Express 20:20582–20598
Pieles G, Geyer SH, Szumska D, Schneider J, Neubauer S, Clarke K, Dorfmeister K, Franklyn A, Brown SD, Bhattacharya S, Weninger WJ (2007) μMRI–HREM pipeline for high-throughput, high-resolution phenotyping of murine embryos. J Anat 211:132–137
Winick H (1987) Synchrotron radiation. Sci Am 257:88–101
Powles-Glover N, De Schaepdrijver L, French J, Stewart J (2014) Comparison of Faxitron™ versus MicroCT imaging of the skeleton of the suckling rat. Reprod Toxicol 48:44–50
Tavella S, Ruggiu A, Giuliani A, Brun F, Canciani B, Manescu A, Marozzi K, Cilli M, Costa D, Liu Y, Piccardi F, Tasso R, Tromba G, Rustichelli F, Cancedda R (2012) Bone turnover in wild type and pleiotrophin-transgenic mice housed for three months in the International Space Station (ISS). PLoS One 7:e33179
Minematsu A, Nishii Y, Imagita H, Takeshita D, Sakata S (2016) Whole-body vibration can attenuate the deterioration of bone mass and trabecular bone microstructure in rats with spinal cord injury. Spinal Cord 54:597–603
Chappard D, Retailleau-Gaborit N, Legrand E, Baslé MF, Audran M (2005) Comparison insight bone measurements by histomorphometry and μCT. J Bone Miner Res 20:1177–1184
Schneider P, Meier M, Wepf R, Müller R (2010) Towards quantitative 3D imaging of the osteocyte lacuno-canalicular network. Bone 47:848–858
Cooper DML, Erickson B, Peele AG, Hannah K, Thomas CDL, Clement JG (2011) Visualization of 3D osteon morphology by synchrotron radiation micro-CT. J Anat 219:481–489
Schneider P, Stauber M, Voide R, Stampanoni M, Donahue LR, Müller R (2007) Ultrastructural properties in cortical bone vary greatly in two inbred strains of mice as assessed by synchrotron light based micro- and nano-CT. J Bone Miner Res 22:1557–1570
Wolf TG, Paqué F, Woop A-C, Willershausen B, Briseño-Marroquín B (2017) Root canal morphology and configuration of 123 maxillary second molars by means of micro-CT. Int J Oral Sci 9:33–37
Kitchen MJ, Buckley GA, Gureyev TE, Wallace MJ, Andres-Thio N, Uesugi K, Yagi N, Hooper SB (2017) CT dose reduction factors in the thousands using X-ray phase contrast. Sci Rep 7:15953
Busse M, Müller M, Kimm MA, Ferstl S, Allner S, Achterhold K, Herzen J, Pfeiffer F (2018) Three-dimensional virtual histology enabled through cytoplasm-specific X-ray stain for microscopic and nanoscopic computed tomography. Proc Natl Acad Sci 115:2293–2298
Tanabe N, Vasilescu DM, Kirby M et al (2018) Analysis of airway pathology in COPD using a combination of computed tomography, micro-computed tomography and histology. Eur Respir J 1701245:51
Huang H, Chen J, Wang Y, Chen XY, Gong DC (2017) Study on multi-density contrast agent fillers of duct casting based on CT three-dimensional reconstruction. J Huazhong Univ Sci Technolog Med Sci 37:300–306
Wagner R, Van Loo D, Hossler F et al (2011) High-resolution imaging of kidney vascular corrosion casts with Nano-CT. Microsc Microanal 17:215–219
Russo FM, Toelen J, Eastwood MP, Jimenez J, Miyague AH, Vande Velde G, DeKoninck P, Himmelreich U, Vergani P, Allegaert K, Deprest J (2016) Transplacental sildenafil rescues lung abnormalities in the rabbit model of diaphragmatic hernia. Thorax 71:517–525. https://doi.org/10.1136/thoraxjnl-2015-207949
Ehling J, Babi kova J, Gremse F, Klinkhammer BM, Baetke S, Knuechel R, Kiessling F, Floege J, Lammers T, Boor P (2016) Quantitative micro-computed tomography imaging of vascular dysfunction in progressive kidney diseases. J Am Soc Nephrol 27:520–532
Metscher BD (2009) MicroCT for comparative morphology: simple staining methods allow high-contrast 3D imaging of diverse non-mineralized animal tissues. BMC Physiol 9:11
Metscher BD (2009) MicroCT for developmental biology: a versatile tool for high-contrast 3D imaging at histological resolutions. Dev Dyn 238:632–640
Dullin C, Ufartes R, Larsson E, Martin S, Lazzarini M, Tromba G, Missbach-Guentner J, Pinkert-Leetsch D, Katschinski DM, Alves F (2017) μCT of ex-vivo stained mouse hearts and embryos enables a precise match between 3D virtual histology, classical histology and immunochemistry. PLoS One 12:e0170597
Missbach-Guentner J, Pinkert-Leetsch D, Dullin C, Ufartes R, Hornung D, Tampe B, Zeisberg M, Alves F (2018) 3D virtual histology of murine kidneys–high resolution visualization of pathological alterations by micro computed tomography. Sci Rep 8:1407
Morales AG, Stempinski ES, Xiao X et al (2016) Micro-CT scouting for transmission electron microscopy of human tissue specimens. J Microsc 263:113–117
Saccomano M, Albers J, Tromba G et al (2018) Synchrotron inline phase contrast μCT enables detailed virtual histology of embedded soft-tissue samples with and without staining. J Synchrotron Radiat https://doi.org/10.1107/S1600577518005489
Bhaduri B, Edwards C, Pham H, Zhou R, Nguyen TH, Goddard LL, Popescu G (2014) Diffraction phase microscopy: principles and applications in materials and life sciences. Adv Opt Photon 6:57–119
Paganin D, Mayo SC, Gureyev TE, Miller PR, Wilkins SW (2002) Simultaneous phase and amplitude extraction from a single defocused image of a homogeneous object. J Microsc 206:33–40
Shikhaliev PM, Fritz SG (2011) Photon counting spectral CT versus conventional CT: comparative evaluation for breast imaging application. Phys Med Biol 56:1905–1930
Porra L, Dégrugilliers L, Broche L, Albu G, Strengell S, Suhonen H, Fodor GH, Peták F, Suortti P, Habre W, Sovijärvi ARA, Bayat S (2018) Quantitative imaging of regional aerosol deposition, lung ventilation and morphology by synchrotron radiation CT. Sci Rep 8:3519
Brun F, Massimi L, Fratini M, Dreossi D, Billé F, Accardo A, Pugliese R, Cedola A (2017) SYRMEP Tomo Project: a graphical user interface for customizing CT reconstruction workflows. Adv Struct Chem Imaging 3:4
Koh S, Suh S, Lee D et al (2006) Phase contrast radiography of Lewy bodies in Parkinson disease. Neuroimage 32:566–569
Noda-Saita K, Yoneyama A, Shitaka Y, Hirai Y, Terai K, Wu J, Takeda T, Hyodo K, Osakabe N, Yamaguchi T, Okada M (2006) Quantitative analysis of amyloid plaques in a mouse model of Alzheimer’s disease by phase-contrast X-ray computed tomography. Neuroscience 138:1205–1213
Connor DM, Benveniste H, Dilmanian FA, Kritzer MF, Miller LM, Zhong Z (2009) Computed tomography of amyloid plaques in a mouse model of Alzheimer’s disease using diffraction enhanced imaging. Neuroimage 46:908–914
Pinzer B, Cacquevel M, Modregger P et al (2012) Imaging brain amyloid deposition using grating-based differential phase contrast tomography. Neuroimage 61:1336–1346
Astolfo A, Lathuiliere A, Laversenne V et al (2016) Amyloid-β plaque deposition measured using propagation-based X-ray phase contrast CT imaging. J Synchrotron Radiat 23:813–819
Schulz G, Waschkies C, Pfeiffer F, Zanette I, Weitkamp T, David C, Müller B (2012) Multimodal imaging of human cerebellum-merging X-ray phase microtomography, magnetic resonance microscopy and histology. Sci Rep 2:826
Schulz G, Weitkamp T, Zanette I, Pfeiffer F, Beckmann F, David C, Rutishauser S, Reznikova E, Muller B (2010) High-resolution tomographic imaging of a human cerebellum: comparison of absorption and grating-based phase contrast. J R Soc Interface 7:1665–1676. https://doi.org/10.1098/rsif.2010.0281
Dyer EL, Roncal WG, Prasad JA et al (2017) Quantifying mesoscale neuroanatomy using X-ray microtomography. eNeuro 4:ENEURO.0195–ENEU17.2017. https://doi.org/10.1523/ENEURO.0195-17.2017
Töpperwien M, Krenkel M, Vincenz D, Stöber F, Oelschlegel AM, Goldschmidt J, Salditt T (2017) Three-dimensional mouse brain cytoarchitecture revealed by laboratory-based x-ray phase-contrast tomography. Sci Rep 7:42847
Zhang M, Sun D, Xie Y et al (2014) Three-dimensional visualization of rat brain microvasculature following permanent focal ischaemia by synchrotron radiation. Br J Radiol 87:20130670
Miao P, Wu Z, Li M et al (2016) Synchrotron radiation X-ray phase-contrast tomography visualizes microvasculature changes in mice brains after ischemic injury. Neural Plast 2016:3258494
Bartels M, Krenkel M, Cloetens P, Möbius W, Salditt T (2015) Myelinated mouse nerves studied by X-ray phase contrast zoom tomography. J Struct Biol 192:561–568
Pfeiffer F, Bunk O, David C, Bech M, le Duc G, Bravin A, Cloetens P (2007) High-resolution brain tumor visualization using three-dimensional x-ray phase contrast tomography. Phys Med Biol 52:6923–6930
Prajapati SI, Kilcoyne A, Samano AK, Green DP, McCarthy SD, Blackman BA, Brady MM, Zarzabal LA, Tatiparthy AK, Sledz TJ, Duong T, Ohshima-Hosoyama S, Giles FJ, Michalek JE, Rubin BP, Keller C (2011) MicroCT-based virtual histology evaluation of preclinical medulloblastoma. Mol Imaging Biol 13:493–499
Lwin T-T, Yoneyama A, Hara A et al (2016) Spontaneous brain tumor imaging of aged rat by crystal X-ray interferometer-based phase-contrast X-ray CT. Acta Radiol Open 5:2058460115626958
Kerckhofs G, Stegen S, van Gastel N et al (2017) Simultaneous three-dimensional visualization of mineralized and soft skeletal tissues by a novel microCT contrast agent with polyoxometalate structure. Biomaterials
Kooi T, Litjens G, van Ginneken B, Gubern-Mérida A, Sánchez CI, Mann R, den Heeten A, Karssemeijer N (2017) Large scale deep learning for computer aided detection of mammographic lesions. Med Image Anal 35:303–312
Henriksen EL, Carlsen JF, Vejborg IM et al (2018) The efficacy of using computer-aided detection (CAD) for detection of breast cancer in mammography screening: a systematic review. Acta Radiol:028418511877091. https://doi.org/10.1177/0284185118770917
Bueno G, Fernández-Carrobles MM, Deniz O, García-Rojo M (2016) New trends of emerging technologies in digital pathology. Pathobiology 83:61–69
Sengle G, Tufa SF, Sakai LY, Zulliger MA, Keene DR (2013) A correlative method for imaging identical regions of samples by micro-CT, light microscopy, and electron microscopy: imaging adipose tissue in a model system. J Histochem Cytochem 61:263–271
Scott AE, Vasilescu DM, Seal KAD, Keyes SD, Mavrogordato MN, Hogg JC, Sinclair I, Warner JA, Hackett TL, Lackie PM (2015) Three dimensional imaging of paraffin embedded human lung tissue samples by micro-computed tomography. PLoS One 10:e0126230
Handschuh S, Baeumler N, Schwaha T, Ruthensteiner B (2013) A correlative approach for combining microCT, light and transmission electron microscopy in a single 3D scenario. Front Zool 10:44
Khimchenko A, Deyhle H, Schulz G, Schweighauser G, Hench J, Chicherova N, Bikis C, Hieber SE, Müller B (2016) Extending two-dimensional histology into the third dimension through conventional micro computed tomography. NeuroImage 139:26–36
Martin S, Lazzarini M, Dullin C, Balakrishnan S, Gomes FV, Ninkovic M, el Hady A, Pardo LA, Stühmer W, del-Bel E (2017) SK3 channel overexpression in mice causes hippocampal shrinkage associated with cognitive impairments. Mol Neurobiol 54:1078–1091
Albers J, Markus MA, Alves F, Dullin C (2018) X-ray based virtual histology allows guided sectioning of heavy ion stained murine lungs for histological analysis. Sci Rep 8:7712
Bidola P, Morgan K, Willner M et al (2017) Application of sensitive, high-resolution imaging at a commercial lab-based X-ray micro-CT system using propagation-based phase retrieval. J Microsc 266:211–220
Hainfeld J, Slatkin D, Focella T, Smilowitz H (2006) Gold nanoparticles: a new X-ray contrast agent. Br J Radiol 79:248–253
Daraee H, Eatemadi A, Abbasi E, Fekri Aval S, Kouhi M, Akbarzadeh A (2016) Application of gold nanoparticles in biomedical and drug delivery. Artif Cells Nanomedicine Biotechnol 44:410–422
Metscher BD, Müller GB (2011) MicroCT for molecular imaging: quantitative visualization of complete three-dimensional distributions of gene products in embryonic limbs. Dev Dyn 240:2301–2308
Krenkel M, Markus A, Bartels M, Dullin C, Alves F, Salditt T (2015) Phase-contrast zoom tomography reveals precise locations of macrophages in mouse lungs. Sci Rep 5:9973
Marinescu M, Langer M, Durand A, Olivier C, Chabrol A, Rositi H, Chauveau F, Cho TH, Nighoghossian N, Berthezène Y, Peyrin F, Wiart M (2013) Synchrotron radiation X-ray phase micro-computed tomography as a new method to detect iron oxide nanoparticles in the brain. Mol Imaging Biol 15:552–559
Rositi H, Frindel C, Langer M, Wiart M, Olivier C, Peyrin F, Rousseau D (2013) Information-based analysis of X-ray in-line phase tomography with application to the detection of iron oxide nanoparticles in the brain. Opt Express 21:27185–27196
Astolfo A, Arfelli F, Schültke E, James S, Mancini L, Menk RH (2013) A detailed study of gold-nanoparticle loaded cells using X-ray based techniques for cell-tracking applications with single-cell sensitivity. Nanoscale 5:3337–3345
Schültke E, Menk R, Pinzer B, Astolfo A, Stampanoni M, Arfelli F, Harsan LA, Nikkhah G (2014) Single-cell resolution in high-resolution synchrotron X-ray CT imaging with gold nanoparticles. J Synchrotron Radiat 21:242–250
Jain S, Hirst D, O’sullivan J (2012) Gold nanoparticles as novel agents for cancer therapy. Br J Radiol 85:101–113
Adam J-F, Nemoz C, Bravin A, Fiedler S, Bayat S, Monfraix S, Berruyer G, Charvet AM, Bas JFL, Elleaume H, Estève F (2005) High-resolution blood–brain barrier permeability and blood volume imaging using quantitative synchrotron radiation computed tomography: study on an F98 rat brain glioma. J Cereb Blood Flow Metab 25:145–153
Tang W, Huang S, Du L et al (2018) Expression of HMGB1 in maternal exposure to fine particulate air pollution induces lung injury in rat offspring assessed with micro-CT. Chem Biol Interact 280:64–69
Ritman EL, Dunsmuir JH, Faridani A et al (1997) Local reconstruction applied to x-ray microtomography. In: Inverse problems in wave propagation. Springer, pp 443–452
van de Kamp T, dos Santos Rolos T, Baumbach T, Krogmann L (2014) Scanning the past–synchrotron X-ray microtomography of fossil wasps in amber. Entomol Heute 26:150–160
Dullin C, Albers J, Tromba G, Andrä M, Ramilli M, Bergamaschi A (2018) MÖNCH detector enables fast and low-dose free-propagation phase-contrast computed tomography of in situ mouse lungs. J Synchrotron Radiat 25:565–569
Grabherr S, Stephan B, Buck U et al (2007) Virtopsy–radiology in forensic medicine. Imaging Decis MRI 11:2–9
Acknowledgements
This work was supported by the members of the X-ray Imaging Study Group within the European Society for Molecular Imaging.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Albers, J., Pacilé, S., Markus, M.A. et al. X-ray-Based 3D Virtual Histology—Adding the Next Dimension to Histological Analysis. Mol Imaging Biol 20, 732–741 (2018). https://doi.org/10.1007/s11307-018-1246-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11307-018-1246-3