Abstract
Introduction
It is challenging to establish the mechanisms involved in the variety of well-defined clinical phenotypes in autism spectrum disorder (ASD) and the pathways involved in their pathogeneses.
Objectives
The aim of the present study was to evaluate the metabolomic profiles of children with ASD subclassified by mental regression (AR) phenotype and with no regression (ANR).
Methods
The present study was a cross-sectional case–control study. Thirty children aged 2–6 years with ASD were included: 15 with ANR and 15 with AR. In addition, a control group of 30 normally developing children was selected and matched to the ASD group by sex and age. Plasma samples were analyzed with a metabolomics single platform methodology based on liquid chromatography-mass spectrometry. Univariate and multivariate analysis, including orthogonal partial least squares-discriminant analysis modeling and Shared-and-Unique-Structures plots, were performed using MetaboAnalyst 4.0 and SIMCA-P 15. The primary endpoint was the metabolic signature profiling among healthy children and autistic children and their subgroups.
Results
Metabolomic profiles of 30 healthy children, 15 ANR and 15 AR were compared. Several differences between healthy children and children with ASD were detected, involving mainly amino acid, lipid and nicotinamide metabolism. Furthermore, we report subtle differences between the ANR and AR groups.
Conclusions
In this study, we report, for the first time, the plasmatic metabolomic profiles of children with ASD, including two different phenotypes based on mental regression status. The use of a liquid chromatography-mass spectrometry platform approach for metabolomics in ASD children using plasma appears to be very efficient and adds further support to previous findings in urine. Furthermore, the present study documents several changes related to amino acid, NAD+ and lipid metabolism that, in some cases, such as arginine and glutamate pathway alterations, seem to be associated with the AR phenotype. Further targeted analyses are needed in a larger cohort to validate the results presented herein.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Background
Autism spectrum disorder (ASD) is a neurodevelopmental disorder characterized by persistent deficits in social communication and social interaction across multiple contexts with repetitive and restricted patterns of behaviors, activities, and interests (American Psychiatric Publishing 2013). There is a large heterogeneous spectrum of features and a wide range of medical comorbidities. The American Psychiatric Association’s Diagnostic and Statistical Manual, Fifth Edition (DSM-5), established the criteria for ASD covering two domains, i.e., social communication difficulties and repetitive and restricted behaviors, and includes abnormal sensory responses as a cardinal symptom (Howes et al. 2018). Symptoms must be present in early development of ASD, but the age of onset and its evolution, i.e., with or without mental regression, varies considerably. There are phenotypes in which some children undergo normal neurodevelopment and then regress in their developmental milestones. Metabolic alterations can underlie several comorbidities associated with ASD, such as seizures and sleep disorders and could contribute to autism severity (Altieri et al. 2011; Gabriele et al. 2014; Gevi et al. 2016).
The diagnosis of ASD is based on reported and observed behavior in keeping with diagnostic classification systems supported by test to exclude other explanations of the behaviors. Therefore, it is difficult to establish the variety of well-defined phenotypes in autism disorders and the pathways involved in their pathogeneses. Therefore, novel techniques, such as metabolomic profiling, can contribute to this objective because of their potential to enable the global analysis of low-molecular-weight metabolites, thus aiding in elucidating systemic perturbations. For instance, in 2010, Yap et al. (2010) reported a perturbation in the tryptophan-nicotinic acid metabolic pathway using an nuclear magnetic resonance-based platform in urine from autistic children. Metabolic alterations related to amino acids, carbohydrates and oxidative stress pathways were then described after urinary analysis using liquid chromatography-mass spectrometry (LC–MS) (Bitar et al. 2018; Mavel et al. 2013; Ming et al. 2012) and nuclear magnetic resonance (Noto et al. 2014). Overall, the plasma and urine metabolomes associate ASD with perturbed metabolic pathways related to tryptophan, nicotinic acid, purines, vitamin B6, and the enhancement of oxidative stress, as well as gut microbiota alterations. (Gevi et al. 2016; Lussu et al. 2017; Mavel et al. 2013; Mussap et al. 2016; Nadal-Desbarats et al. 2014).
Novel changes in several plasma metabolites in ASD were recently reported (West et al. 2014). Interestingly, such findings, including alterations in the aspartate, citrate, creatinine, DHEA-S, isoleucine, glutamate and glutarate metabolic pathways, had not been previously reported in the metabolomic profiling of urine (West et al. 2014). Hence, metabolomic analysis in plasma appears to be a suitable tool for further investigation of the disease mechanisms and has the potential to identify metabolic profiles within ASD. Thus, the aim of the present study was to evaluate the metabolomic profile of children with ASD subclassified by mental regression phenotype (AR) and non-mental regression (ANR) in plasma samples with an analytical platform methodology based on LC–MS.
2 Subjects and methods
The present study was a cross-sectional case–control study and was approved by the Clinical Research and Bioethics Committee at Reina Sofia University Hospital respecting the fundamental principles established in the Declaration of Helsinki. The selected subjects were incorporated into the study after all inclusion criteria were fulfilled and informed written consent from the children’s legal guardians was obtained.
2.1 Participants
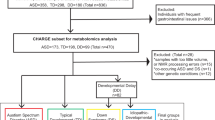
Fifty-seven ASD children were initially selected for this study (the recruitment started during 2015 in the Department of Psychology, Reina Sofia University Hospital, Cordoba, Spain). Of those, three children were excluded for not meeting the diagnostic criteria for ASD during the subsequent follow-up interviews at 18 months. Such interview is a follow-up to verify if the individuals continue meeting the DSM-5 Criteria.
All ASD children selected were between 2 and 6 years old with an agreed clinical diagnosis using the criteria of the International Classification Disease 10th Edition for ASD (World Health Organization 1992) and DSM-5 (American Psychiatric Publishing 2013). The diagnosis was confirmed by scores above the cut-off points of two tests, the Autism Diagnostic Observation Schedule test, with revised algorithms, and the Pervasive Developmental Disorders (Behavior Inventory) (PDDBI), the latter as a means of obtaining a measure of the severity of ASD. The ASD group was also classified according to whether the children presented developmental delay (a score lower than 70 in the cognitive quotient of the Battelle developmental test) or not. Within the ASD group, there were 20 children classified as AR and 32 classified as ANR: two children could not be clearly classified into either of these subgroups. The subdivision (ANR and AR) was based on the presence or absence of developmental regression during the first 2 years of life, which was assessed by a five-item questionnaire following the guide used by the ADI-R clinical interview for the evaluation of this process (Kim and Lord 2012). ASD children who obtained a score equal to or greater than three were included in the mental regression group and those with a score of less than three were included in the non-mental regression group.
ASD children presenting with other known neurological, metabolic or genetic diagnoses were excluded, as were children with medical treatment for autism-related behavioral comorbidities that may interfere with the results, such as sedatives, muscle relaxants or similar.
For the present study, we selected a subsample of 30 ASD subjects (15 ANR and 15 AR) choosing those with higher scores towards one group or another in the tests thus eliminating those with more doubtful data.
Additionally, a control group of 30 normally developing children was selected and matched to the ASD group by sex and age. Children in the control group were chosen from those who came to the hospital for pre-anesthesia for minor surgery (mainly hernias). The clinical and analytical absence of illness was confirmed in this group of healthy children after normal results. Table 1 shows the general characteristics of the selected ASD subgroups and healthy children (HC) participants.
2.2 Blood sampling
Blood samples (6 ml) were drawn following a 12-h fast. In all the children with ASD, the blood test was carried out after confirming the diagnosis, at the recruitment. In HC, all the analysis were realized before the anesthesia and surgery intervention. Blood and general biochemical analyses were performed to confirm the absence of other diseases, and other plasma aliquots were stored at − 80 °C.
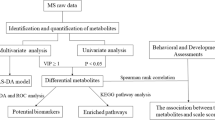
2.3 Metabolomics analysis
The major outcome of the study was to investigate differential metabolomic signatures among groups, therefore, we considered a broad analytical platform for such aim. Metabolon Inc. (Durham, NC, USA) platform have proven reliable and fit our needs due to its broad coverage. All metabolomics analyses were developed by Metabolon, Inc. (Durham, NC, USA) following methods previously described (Evans et al. 2009, 2014). Details of the analytical methods, data extraction, preprocessing and compound identification are listed in eMethods 1 in Additional File 1.
2.4 Statistical analysis
2.4.1 Univariate analysis
We used ANOVA tests to identify biochemical compounds that differed significantly between experimental groups. We performed a log transformation (to reduce the heteroscedasticity) and due to the difficulty of this method to handle zero or missing values we imputed missing values, if any, using a value corresponding to half of the minimum positive of each variable. Furthermore, to correct for multiple testing and false positives we used a false discovery rate cut-off of q < 0.1 (more in eMethods 1 in Additional File 1).
2.4.2 Multivariate analysis
2.4.2.1 Data processing
The metabolomics dataset was preprocessed using MetaboAnalyst v4.0 (Xia et al. 2015) (Quebec, Canada). All missing values were replaced using a value corresponding to half of the minimum positive of each variable. Next, the variables were log-transformed and unit variance-scaled. Data were introduced into SIMCA-P (SIMCA-P 15.0, Umetrics, Umeå, Sweden) for multivariate analysis using unsupervised and supervised models.
2.4.2.2 PCA and OPLS-DA
Principal component analysis (PCA) and orthogonal partial least-squares-discriminant analysis (OPLS-DA) were carried out to visualize the metabolic alterations in HC and ASD children and between the AR and ANR groups. PCA modeling was used for an initial exploration of the data with the main purpose of detecting potential outliers and patterns. OPLS-DA models were built to identify patterns and the features that discriminate between the different groups, e.g., ASD vs HC and ANR vs AR. In these discriminant analyses, the default seven-round cross-validation in the SIMCA-P software package was applied. Furthermore, the cross-validation ANOVA (CV-ANOVA) was calculated to assess the reliability of the models (further details in eMethods1). Also 100-fold permutation tests were calculated and its plots and together with the observed vs. predicted plots are included in the supplemental material (eFigure 3). We also obtained the so-called variable importance in the projection (VIP) values that were relevant for group discrimination. We selected features with a VIP > 1.5 to integrate the pathway analysis. Furthermore, the advantage of using OPLS-DA is the capability of this model to decompose the data into “predictive” information related to the response of Y (in our study, HC vs ASD, ANR or AR) and “orthogonal” structured information not related to the response (and that could be related to other factors such as technical or biological variation).
To improve the interpretation, shared-and-unique (SUS)-plots were built using the p(corr) of two different models. Thus, it is possible to extract metabolites related to a specific model and to identify shared features between both models either in a similar trend or in the inverse direction (more details are provided in eFigure 1 in Additional File 1).
3 Results
3.1 General characteristics
Table 1 also shows the results of the Battelle, CARS and PDDBI psychological tests for both AR and ANR children, which were significantly different among groups.
3.2 Metabolite identification using an untargeted approach in a UPLC–MS/MS platform
Plasma metabolic profiling was established to explore metabolic patterns associated with ASD and the mental regression status. Seven-hundred fifty metabolites were detected: 295 in C18-negative mode, 75 in negative HILIC mode, 189 in early positive mode and 191 in late positive mode. A summary of the compounds that achieved statistical significance (p ≤ 0.05) between the ASD and HC groups is shown in eTable 1 in Additional File 2.
3.2.1 Univariate analysis: single differences between classes
3.2.1.1 HC versus ASD
ANOVA of the HC and ASD groups showed several metabolites whose concentrations differed significantly (eTable 1 in Additional File 2; p < 0.001 and FDR < 0.1). Such metabolites corresponded mainly to the malate-aspartate shuttle, the urea cycle, ammonia recycling, the glucose-alanine cycle, beta-alanine metabolism, and the aspartate and tryptophan metabolic pathways (according to KEGG pathway analysis). A heatmap (including the top 25 significant metabolites) in which it was possible to highlight the differences between the two groups can be seen in Fig. 1.
3.2.1.2 ANR versus AR
When comparing the ANR and AR subgroups, seven compounds (decanoylcarnitine C10, arachidate, laurate, octanoylcarnitine, quinate, 7-methylurate and myristate) were significantly different (all p < 0.001 and FDR < 0.1). These metabolites were related mainly to fatty acid metabolism, except for quinate and 7-methylurate.
3.2.2 Multivariate analysis
3.2.2.1 PCA
It was not possible to identify a valid separation between the HC and ASD groups using this approach (Fig. 2a). When labelling the samples as HCs, ANR and AR we did not detect any clustering (Fig. 2b (PCA model: R = 0.66; Q = 0.0708)).
a PCA scores plot including samples from HC and ASD. HC healthy children, ASD autistic children. b PCA scores plot including samples from HC and autistic children (labelled as ANR and AR). HC healthy children, AR autistic mental-regression children, ANR autistic non-mental regression children. c OPLS-DA model score plot showing separation by condition, including ASD children and HCs. HC healthy children, ASD autistic children
3.2.2.2 OPLS-DA modeling
Four different models were built. The first compared the HC vs ASD groups (Fig. 2c). The other models built were as follows: HC vs ANR, HC vs AR and ANR vs AR (non-significant). Table 2 summarizes the predictive abilities of each constructed model, and several lists of VIP (total, predictive and orthogonal) features are included in eTables 2–4 in Additional File 2.
The SUS-plot (eFigure 1 in Additional File 1), which combines the HC vs ANR and HC vs AR models (built in binary mode using the HC group as a reference), helps identify two metabolites shared by both models in a decreasing trend in both subgroups (glutamate and aspartate). 4-Methyl-oxopentanoate was found in both models but in an increasing trend. Furthermore, three metabolites were identified in relation to ANR in an increasing trend: 3-methylxanthine, 7-methylurate and 3-methylhistidine. Finally, we identified a decrease in laurate as the only marker specific to the AR group.
3.3 Pathway enrichment analysis constructed using OPLS-DA predictive and orthogonal components
Two lists of VIP features were used for the pathway enrichment analysis using MetaboAnalyst 4.0: one was built from the predictive component (related to the class) and the other was built from the orthogonal components (not related to the class but to orthogonal variation). Due to limitations of the database (KEGG pathways) within the MetaboAnalyst platform, not all the metabolites were recognized. Additionally, an over-representation analysis was performed without significant results after FDR cutoff (Figure included as eFigure 3).
3.3.1 HC versus ASD
Ninety-one metabolites were selected from the predictive component (eTable 2); however, only 77 were recognized. The integration of these metabolites showed that five canonical pathways were significantly modified in the ASD group compared with HCs, namely, aminoacyl-tRNA biosynthesis, alanine, aspartate and glutamate metabolism, galactose and nicotinate and nicotinamide metabolism and panthothenate and CoA (all with raw p values ≤ 0.01 and FDR < 0.1). The inclusion of compounds into the orthogonal component revealed only a change in aminoacyl-tRNA biosynthesis, which did not correlate with the ASD condition (p value ≤ 0.01 and FDR < 0.1).
3.3.2 HC versus ANR
The pathway enrichment analysis included 88 metabolites from the predictive component and 80 from the orthogonal component (eTable 3). Only caffeine metabolism in the first and nitrogen metabolism in the latter were significantly affected (raw p values ≤ 0.01 and FDR < 0.1).
3.3.3 HC versus AR
Ninety-four compounds from the predictive component and 58 from the orthogonal components were analyzed (eTable 4); Four canonical pathways reflected a significant change related to the predictive component, namely, aminoacyl-tRNA biosynthesis; alanine, aspartate and glutamate metabolism; panthothenate; and CoA and fatty acid biosynthesis (raw p values ≤ 0.01 and FDR < 0.1). No pathway was associated with the orthogonal component.
4 Discussion
In this study, we report, for the first time, the plasmatic metabolomic profiling of ASD children subclassified by mental regression (AR) phenotype and with no regression (ANR). With the use of an LC–MS platform including C18 and HILIC columns, we covered a broad range of metabolites, which was complemented with comprehensive statistical and pathway enrichment analyses. We present several alterations observed in ASD compared with HCs involving mainly amino acid, lipid and nicotinamide metabolism. However, we were able to distinguish only subtle significant differences when isolating the ASD subgroups.
Due to the complexity of ASD, rather than focusing on single biomarkers, which might result in inaccurate results, the strength of our research lies in the integration of the most discriminant metabolites (and the exclusion of the orthogonal metabolites) in their respective pathways, which was provided by the OPLS-DA modelling. Nevertheless, the use of univariate and multivariate analysis should be considered as complementary and confirmatory; i.e. laurate that was significantly different between ANR and AR as shown in the univariate analysis, also appeared to be as a significant discriminator in the SUS-plot (built from the multivariate modeling).
4.1 Endogenous metabolites
Glutamate functions as a major excitatory molecule in the brain, and alterations in its metabolism can lead to imbalances in the excitation/inhibition equilibrium of neural networks, which may lead to autistic-like symptoms. Herein, we report similar decrease in glutamate in both the ANR and AR groups compared with the HC group (in both univariate and multivariate analysis). Although the relative concentrations of aspartate and glutamate were modified in both groups, other metabolites were specific within each group, as shown by both statistical analyses, such as gamma carboxyglutamate, N-Carboxyethyl-gamma-aminobutyric acid and 4-hydroxyglutamate in the ANR group and N-acetyl-aspartyl-glutamate (NAAG) in the AR group. Alterations of these metabolic pathways in ASD have been previously reported in urine but in an inverse manner (Kałużna-Czaplińska et al. 2017; Rolf et al. 1993; Rossignol and Frye 2012; Yap et al. 2010) and may be related with the pathogenesis of the disorder and disease severity (Alabdali et al. 2014). However, they may also be associated with food restrictions, such as diets low in protein or lacking certain types of food or impaired digestion. Also, the restriction might lead to the substitution with other types of foods thus this should be analyzed thoroughly (Kałużna-Czaplińska et al. 2017). Interestingly, using the SUS-plot, we identified metabolites specific to each group that although have been proposed as biomarkers of intake of dietary products, such as meat (Schmedes et al. 2016) or cocoa (Garcia-Aloy et al. 2015) could be related also to some metabolic alterations and need to be studied in detail in further studies.
Some biochemicals comprising the malate-aspartate shuttle were decreased in the entire ASD cohort (as shown in both analyses), suggesting deregulation in this mitochondrial process. The malate-aspartate shuttle is a crucial system that supports oxidative phosphorylation and ATP production (Napolioni et al. 2011). We should note that changes in the activity of aspartate-glutamate carriers have been detected in gray matter obtained from autistic individuals (Palmieri et al. 2010). Taken together, the imbalance in glutamate metabolism may also originate from changes in malate-aspartate shuttle activity.
Arginine, N-acetylarginine, homoarginine and n-alpha-acetylornithine discriminated the ASD cohort, and particularly the AR group, from the control group (multivariate analysis, univariate analysis only shows arginine as significant). In this regard, urinary (Diémé et al. 2015) and plasmatic (Delwing et al. 2008) arginine have been reported to be increased in ASD children. An excessive amount of arginine and its compounds might increase oxidative stress by increasing nitric oxide (NO). As reviewed by Rossignol et al. it seems that ASD patients might be both, more vulnerable to NO and present higher concentrations of NO at baseline, factors that could act synergistically to cause significant mitochondrial impairment (Rossignol and Frye 2012). Our findings suggest that AR subjects have a more dysregulated NO pathway, and such alterations may be related with the regression.
The branched-chain amino acids (BCAAs), valine, isoleucine, and leucine, are important constituents of proteins but are also readily degraded into carbon skeletons that may enter anabolic pathways (i.e., gluconeogenesis or fatty acid synthesis) or pathways associated with energy generation (i.e., the TCA cycle). In the present study, we found a possible alteration in the BCAA metabolism in ASD, which was supported by an accumulation of several alpha-keto derivates, such as 3-methyl-2-oxobutyrate, 3-methyl-2-oxovalerate, and 4-methyl-2-oxopentanoate. These accumulations indicate that these compounds exhibit reduced incorporation into the TCA cycle, thus contributing to alterations in anabolic processes and/or energy generation. These changes point towards a dysregulation in glutamate homeostasis, as the BCAA aminotransferase utilizes alpha-ketoglutarate to transfer the amino group and forms glutamate, both of which were decreased in the ASD cohort (univariate and multivariate analyses). Moreover, in connection with these alterations, we detected an increase in carnitine-conjugated intermediates (i.e., isovalerylcarnitine and isobutyrylcarnitine) in the ASD cohort, and acylcarnitines decanoylcarnitine and octanoylcarnitine were significantly higher in the ANR group than the AR group as shown in the univariate and multivariate analyses. It is known that the inhibition of the TCA cycle may result in an elevation of TCA cycle intermediates but also impairs fatty acid β-oxidation. Therefore, these abnormalities may be associated with mitochondrial dysfunction in children with ASD (Frye et al. 2013; Rossignol and Frye 2012).
In addition, the association between alterations in urinary tryptophan metabolism in autistic children (Kałużna-Czaplińska et al. 2017) entails numerous irregularities, including the serotonin pathways. In this regard, we found that tryptophan was significantly higher in both subgroups (ANR and AR, both statistical analyses) than in HCs. Furthermore, other tryptophan-associated metabolites, such as kynurenine, 5-bromotryptophan, 3-indoxyl sulfate, indolelactate and 6-hydroxyindole sulfate, were elevated in the ANR group. Kynurenine is produced by indoleamine 2,3-dioxygenase in response to inflammatory stimuli. Thus, an increase in the kynurenine level may reflect increased inflammation in the ASD group. Changes in NAD+ metabolism were noted, with a significant decline in the nicotinamide levels and increases in 1-methyl-nicotinamide and N-methyl-2-pyridone-5-carboxamide (with a VIP = 1.46) in the ASD cohort. Remarkably, similar changes, but in urine, have been reported (Adams et al. 2011; Yap et al. 2010). Nicotinamide is a key intermediate in the synthesis of NAD+, a coenzyme that plays a vital role in redox reactions by accepting electrons and forming NADH as part of β-oxidation, glycolysis, and the TCA cycle. In ASD patients, a decrease in the conversion of tryptophan to melatonin could lead to an increase in nicotinic synthesis, thus increasing the demand for methylation and increasing the susceptibility to oxidative stress (Mussap et al. 2016). In fact, n-methyl nicotinamide and N-methyl-2-pyridone-5-carboxamide have been implicated in the pathogenesis of Parkinson’s disease (Fukushima et al. 1995; Willets et al. 1993).
4.2 Lipid metabolism
Evidence has shown that abnormalities in lipid metabolism may play a role in the pathogenesis of ASD and can be connected with metabolic and dietary alterations in the regressive form of autism (El-Ansary et al. 2011; Goldani et al. 2014). The lysolipids 1-palmitoyl-glycerol-phosphatidyl-etholamine (GPE) and 1-stearoyl-GPE were decreased in plasma obtained from autistic individuals (univariate and multivariate analysis, eTables1, 2). These are degradation products derived from phospholipids and are formed when phospholipases remove fatty acid moieties from the Sn-1 and Sn-2 positions of phospholipids (Khaselev and Murphy 2000). Changes in triacylglycerol metabolism were also noted based on mental regression status, with an accumulation of several species of mono- and diacylglycerols in the AR group compared to the ANR group (univariate analysis, eTable 1). Mono- and diacylglycerols are intermediates in triacylglycerol metabolism; they are formed when lipases remove fatty acids from the glycerol backbone of triacylglycerols. Moreover, the univariate analysis (eTable 1) revealed that there was a marked decrease in both long- and medium-chain fatty acids in samples from the AR group. These alterations may indicate differences in triacylglycerol and phospholipid metabolism, fatty acid utilization, and beta-oxidation. We should note that the changes in free fatty acids were more pronounced in the AR subgroup and, in some cases, showed significant changes between the AR and ANR groups (i.e.,myristate and palmitate) in the univariate analysis, and the change in laurate appears to be specific to the AR group, as reflected in the univariate and the SUS-plot (the OPLS-DA model comparing AR and ANR was not analyzed due to the lack of significance and thus, invalid). These results agree with the report by El-Ansary (El-Ansary et al. 2011) in which they concluded that impaired fatty acid profiles could be used as highly reliable diagnostic biomarkers in a Saudi autistic cohort and may reflect mitochondrial dysfunction as a putative causative mechanism (Pastural et al. 2009).
Several sphingomyelins discriminated the ASD cohort (eTable 2) from the HCs in the predictive component. However, due to limitations in the MetaboAnalyst module, it was not possible to integrate them into their respective pathways. Nevertheless, an alteration in sphingolipid metabolism, in particular sphingomyelins, has been associated with autism and is due to an abnormal developmental trajectory of white matter (Barnea-Goraly et al. 2004; Mak-Fan et al. 2013; Wang et al. 2016). Nonetheless, we did not observe differences among the subgroups.
Overall, the pathway enrichment analysis showed several routes modified in the ASD cohort compared with HCs. Nonetheless, when analyzing the subgroups, these routes were not maintained; the ANR group showed fewer changes. We speculate that differences in the AR group were strong enough to make significant differences in the ASD cohort and therefore these differences may be associated with the regression in the disease. However, we could not discard that some changes may be related to lifestyle (i.e., diet, physical activity, etc.) although we used a multivariate model which helps to isolate the effect of features that are not related to the class studied (in this case, ASD or its subgroups).
The global approach that we developed in the present study seems to be very effective and provides broad coverage; however, it is important to address several limitations. Although it was possible to detect significant differences between the groups and using cross-validation methods in the multivariate analysis, the sample size was small, and a larger number of subjects should be included in further studies to detect more subtle changes, particularly when comparing the AR and ANR subgroups. Differences between the univariate and multivariate analysis show slightly differences that might be due to the restrictive cutoffs utilized on each type of analysis, however, the use of both approaches help us to confirm some changes with confidence when they appear in both analyses.
Moreover, it is necessary to perform targeted analyses to make a proper quantification of the metabolites and their impact through the different metabolic pathways and to validate these findings reported in an external cohort.
Our main interest is to continue this investigation including data from the microbiome and additional food intake and lifestyle questionnaires to identify the changes relates to the latter or those linked to the disease or a specific phenotype. In the future, if a genetic or molecular profile in patients could be identified, patients could be stratified according to it and subsequently compared regarding cognitive, neuroimaging and biochemical measures to treat individuals with ASD more efficiently and with a more personalized approach (Howes et al. 2018).
5 Conclusion
In summary, the use of an LC-MS platform approach, including C18 and HILIC columns, for metabolomics in ASD children using plasma appears to have a broad coverage of the metabolome and adds further support to previous findings in urine, such as changes in tryptophan and nicotinic acid metabolic pathway. Furthermore, the present study documents several changes related to amino acid, NAD+ and lipid metabolism in plasma. In some cases, such as in the arginine and the glutamate pathways and free fatty acids, alterations may be associated with mental regression in ASD. Further targeted analyses, including a systems biology approach with the study of genome, transcriptome and the study of the microbiome, including a larger cohort are needed to validate the results presented herein. Research regarding potential mitochondrial dysfunction in a subset of ASD subjects is guarantee.
Data availability
The metabolomics datasets used and/or analyzed during the current study are included as Supplemental Material and any additional data is available from the corresponding author on reasonable request.
References
Adams, J. B., Audhya, T., McDonough-Means, S., Rubin, R. A., Quig, D., Geis, E., et al. (2011). Nutritional and metabolic status of children with autism vs. neurotypical children, and the association with autism severity. Nutrition & Metabolism, 8(1), 34. https://doi.org/10.1186/1743-7075-8-34.
Alabdali, A., Al-Ayadhi, L., & El-Ansary, A. (2014). Association of social and cognitive impairment and biomarkers in autism spectrum disorders. Journal of Neuroinflammation, 11(1), 4. https://doi.org/10.1186/1742-2094-11-4.
Altieri, L., Neri, C., Sacco, R., Curatolo, P., Benvenuto, A., Muratori, F., et al. (2011). Urinary p-cresol is elevated in small children with severe autism spectrum disorder. Biomarkers. https://doi.org/10.3109/1354750x.2010.548010.
American Psychiatric Publishing, I. (2013). Diagnostic and statistical manual of mental disorders: DSM-5™ (5th ed.). Arlington: American Psychiatric Publishing Inc.
Barnea-Goraly, N., Kwon, H., Menon, V., Eliez, S., Lotspeich, L., & Reiss, A. L. (2004). White matter structure in autism: Preliminary evidence from diffusion tensor imaging. Biological Psychiatry, 55(3), 323–326. https://doi.org/10.1016/j.biopsych.2003.10.022.
Bitar, T., Mavel, S., Emond, P., Nadal-Desbarats, L., Lefèvre, A., Mattar, H., et al. (2018). Identification of metabolic pathway disturbances using multimodal metabolomics in autistic disorders in a Middle Eastern population. Journal of Pharmaceutical and Biomedical Analysis, 152, 57–65. https://doi.org/10.1016/j.jpba.2018.01.007.
Delwing, D., Delwing, D., Bavaresco, C. S., & Wyse, A. T. S. (2008). Protective effect of nitric oxide synthase inhibition or antioxidants on brain oxidative damage caused by intracerebroventricular arginine administration. Brain Research, 1193, 120–127. https://doi.org/10.1016/j.brainres.2007.11.052.
Diémé, B., Mavel, S., Blasco, H., Tripi, G., Bonnet-Brilhault, F., Malvy, J., et al. (2015). Metabolomics study of urine in autism spectrum disorders using a multiplatform analytical methodology. Journal of Proteome Research, 14(12), 5273–5282. https://doi.org/10.1021/acs.jproteome.5b00699.
El-Ansary, A. K., Ben Bacha, A. G., & Al- Ayahdi, L. Y. (2011). Plasma fatty acids as diagnostic markers in autistic patients from Saudi Arabia. Lipids in Health and Disease, 10(1), 62. https://doi.org/10.1186/1476-511X-10-62.
Evans, A. M., Bridgewater, B. R., Liu, Q., Mitchell, M. W., Robinson, R. J., Dai, H., et al. (2014). High resolution mass spectrometry improves data quantity and quality as compared to unit mass resolution mass spectrometry in high-throughput profiling metabolomics. Journal of Postgenomics Drug & Biomarker Development. https://doi.org/10.4172/2153-0769.1000132.
Evans, A. M., DeHaven, C. D., Barrett, T., Mitchell, M., & Milgram, E. (2009). Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Analytical Chemistry, 81(16), 6656–6667. https://doi.org/10.1021/ac901536h.
Frye, R. E., Melnyk, S., & MacFabe, D. F. (2013). Unique acyl-carnitine profiles are potential biomarkers for acquired mitochondrial disease in autism spectrum disorder. Translational Psychiatry, 3(1), e220. https://doi.org/10.1038/tp.2012.143.
Fukushima, T., Tawara, T., Isobe, A., Hojo, N., Shiwaku, K., & Yamane, Y. (1995). Radical formation site of cerebral complex I and Parkinson’s disease. Journal of Neuroscience Research, 42, 385–390.
Gabriele, S., Sacco, R., Cerullo, S., Neri, C., Urbani, A., Tripi, G., et al. (2014). Urinary p-cresol is elevated in young French children with autism spectrum disorder: A replication study. Biomarkers. https://doi.org/10.3109/1354750x.2014.936911.
Garcia-Aloy, M., Llorach, R., Urpi-Sarda, M., Jáuregui, O., Corella, D., Ruiz-Canela, M., et al. (2015). A metabolomics-driven approach to predict cocoa product consumption by designing a multimetabolite biomarker model in free-living subjects from the PREDIMED study. Molecular Nutrition & Food Research, 59(2), 212–220. https://doi.org/10.1002/mnfr.201400434.
Gevi, F., Zolla, L., Gabriele, S., & Persico, A. M. (2016). Urinary metabolomics of young Italian autistic children supports abnormal tryptophan and purine metabolism. Molecular Autism, 7(1), 47. https://doi.org/10.1186/s13229-016-0109-5.
Goldani, A. A. S., Downs, S. R., Widjaja, F., Lawton, B., & Hendren, R. L. (2014). Biomarkers in autism. Frontiers in Psychiatry, 5, 100. https://doi.org/10.3389/fpsyt.2014.00100.
Howes, O. D., Rogdaki, M., Findon, J. L., Wichers, R. H., Charman, T., King, B. H., et al. (2018). Autism spectrum disorder: Consensus guidelines on assessment, treatment and research from the British Association for Psychopharmacology. Journal of Psychopharmacology. https://doi.org/10.1177/0269881117741766.
Kałużna-Czaplińska, J., Jóźwik-Pruska, J., Chirumbolo, S., & Bjørklund, G. (2017). Tryptophan status in autism spectrum disorder and the influence of supplementation on its level. Metabolic Brain Disease, 32(5), 1585–1593. https://doi.org/10.1007/s11011-017-0045-x.
Khaselev, N., & Murphy, R. C. (2000). Structural characterization of oxidized phospholipid products derived from arachidonate-containing plasmenyl glycerophosphocholine. Journal of Lipid Research, 41(4), 564–572.
Kim, S. H., & Lord, C. (2012). New autism diagnostic interview-revised algorithms for toddlers and young preschoolers from 12 to 47 months of age. Journal of Autism and Developmental Disorders, 42(1), 82–93. https://doi.org/10.1007/s10803-011-1213-1.
Lussu, M., Noto, A., Masili, A., Rinaldi, A. C., Dessì, A., De Angelis, M., et al. (2017). The urinary 1H-NMR metabolomics profile of an italian autistic children population and their unaffected siblings. Autism Research, 10(6), 1058–1066. https://doi.org/10.1002/aur.1748.
Mak-Fan, K. M., Morris, D., Vidal, J., Anagnostou, E., Roberts, W., & Taylor, M. J. (2013). White matter and development in children with an autism spectrum disorder. Autism, 17(5), 541–557. https://doi.org/10.1177/1362361312442596.
Mavel, S., Nadal-Desbarats, L., Blasco, H., Bonnet-Brilhault, F., Barthélémy, C., Montigny, F., et al. (2013). 1H-13C NMR-based urine metabolic profiling in autism spectrum disorders. Talanta, 114, 95–102. https://doi.org/10.1016/j.talanta.2013.03.064.
Ming, X., Stein, T. P., Barnes, V., Rhodes, N., & Guo, L. (2012). Metabolic perturbance in autism spectrum disorders: A metabolomics study. Journal of Proteome Research, 11(12), 5856–5862. https://doi.org/10.1021/pr300910n.
Mussap, M., Noto, A., & Fanos, V. (2016). Metabolomics of autism spectrum disorders: Early insights regarding mammalian-microbial cometabolites. Expert Review of Molecular Diagnostics, 16(8), 869–881. https://doi.org/10.1080/14737159.2016.1202765.
Nadal-Desbarats, L., Aïdoud, N., Emond, P., Blasco, H., Filipiak, I., Sarda, P., et al. (2014). Combined 1H-NMR and 1H-13C HSQC-NMR to improve urinary screening in autism spectrum disorders. Analyst, 139(13), 3460–3468. https://doi.org/10.1039/c4an00552j.
Napolioni, V., Persico, A. M., Porcelli, V., & Palmieri, L. (2011). The mitochondrial aspartate/glutamate carrier AGC1 and calcium homeostasis: Physiological links and abnormalities in autism. Molecular Neurobiology, 44(1), 83–92. https://doi.org/10.1007/s12035-011-8192-2.
Noto, A., Fanos, V., Barberini, L., Grapov, D., Fattuoni, C., Zaffanello, M., et al. (2014). The urinary metabolomics profile of an Italian autistic children population and their unaffected siblings. The Journal of Maternal-Fetal & Neonatal Medicine, 27(sup2), 46–52. https://doi.org/10.3109/14767058.2014.954784.
Palmieri, L., Papaleo, V., Porcelli, V., Scarcia, P., Gaita, L., Sacco, R., et al. (2010). Altered calcium homeostasis in autism-spectrum disorders: Evidence from biochemical and genetic studies of the mitochondrial aspartate/glutamate carrier AGC1. Molecular Psychiatry, 15(1), 38–52. https://doi.org/10.1038/mp.2008.63.
Pastural, É., Ritchie, S., Lu, Y., Jin, W., Kavianpour, A., Khine Su-Myat, K., et al. (2009). Novel plasma phospholipid biomarkers of autism: Mitochondrial dysfunction as a putative causative mechanism. Prostaglandins Leukotrienes and Essential Fatty Acids, 81(4), 253–264. https://doi.org/10.1016/j.plefa.2009.06.003.
Rolf, L. H., Haarmann, F. Y., Grotemeyer, K. H., & Kehrer, H. (1993). Serotonin and amino acid content in platelets of autistic children. Acta Psychiatrica Scandinavica, 87(5), 312–316.
Rossignol, D. A., & Frye, R. E. (2012). Mitochondrial dysfunction in autism spectrum disorders: A systematic review and meta-analysis. Molecular Psychiatry, 17(3), 290–314. https://doi.org/10.1038/mp.2010.136.
Schmedes, M., Aadland, E. K., Sundekilde, U. K., Jacques, H., Lavigne, C., Graff, I. E., et al. (2016). Lean-seafood intake decreases urinary markers of mitochondrial lipid and energy metabolism in healthy subjects: Metabolomics results from a randomized crossover intervention study. Molecular Nutrition & Food Research, 60(7), 1661–1672. https://doi.org/10.1002/mnfr.201500785.
Wang, H., Liang, S., Wang, M., Gao, J., Sun, C., Wang, J., et al. (2016). Potential serum biomarkers from a metabolomics study of autism. Journal of Psychiatry and Neuroscience, 41(1), 27–37. https://doi.org/10.1503/jpn.140009.
West, P. R., Amaral, D. G., Bais, P., Smith, A. M., Egnash, L. A., Ross, M. E., et al. (2014). Metabolomics as a tool for discovery of biomarkers of autism spectrum disorder in the blood plasma of children. PLoS ONE, 9(11), e112445. https://doi.org/10.1371/journal.pone.0112445.
Willets, J. M., Lunec, J., Williams, A. C., & Griffiths, H. R. (1993). Neurotoxicity of nicotinamide derivatives: Their role in the aetiology of Parkinson’s disease. Biochemical Society Transactions, 21(Pt 3), 299S.
World Health Organization. (1992). The ICD-10 classification of mental and behavioural disorders. International Classification, 10, 1–267. https://doi.org/10.1002/1520-6505(2000)9:5%3c201:AID-EVAN2%3e3.3.CO;2-P.
Xia, J., Sinelnikov, I. V., Han, B., & Wishart, D. S. (2015). MetaboAnalyst 3.0—Making metabolomics more meaningful. Nucleic Acids Research. https://doi.org/10.1093/nar/gkv380.
Yap, I. K. S., Angley, M., Veselkov, K. A., Holmes, E., Lindon, J. C., & Nicholson, J. K. (2010). Urinary metabolic phenotyping differentiates children with autism from their unaffected siblings and age-matched controls. Journal of Proteome Research, 9(6), 2996–3004. https://doi.org/10.1021/pr901188e.
Acknowledgements
We gratefully acknowledge the patients, controls and family members who participated in this study.
Funding
ODRH has received funding from the European Union Seventh Framework Programme (FP7-PEOPLE-2013-COFUND) under grant agreement n° 609020 - Scientia Fellows.
Author information
Authors and Affiliations
Contributions
ODRH conducted the metabolomics data analyses, analyzed and interpreted the biochemical data and wrote the manuscript. AGF, MJTA, collected the plasma samples, interpreted the data. MGC and AG designed the study. JLPN, KFR, PMB were responsible of the clinical assessments and interpreted the data. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
The present study was a cross-sectional case–control study case–control study and was approved by the Clinical Research and Bioethics Committee at Reina Sofia University Hospital respecting the fundamental principles established in the Declaration of Helsinki of 1964.
Informed consent
Informed consent from the children’s legal guardians was obtained.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rangel-Huerta, O.D., Gomez-Fernández, A., de la Torre-Aguilar, M.J. et al. Metabolic profiling in children with autism spectrum disorder with and without mental regression: preliminary results from a cross-sectional case–control study. Metabolomics 15, 99 (2019). https://doi.org/10.1007/s11306-019-1562-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-019-1562-x