Abstract
Using a novel approach combining four complementary metabolomic and mineral platforms with genome-wide genotyping at 1536 single nucleotide polymorphism (SNP) loci, we have investigated the extent of biochemical and genetic diversity in three commercially-relevant waxy rice cultivars important to food production in the Lao People’s Democratic Republic (PDR). Following cultivation with different nitrogen fertiliser regimes, multiple metabolomic data sets, including minerals, were produced and analysed using multivariate statistical methods to reveal the degree of similarity between the genotypes and to identify discriminatory compounds supported by multiple technology platforms. Results revealed little effect of nitrogen supply on metabolites related to quality, despite known yield differences. All platforms revealed unique metabolic signatures for each variety and many discriminatory compounds could be identified as being relevant to consumers in terms of nutritional value and taste or flavour. For each platform, metabolomic diversity was highly associated with genetic distance between the varieties. This study demonstrates that multiple metabolomic platforms have potential as phenotyping tools to assist breeders in their quest to combine key yield and quality characteristics. This better enables rice improvement programs to meet different consumer and farmer needs, and to address food security in rice-consuming countries.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Since the turn of the century, many rice-producing countries have gained access to export markets, bringing significant income to their countries (Dodsworth 1997; Reyes 1997). This is due to both economic development and yield benefits delivered by the Green Revolution. The rice of the Green Revolution was of the non-waxy type, meaning that it contains amylose. In the Lao People’s Democratic Republic (PDR), waxy rices, which do not contain amylose, are the staple food (Bounphanousay et al. 2000). As a result, the Lao PDR gained no benefit from the Green Revolution, and continuing to grow low-yielding traditional varieties significantly compromised food security until the late 1990s. At this time, breeding programs tailored to waxy rices led to the release of improved varieties such as Thadokkham 1 (TDK1) and Thasano 1 (TSN1) (Inthapanya et al. 2006). Despite higher yield and acceptable quality, these cultivars do not have the same traits of quality, or command the same price premium, as the two most popular traditional varieties, Kai Noy Leuang (KNL) and Hom Nang Nouane (HNN). As the Lao PDR negotiates its entry into the World Trade Organisation, export opportunities are emerging for its prized traditional waxy varieties of rice. If the quality of the two traditional varieties can be combined with the agronomic adaptation of high-yielding improved varieties, such as TDK1 or TSN1, this could have considerable positive economic and social impact on poor Lao rice farmers.
In order to combine quality with yield, it is necessary to understand important quality traits, be able to select for them in a breeding program, and to find a suitable agronomically adapted background. A previous study indicates that TSN1, which is responsive to fertilisers, yields twice as much as the traditional varieties, and has better milling quality than TDK1 (Boualaphanh et al. 2011), is such a suitable background. However, identifying quality traits is complex. Current tools of quality evaluation have evolved around the more commonly consumed non-waxy types of rice, and particularly centre around the effects of amylose (Fitzgerald et al. 2009). These tools, therefore, are potentially inappropriate for waxy types.
Over the past few years, genotyping technology has advanced rapidly, to the point where identification of important loci has become significantly easier (Newton-Cheh and Hirschhorn 2005; Shirasawa et al. 2006; Raghavan et al. 2007; Hyten et al. 2009; Lee et al. 2009; Wright et al. 2010) when an appropriate mapping population and accurate phenotyping tools are available (Tarpley and Roessner 2007). Aside from sensory panels, there is no phenotyping tool that adequately describes the taste of rice (Champagne et al. 2010). In the last decade, technologies for the detection and identification of small molecules in plant tissue has undergone rapid development (Hall 2006) to the point where these ‘metabolomic’ tools could be utilised to reveal biochemically important metabolites that affect the taste and nutritional value of the foods we eat (Hall et al. 2008).
The objectives of the present study are to develop a unique multi-platform metabolomics and ionomics approach to (i) identify differences in primary metabolites, volatile compounds and mineral elements between the grains of TSN1, KNL and HNN grown at four different N fertiliser regimes; (ii) determine the effect of N on the metabolic signature of each variety; and (iii) determine whether differences in grain metabolites correlate with genetic differences between the varieties and with known differences in taste and flavour that Lao rice-consumers prize. The over-arching objective is to explore the possibility of developing a new generation of selection tools for rice improvement programs, which could also have broader application for all grain crop improvement programs.
2 Materials and methods
2.1 Plant materials
Three varieties of rice, Oryza sativa L., were used in this study. TSN1 is in the indica germplasm class and is an improved variety with valuable traits of agronomic adaptation, HNN is a traditional landrace and is also indica, and KNL is a traditional landrace and in the tropical japonica germplasm class. Both traditional varieties have valuable traits of quality. Twenty five seedlings of TSN1, HNN and KNL were planted as subplots in a split plot design within four main plots at the Agriculture Research Centre, Vientiane, Lao PDR. N was applied at either 0, 30, 60 or 90 kg/ha to each main plot, and 30 kg/ha each of P and K were applied to the four main plots (Boualaphanh et al. 2011). Each variety was replicated three times in each main plot. Grain from each subplot was harvested at maturity and sun-dried. Paddy from each sample was dehulled (Satake Rice Machine, Tokyo, Japan), brown grain was polished (Grainman 60-230-60-2AT, Grain Machinery Mfg. Corp., Miami, FL, USA), and the milled rice from each subplot (three varieties and three replicates) was then dispatched to collaborators for profiling of metabolites and mineral elements.
2.2 SNP genotyping
DNA was extracted from leaves of the three varieties exactly as previously described (Cuevas et al. 2010). The DNA extracts were quantified to 50 ng/ul prior to genotyping with Nanodrop 1000 (Thermo Scientific, Wilmington, DE, USA). Single nucleotide polymorphism (SNP) genotyping was carried out at 1536 loci (Zhao et al. 2010) using a BeadXpress (Illumina, San Diego, CA, USA). SNP calls were analysed using Alchemy software (Wright et al. 2010). SNP maps were generated and genetic distances were calculated using the software GGT 2.0: Graphical Genotyping (van Berloo 2008). Euclidean distance was used as a similarity coefficient to determine the genetic distance between each variety.
2.3 Metabolite and elemental profiling
Milled rice of each variety, replicate and N treatment was ground in liquid nitrogen and stored at −80°C. Primary metabolites in polar extracts were profiled by 1H-NMR and GC-EI-TOF-MS. Volatile compounds were measured by GC-MS of headspace sampled by SPME and mineral elemental analysis was done by ICP-MS. The mineral data were autoscaled with full cross validation, and the three metabolite datasets were mean-centred and Pareto scaled using either Amix software v. 3.5 (Bruker BioSpin GmbH, Rheinstetten, Germany) or SIMCA-P 11.0 (Umetrics AB, Umeå, Sweden) (Eriksson et al. 2006) for subsequent analysis by PCA. The tendencies highlighted with PCA for the discriminant compounds were verified using univariate analyses (Analysis of Variance, ANOVA).
2.3.1 1H-NMR profiling of polar compounds
1H-NMR profiling was performed on ethanolic extracts after precipitation of residual starch. Two replicates for each ground rice flour sample were extracted as previously described for fruit and Arabidopsis (Moing et al. 2004), with modifications. Fifty mg of lyophilised powder were extracted successively with 2 ml of ethanol/water mixtures: 80/20, 50/50 (v/v) and pure water (4 ml) for 15 min at 80°C. After centrifugation, the supernatants were combined, dried under vacuum and lyophilised. The lyophilised extracts were mixed with 500 μl of 100 mM potassium phosphate buffer pH 6.0, 1 mM ethylene diamine tetraacetic acid disodium salt (EDTA), in D2O, titrated with KOD solution to pH 6 when necessary. In order to precipitate residual starch, nine volumes of absolute ethanol (4.5 ml) were added to rice extracts (500 μl), which were then mixed vigorously for 30 s. Samples were stored at 4°C for 24 h and then centrifuged at 30,000×g for 20 min. The supernatant was then collected and dried under vacuum. Dried extracts were solubilised with 500 μl of D2O and 5 μl of solution of sodium salt of deuterated trimethylsilylpropionic acid (TSP-d4, 0.01% final concentration) were added. The mixture was centrifuged at 10,000×g for 5 min at room temperature. The supernatant was then transferred into a 5 mm NMR tube for acquisition.
1H-NMR spectra were recorded on a Bruker™ Avance Spectrometer (Bruker BioSpin, Wissembourg, France) at 500.162 MHz and 300 K using a 5 mm Broad Band Inverse probe (BBI). Spectra were acquired using a classical mono-dimensional liquid pulse sequence set with a 90° pulse, 32 K data points, a spectral width of 6000 Hz, 20 s relaxation delay and 64 scans. The acquired spectra were line broadened (0.3 Hz), and manually phased using TOPSPIN v1.3 software (Bruker BioSpin, Wissembourg, France) and manually baseline corrected. They were then aligned with the TSP-d4 signal at δ 0.00 ppm. Before statistical analysis, data reduction of the entire spectra (10–0.5 ppm with exclusion of 4.9–4.5 ppm leading to 456 integrated buckets of 0.02 ppm width) was done, followed by normalisation against total intensity. Attribution of each significant bucket to one compound, or a mixture of compounds, was performed using comparison to a library of authentic standards and standard spiking. The 1H-NMR spectra were converted into JCAMP-DX (the Joint Committee on Atomic and Molecular Physical data—Data Exchange format) standard exchange format and have been deposited, with associated metadata and compound list, into the Metabolomics Repository of Bordeaux MeRy-B (http://www.cbib.u-bordeaux2.fr/MERYB/projects/query_exp.php?project_id=37).
2.3.2 Derivatised GC-MS analysis of polar compounds
Metabolite profiling of polar compounds was performed as detailed previously (Allwood et al. 2009) using gas chromatography coupled to electron impact ionization/time-of-flight mass spectrometry (GC-EI/TOF-MS). An Agilent 6890N24 gas chromatograph (Agilent Technologies, Böblingen, Germany) was equipped with a FactorFour VF-5 ms capillary column, 30-m length, 0.25-mm inner diameter, 0.25-μm film thickness (Varian-Agilent Technologies, Böblingen, Germany), which was connected to a Pegasus III time-of-flight mass spectrometer (LECO, St. Joseph, MI, USA). Fifty mg ground rice flour samples were standardised by 30 μl of 0.02 mg ml−1 13C6-sorbitol in water and subjected to a two-step methanol (15 min at 70°C) and chloroform (5 min at 37°C) extraction. The polar metabolite fraction was obtained by liquid partitioning into water/methanol using a final methanol/water/chloroform solvent phase system (330:400:230, v/v/v). 80 μl aliquots of the polar metabolite fraction were dried by vacuum concentration and chemically derivatised by sequential methoxyamination and trimethylsilylation prior to GC-EI/TOF-MS analysis (Allwood et al. 2009). Retention indices (RIs) were calibrated by addition of a C10, C12, C15, C18, C19, C22, C28, C32, and C36 n-alkane mixture to each sample immediately prior to splitless GC injection.
GC-EI/TOF-MS chromatograms were acquired, visually controlled, baseline corrected and exported in NetCDF file format using ChromaTOF software (Version 4.22; LECO, St. Joseph, USA). GC-MS chromatography data were converted into a RI-aligned standardized numerical data matrix using the TagFinder software (Luedemann et al. 2008; Allwood et al. 2009). Compounds were identified within the TagFinder software by mass spectral and RI matching to the reference collection of the Golm Metabolome Database (GMD, http://gmd.mpimpgolm.mpg.de/; Hummel et al. 2010). Guidelines for this manually supervised metabolite identification process were the presence of at least three specific mass fragments per compound and a retention index deviation <1.0% (Strehmel et al., 2008). All mass features of an experiment were normalised to sample weight and to internal standard prior to statistical analysis.
2.3.3 Headspace GC-MS analysis of volatile compounds
Headspace volatiles were collected by SPME using a 65-mm polydimethylsiloxane-divinylbenzene fibre (Supelco, Bellefonte, USA) as described in detail (Verhoeven et al. 2011). The volatile compounds were thermally desorbed at 250°C by inserting the fiber for 1 min into the GC injection port (GC 8000, Fisons Instruments, Cheshire, UK). The released compounds were transferred onto the analytical column (HP-5, 30 m × 0.25 mm ID, 1.05 μm – film thickness) in splitless mode. The temperature program started at 45°C (2-min hold) and rose 5°C min−1 to 250°C (5-min hold). The column effluent was ionised by electron impact (EI) ionisation at 70 eV (MD800 electron impact MS, Fisons Instruments, Cheshire, UK). Mass scanning was done from 35 to 400 m/z with a scan time of 2.8 scans s−1. GC-MS raw data were processed by using MetAlign software (Lommen 2009) to extract and align the mass signals (s/n ≥ 3). Mass signals that were below s/n of 3 were randomized between 2.4 and 3 times the calculated noise value. Mass signals that were present in ≤6 samples were discarded. Signal redundancy per metabolite was removed by means of clustering and mass spectra were reconstructed (Tikunov et al. 2005). Metabolites were identified by matching the mass spectra of obtained metabolites to authentic reference standards and the NIST08, Wiley, and Wageningen Natural compounds spectral library and by comparison with RIs in the literature (Strehmel et al. 2008).
2.3.4 ICP-MS analysis of mineral elements
The ground rice samples were digested in 100 ml closed vessels in a microwave oven (Multiwave 3000, Anton Paar, Graz, Austria) for 50 min at 210°C with a maximum pressure of 40 bar. The digestion medium consisted of 250 mg dry sample, 5 ml 65% ultrapure HNO3 (J.T. Baker Instra-Analysed Reagent) and 5 ml 15% H2O2 (30% Extra-Pure, Riedel de Häen, Selze, Germany). After digestion, the samples were diluted to 3.5% v/v HNO3 with ultrapure water (Milli-Q Element, Millipore, Massachusetts, USA) (Hansen et al. 2009). Multi-elemental analysis was performed using ICP-MS (Agilent 7500ce, Agilent Technologies, Manchester, UK) tuned in standard mode. The plasma power was operated at 1500 W and the argon carrier and make-up gases were set at 0.82 and 0.17 l min−1, respectively. Sample uptake was maintained at approximately 0.6 ml min−1 by a perfluoroalkoxy micro-flow nebulizer. Elimination of spectral interferences was obtained by the use of an octopole ion guide with the cell gasses helium or hydrogen as described previously (Laursen et al. 2009). Seven replicates of certified reference material NIST 8436 (durum wheat, particle size <200 μm; National Institute of Standards and Technology, Gaithersburgh, MD, USA) were included to validate digestion efficiency and analytical accuracy. Only element concentrations deviating less than ±10% from the certified mean reference values were accepted. Limit of detection (LOD) was determined as three times the standard deviation of minimum 7 blanks and only data above LOD was included for chemometrics.
2.4 Association analysis
In order to determine the extent that phenotypic divergence reflects genetic divergence, the genetic distance between each variety was compared to the distance between each genotype cluster from each phenotyping platform. For each set of metabolite or mineral element profiling data, mean scores were calculated for each of the three varieties on the first two principal components of the PCA. Genetic distances between varieties were determined as the Euclidean distance calculated from 1536 SNPs assayed on one plant per variety (GGT 2.0 software (van Berloo 2008), and converted to principal coordinates. Procrustes Analysis was used to rotate and scale the variety mean scores on the metabolite principal components to give the best fit to the genetic principal coordinates. The principal coordinates were then used to generate triangles whereby each vertex of the triangle is a variety, and the length of each side is equal to the scaled Euclidean distance.
3 Results
3.1 Genetic differences between varieties
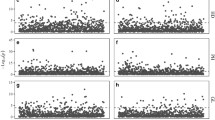
The three varieties studied include an improved indica variety, TSN1, a traditional indica variety, HNN, and KNL which is a traditional variety from the tropical japonica class. Genome-wide genotyping of SNPs at 1536 loci (Fig. 1) shows that the genetic similarity between the traditional and improved indica is about 80%, whereas KNL differs from both the indica varieties by about 50% at these SNP loci. However, it can be seen in Fig. 1b that the differences between KNL and HNN are not at the same loci as the differences between KNL and TSN1. The Euclidean distance between each genotype reflects the genetic similarity and is shown in Table 3.
Genetic differences between the rice varieties. a Genetic variability between TSN1, KNL and HNN rice varieties at 1536 SNP loci across all 12 chromosomes. Different shades of gray indicate differences in nucleic acid. At these loci, the genetic similarity between TSN1 and KNL is 53.2%, and between TSN1 and HNN is 80.7% and between KNL and HNN is 53.6%. b The number of SNP loci on each chromosome (chrom) that are uniquely polymorphic to one variety, when that same locus is monomorphic for the other two varieties
3.2 Discriminating between the quality of each variety
A set of standard quality evaluation data, usually obtained to indicate cooking and sensory quality, was previously determined for each of the three varieties grown in each N treatment (Boualaphanh et al. 2011). The data include tests of gelatinisation temperature; data derived from viscosity curves, in this case breakdown, setback and retrogradation (Fitzgerald et al. 2003); and hardness and stickiness of the cooked grains. Principal Components Analysis (PCA) of this dataset of six quality evaluation variables shows that the first two principal components together explain 98% of the variability. Examination of the score plot (Fig. 2) shows two distinct clusters, but clustering is not on the basis of either genotype or N treatment.
PCA scores plots of data from six routine quality evaluation of grain traits of the three rice varieties cultivated under four different N regimes showing two clusters unrelated to N or genotype. Genotypes: HNN, squares; KNL, circles; TSN1, triangles. N regime: 0 kg N ha−1, unfilled; 30 kg N ha−1, light grey; 60 kg N ha−1, dark grey; 90 kg N ha−1, black
3.3 Discriminating between the grain metabolome of each variety
Four metabolite or elemental profiling technologies were applied to the analysis of polished grains from the three varieties grown under different nitrogen fertiliser regimes in order to try and separate the varieties, and find compounds and minerals that could explain the known flavour differences.
First, 14 mineral elements detected by Inductively Coupled Plasma Mass Spectroscopy (ICP-MS) were quantified in the polished grains of each variety from each N treatment. Observation of the PCA scores plot shows that all samples of KNL formed one distinct cluster, though significant overlap was seen between HNN and TSN1 (Fig. 3a). PC1 explains 47% of the variation and PC2 explains 21%. No clustering or sub-clustering was seen on the basis of N treatment. Several macro- and micronutrient minerals differed significantly between genotypes, with most significant differences between KNL and the other two varieties (Table SI).
Phenotypic variability between rice varieties. a Mineral nutrients determined by ICP-MS, (b) polar metabolite profiling with 1H-NMR fingerprints, (c) polar metabolites determined by GC-TOF-MS, (d) volatiles determined by SPME GC-MS. Genotypes: HNN, squares; KNL, circles; TSN1, triangles. N regime: 0 kg N ha−1, unfilled; 30 kg N ha−1, light grey; 60 kg N ha−1, dark grey; 90 kg N ha−1, black
PCA analysis of proton Nuclear Magnetic Resonance (1H-NMR) signatures of polar extracts showed that the samples clustered on the basis of genotype (Fig. 3b). The major metabolites contributing to separation of TSN1 along PC1 were sucrose and raffinose on the negative side and mainly lipids, betaine and choline on the positive side (Table 1). PC2, however, explaining 18% of the total variability, separated KNL and HNN from TSN1 (Fig. 3b). The major metabolites contributing to this separation along PC2 were choline on the negative side and lipids on the positive side (Tables 1, SII). A clear separation between the lowest and highest N levels in the PC1 × PC2 plane could be observed within the TSN1 cluster, but the effect of N was not clearly seen for the other two genotype clusters (Fig. 3b). The relative intensities of selected spectra regions corresponding to the compounds discriminating the three varieties in Table 1 and their ANOVA data are listed in Table SII.
Since detection of primary compounds is more sensitive by gas chromatography-electro impact-time of flight mass spectroscopy (GC-EI-TOF-MS) than 1H-NMR, the data in Fig. 3b were complemented with GC-EI-TOF-MS of polar extracts. PCA was performed using 43 variables, and samples of each variety clustered together (Fig. 3c). PC1 accounted for 50% of the variation, and along that axis, samples of KNL were separated from those of TSN1 and HNN. The PC2 axis, accounting for 25% of the variation, separated the TSN1 samples from the other two varieties (Fig. 3c). ANOVA showed that the main compounds driving the separation along PC1 were cysteine, 5-oxoproline, ribonic acid, glycerol, threonine, and putrescine, tyrosine and trehalose (Table 1, SIII). The major metabolites that separated TSN1 along the PC2 axis were threitol, arabinonic acid, proline, azelaic acid, glycerol, fumarate, and allantoin (Table 1, SIII). Many of the other compounds leading to the separation along the PC axes were identified as sugar alcohols and hexose sugars (Table 1). A selection of the most discriminating compounds and the ANOVA data are shown in Table SIII.
Volatile compounds are also major determinants of taste and flavour and these were determined for each variety using GC-MS. The PCA scores plot shows that volatile compounds determined by solid phase micro extraction (SPME) GC-MS clearly separated the three varieties (Fig. 3d). PC1 explained 34% of total variability, and TSN1 was separated along the PC1 axis from HNN and KNL. PC2 explained 19% of total variability and clearly separated the samples of HNN from the other two varieties (Fig. 3d). Four significant principal components were extracted, cumulatively explaining 77% of variation. The three varieties were separated by 54 compounds (Table 2), though not all could be identified by the spectral libraries. The major discriminating compound was 2 acetyl-1-pyrroline (2AP). TSN1 had undetectable levels of 2AP while the other two varieties both contained 2AP. The elevated concentration of 2AP in KNL contributed to the separation of those samples from HNN. HNN grains are characterised by ketones, KNL grains by 2AP and several alcohols, and TSN1 grains, by only three volatiles compounds that could be identified, such as butane 2,3 diol (Table 2) and twelve that could not. Interestingly, several of the discriminating volatile compounds unique to each variety have quite low odour thresholds and unique flavour notes (Table 2), and the relative importance of each is shown in Table SIV.
3.4 Relating the biochemical phenotype of the grain with the genome
For each metabolite and mineral element profiling platform, Euclidean distances between varieties were calculated from Fig. 3, using the first two principal components (Table 3). Based on 1536 SNP loci of the three varieties, the genetic distance between HNN and TSN1 was smallest, and was similar between KNL and HNN and between KNL and TSN1 (Table 3). The residual sum of squares from the Procrustes rotation of the association between Euclidean distances based on the genome and metabolome or ionome shows that each biochemical profile associates with genotype (Table 3). Comparing the triangles generated from each set of normalised coordinates shows that mineral elements, polar metabolites and volatile compounds all associate very well with the genetic distance between each (Fig. 4). All platforms showed a smaller distance between HNN and TSN1, and a larger distance between KNL and the other two varieties.
Association between genetic and phenotypic differences between rice varieties. Comparison of standardised coordinates of the Procrustes rotation (Table 3) of Euclidean distances separating genotypes in Fig. 1a and the metabolomic and elemental data from Fig. 3, plotted as triangles showing excellent agreement between genotype and all metabolic profiles of each variety
4 Discussion
In many rice-consuming countries, flavour of the rice is considered just as important as texture (Schiller et al. 2006). For example, the rices used in this study differ in taste (Bounphanousay 2007), and the two traditional varieties (HNN and KNL) command a 20% price premium in Lao markets over TSN1. Standard tools used to measure grain quality could not differentiate the three varieties (Fig. 2), indicating a need to bring new science to rice quality. In the present study, a unique combination of analytical metabolomics and elemental profiling platforms has been used to determine volatile and polar primary metabolites and minerals in polished grains. The data obtained were analysed to determine if (i) unique profiles existed for each variety that might explain the differences in flavour, (ii) N fertiliser affected these profiles, and (iii) associations could be found between the metabolome of each variety and genotype.
The three varieties were well separated by all metabolite and elemental platforms (Fig. 3a–d), suggesting that each variety has a unique metabolite signature. Most of the detected primary polar metabolites were present in all three varieties (Table 1), but at differing levels, so discrimination between the varieties from these platforms is mainly quantitative, which is possibly expected with primary metabolism. By contrast, Table 2 shows that particular volatile compounds were often unique to each variety, suggesting that these contribute more than the polar primary metabolites to the uniqueness of each metabolic signature.
No consistent effect of N regime was seen for any of the datasets (Figs. 2, 3a–d), and in all cases, clustering was based on genotype. Some association with N was seen within the TSN1 genotype cluster for volatile and primary compounds, but not for the other two varieties. Previous data on the same samples (Boualaphanh et al. 2011) are consistent with these findings. The yield components mostly affected by N were increased tiller number and grain number per panicle, and no change in grain weight or protein content (Boualaphanh et al. 2011). Traditional varieties are not usually responsive to N, so it is therefore unsurprising that N did not induce consistent variation in metabolites.
The three varieties show distinct genetic differences (Fig. 1a). The two indica varieties share the most genetic similarity, but both indica varieties share only about 50% homology with the tropical japonica variety, though the differences are not at the same loci (Fig. 1b). These genetic differences are consistent with known groupings within domesticated rice (Garris et al. 2005). Table 3 and Fig. 4 show that metabolomes and the ionome associated excellently with genetic distances, indicating that (i) discriminating compounds follow genetic differences between germplasm classes, (ii) sufficient polymorphic loci existed to associate with metabolites, and (iii) metabolite data from each platform can be associated with mapping populations, using discriminating SNPs to find candidate loci. In the future, this will enable discovery of the genes responsible for the synthesis of important taste, flavour and nutritional compounds, such as those detected in the present paper.
4.1 Nutritionally valuable compounds
Many of the metabolites that were detected in the milled grains have attributed nutritional benefits (Table 1). Examples are GABA, found in all varieties, which is involved in a host of cortical functions in mammals (Sanacora et al. 1999), and is involved in signalling and osmotic regulation in plants (Bouché and Fromm 2004). Choline was detected in all three varieties, and is a methyl donor involved in brain health and liver function (Zeisel et al. 1991). Glucosamine was found in HNN and is known to contribute to the formation of cartilage and is used to alleviate and prevent pain due to arthritis and bone stress (Hughes and Carr 2002). Azelaic acid, found in HNN, is an organic compound that contributes to the health of hair and skin (Nguyen and Bui 1995). A significant amount of lysine was detected in extracts of TSN1, and none in the other two (Table 1, SIII). Lysine, an essential amino acid, has multiple physiological functions (Baker 2005). Cereal proteins are low in lysine, and efforts to increase lysine centre around altering the balance of storage proteins that accumulate (Shih 2004) but perhaps non-protein lysine, such as that found in TSN1, reveals an additional mechanism for increasing lysine in cereals.
Improving the amount of Zn in polished rice is the subject of a large international effort under Harvestplus (www.harvestplus.com) to address hidden hunger. In the present study, the higher concentration of Zn in KNL discriminated it from the other two varieties (Table SI). Consistent with this, KNL was found to have high Zn content in a study using 56 Lao varieties (Bounphanousay 2007). The concentration of Zn in KNL approaches the ambitious targets set by Harvestplus. Consequently, KNL would be a useful starting point for further varietal improvement where sensory properties are to be combined with enhanced nutritional properties.
All three varieties contain hexose sugars such as tagatose and psicose, and sugar alcohols such as xylitol and erythritol (Table 1). These sweeteners do not induce a blood sugar response, which could explain some of the variability found in the glycaemic index of different rices (Wheeler and Pi-Sunyer 2008) and be useful in the management of diseases like Type 2 diabetes (Association 2004).
It has generally been assumed that polished rice provides mostly calories, and that the bran layer contains all the compounds with potential nutritional benefit (Butsat and Siriamornpun 2010; Shen et al. 2009; Heinemann et al. 2008; Yu et al. 2007; Sakamoto et al. 2007; Sharif and Butt 2006; Yokoyama 2004). This is the first demonstration of the extensive biochemical diversity that occurs in polished rice grains (Tables 1, 2), many of which have a role in human physiology and health.
4.2 Compounds of taste and flavour
Consumers have difficulty describing the taste of rice, so testing for taste is generally not part of rice quality programs (Fitzgerald et al. 2009; Champagne et al. 2010). However, a recent sensory study, using popular varieties grown throughout South and Southeast Asia, revealed a characteristic flavour profile for those varieties commonly grown and consumed in Southeast Asia consisting of sweet, floral, grassy and dairy notes (Champagne et al. 2010). KNL and HNN were found to contain polar and volatile compounds that give sweet, floral, fruity and grassy flavours (Tables 1, 2). HNN contained a number of ketones, many of which are used commercially in low concentrations to impart fruity, nutty, floral and butter/dairy aromas and flavours in foods. KNL contained compounds that are commercially used in low concentrations to impart grassy fresh aroma notes. Several of the volatile compounds detected have an odour threshold detectable by humans (Table 2), based on the detection limit of the GC-MS. The strongest of these is 2AP, but some of the other compounds also have low odour thresholds (Table 2). KNL and HNN can be differentiated by consumers in taste trials (Bounphanousay 2007), and the data presented here suggest that each has different compounds that could contribute to flavours that humans can detect. They both also contain unpleasant compounds (off-flavours) such as putrescine, which was first found in putrefying flesh (Olle 1986), but which is considered to be a precursor in the pathway of aroma in rice (Bradbury et al. 2008). Putrescine was not found in TSN1 (Table 1), consistent with a possible role in 2AP synthesis in the two fragrant rices (Bradbury et al. 2008). However TSN1 contained the highest levels of succinate, fumarate and malate (Table 1), which could indicate oxidation of lipids, leading to aromas of rancidity.
Further analyses are required to correlate the current findings with the quality of cooked rice. The presence of amino acids and reducing sugars among the primary metabolites in the raw grains of each variety leads to the possibility that Maillard reaction products may also be produced during cooking (De Kimpe and Keppens 1996) leading to additional contributions to flavour. Cysteine was found in KNL (Table 1), and this amino acid is an efficient precursor of α-acetyl N heterocycles, which have a very low odour threshold and contribute to the roasted aroma of cooked rice (Kerler et al. 1997).
Three volatile compounds found in TSN1 could be identified (Table 2), and another 12 could not, and this was also found for the other two varieties. Therefore more research is needed in order to build a more complete picture of flavour and fragrance of rice. Nevertheless, this study is the first to show that metabolomic profiling can be a valuable new tool which leads to unique signatures. The generic nature of the approaches also means that such knowledge is not just restricted to the varieties used here. Instead our study shows feasibility and benefit of metabolomic analyses from which all rice and crop improvement programs should profit. The identification of important compounds in rice should be the subject of further work tuned to find genes that lead to their synthesis and accumulation. An expanded palette of both DNA and chemical markers for such compounds would greatly enhance the capacity of rice improvement programs to select actively for delicious and nutritious rice.
References
Allwood, J. W., Erban, A., de Koning, S., Dunn, W., Luedemann, A., Lommen, A., et al. (2009). Inter-laboratory reproducibility of fast gas chromatography-electron impact-time of flight mass spectrometry (GC-EI-OF/MS) based plant metabolomics. Metabolomics, 5, 479–496.
Association, A. D. (2004). Nutrition principles and recommendations in diabetes. Diabetes Care, 27(suppl 1), s36. doi:10.2337/diacare.27.2007.S36.
Baker, D. H. (2005). Comparative nutrition and metabolism: Explication of open questions with emphasis on protein and amino acids. Proceedings of the National Academy of Sciences, 102(50), 17897–17902. doi:10.1073/pnas.0509317102.
Boualaphanh, C., Calingacion, M., Jothityangkoon, D., Sanitchon, J., Cuevas, R., & Fitzgerald, M. (2011). Yield and quality of non-aromatic and aromatic Lao rice in response to nitrogen fertilizer. Science Asia, 37, 89–97. doi:10.2306/scienceasia1513-1874.2011.37.089.
Bouché, N., & Fromm, H. (2004). GABA in plants: Just a metabolite? Trends in Plant Science, 9(3), 110–115.
Bounphanousay, C. (2007). Use of phenotypic characters and DNA profiling for classification of the genetic diversity in black glutinous rice of the Lao PDR. Thailand: Khon Kaen University.
Bounphanousay, C., Appa Rao, S., Kanyavong, K., Sengthong, B., Schiller, J. M., & Jackson, M. T. (2000). Conservation of Lao rice germplasm at the International Rice Genebank, Part 5 (p. 22). Los Banos, Philippines: IRRI.
Bradbury, L., Gillies, S., Brushett, D., Waters, D., & Henry, R. (2008). Inactivation of an aminoaldehyde dehydrogenase is responsible for fragrance in rice. Plant Molecular Biology, 68(4), 439–449.
Butsat, S., & Siriamornpun, S. (2010). Antioxidant capacities and phenolic compounds of the husk, bran and endosperm of Thai rice. Food Chemistry, 119(2), 606–613.
Champagne, E. T., Bett-Garber, K. L., Fitzgerald, M. A., Grimm, C., Lea, J., Ohtsubo, K., et al. (2010). Important sensory properties differentiating premium rice varieties. Rice, 3, 270–281.
Cuevas, R., Daygon, V., Corpuz, H., Waters, D. L. E., Reinke, R. F., & Fitzgerald, M. A. (2010). Melting the secrets of gelatinisation temperature. Functional Plant Biology, 37, 439–447.
De Kimpe, N., & Keppens, M. (1996). Novel syntheses of the major flavor components of bread and cooked rice. Journal of Agricultural and Food Chemistry, 44(6), 1515–1519.
Dodsworth, J. (1997). How Indochina’s economies took off. Finance and Development, 34(1), 20–23.
Eriksson, L., Johansson, E., Kettaneh-Wold, N., Wikström, C., Trygg, J., & Wold, S. (2006). Multi- and megavariate data analysis; part I: Basic principles and applications (2nd ed.). Umea, Sweden: Umetrics Academy.
Fitzgerald, M. A., Martin, M., Ward, R. M., Park, W. D., & Shead, H. J. (2003). Viscosity of rice flour: A rheological and biological study. Journal of Agricultural and Food Chemistry, 51(8), 2295–2299.
Fitzgerald, M. A., McCouch, S. R., & Hall, R. D. (2009). More than just a grain of rice, the global quest for quality [Review]. Trends in Plant Science, 14(3), 133–139. doi:10.1016/j.tplants.2008.12.004.
Garris, A. J., Tai, T. H., Coburn, J., Kresovich, S., & McCouch, S. R. (2005). Genetic structure and diversity in Oryza sativa L. Genetics, 169, 1631–1638.
Hall, R. D. (2006). Plant metabolomics: From holistic hope, to hype, to hot topic. New Phytologist, 169(3), 453–468.
Hall, R. D., Brouwer, I. D., & Fitzgerald, M. A. (2008). Plant metabolomics and its potential application for human nutrition [Review]. Physiologia Plantarum, 132, 162–175. doi:10.1111/j.1399-3054.2007.00989.x.
Hansen, T., Laursen, K., Persson, D., Pedas, P., Husted, S., & Schjoerring, J. (2009). Micro-scaled high-throughput digestion of plant tissue samples for multi-elemental analysis. Plant Methods, 5, 12.
Heinemann, R. J. B., Xu, Z., Godber, J. S., & Lanfer-Marquez, U. M. (2008). Tocopherols, tocotrienols, and γ-oryzanol contents in japonica and indica subspecies of rice (Oryza sativa L.) cultivated in Brazil. Cereal Chemistry, 85(2), 243–247.
Hughes, R., & Carr, A. (2002). A randomized, double-blind, placebo-controlled trial of glucosamine sulphate as an analgesic in osteoarthritis of the knee. Rheumatology, 41(3), 279–284. doi:10.1093/rheumatology/41.3.279.
Hummel, J., Strehmel, N., Selbig, J., Walther, D., & Kopka, J. (2010). Decision tree supported substructure prediction of metabolites from GC-MS profiles. Metabolomics, 6, 322–333.
Hyten, D. L., Smith, J. R., Frederick, R. D., Tucker, M. L., Song, Q., & Cregan, P. B. (2009). Bulked segregant analysis using the GoldenGate Assay to locate the Rpp3 locus that confers resistance to soybean rust in soybean. Crop Science, 49(1), 265–271. doi:10.2135/cropsci2008.08.0511.
Inthapanya, P., Boualaphan, C., Hatsadong, & Schiller, J. M. (2006). The history of lowland rice variety improvements in Laos. In J. M. Schiller, M. B. Chanphengxay, B. Linquist, & S. Appa Rao (Eds.), Rice in Laos (pp. 325–358). Manila: IRRI.
Kerler, J., Vander Ven, J. G. M., & Weenen, H. (1997). α-Acetyl-N-heterocycles in the Maillard reaction [Review]. Food Reviews International, 13(4), 553–575.
Laursen, K., Hesselhøj Hansen, T., Persson, D., Schjoerring, J., & Husted, S. (2009). Multi-elemental fingerprinting of plant tissue by semi-quantitative ICP-MS and chemometrics. Journal of Analytical Atomic Spectroscopy, 24, 1198–1207.
Lee, G.-A., Koh, H.-J., Chung, H.-K., Dixit, A., Chung, J.-W., Ma, K.-H., et al. (2009). Development of SNP-based CAPS and dCAPS markers in eight different genes involved in starch biosynthesis in rice. Molecular Breeding, 24(1), 93–101.
Lommen, A. (2009). MetAlign: Interface-driven, versatile metabolomics tool for hyphenated full-scan mass spectrometry data preprocessing. Analytical Chemistry, 81(8), 3079–3086.
Luedemann, A., Strassburg, K., Erban, A., & Kopka, J. (2008). TagFinder for the quantitative analysis of gas chromatography—mass spectrometry (GC-MS) based metabolite profiling experiments. Bioinformatics, 24(5), 732–737.
Moing, A., Maucourt, M., Renaud, C., Gaudillère, M., Brouquisse, R., Lebouteiller, B., et al. (2004). Quantitative metabolomic profiling by 1-dimensional 1H-NMR analyses: Application to plant genetics and functional genomics. Functional Plant Biology, 31, 889–902.
Newton-Cheh, C., & Hirschhorn, J. N. (2005). Genetic association studies of complex traits: Design and analysis issues [Review]. Mutation Research-Fundamental and Molecular Mechanisms of Mutagenesis, 573(1–2), 54–69.
Nguyen, Q. H., & Bui, T. P. (1995). Azelaic acid—pharmacokinetic and pharmacodynamic properties and its therapeutic role in hyperpigmentary disorders and acne. International Journal of Dermatology, 34(2), 75–84.
Olle, H. (1986). Putrescine, spermidine, and spermine. Physiology, 1(1), 12–15.
Raghavan, C., Naredo, M. E. B., Wang, H., Atienza, G., Liu, B., Qiu, F., et al. (2007). Rapid method for detectin SNPs on agarose gels and its application in candidate gene mapping. Molecular Breeding, 19(2), 87–101. doi:10.1007/s11032-00609046-x.
Reyes, R. A. (1997). The role of the state in Laos’ economic management. In Laos’ dilemmas and options (pp. 48–60). Vientiane: Institute of Southeast Asian Studies.
Sakamoto, S., Hayashi, T., Hayashi, K., Murai, F., Hori, M., Kimoto, K., et al. (2007). Pre-germinated brown rice could enhance maternal mental health and immunity during lactation. European Journal of Nutrition, 46(7), 391–396.
Sanacora, G., Mason, G. F., Rothman, D. L., Behar, K. L., Hyder, F., Petroff, O. A. C., et al. (1999). Reduced cortical {gamma}-aminobutyric acid levels in depressed patients determined by proton magnetic resonance spectroscopy. Archives of General Psychiatry, 56(11), 1043–1047. doi:10.1001/archpsyc.56.11.1043.
Schiller, J. M., Appa Rao, S., Inthapanya, P., & Hatsadong, (2006). Glutinous rice in Laos. In J. M. Schiller, M. B. Chanphengxay, B. Linquist, & S. Appa Rao (Eds.), Rice in Laos (pp. 197–214). Los Banos, Philippines: International Rice Research Institute.
Sharif, K., & Butt, M. S. (2006). Preparation of fiber and mineral enriched pan bread by using defatted rice bran. International Journal of Food Properties, 9(4), 623–636.
Shen, Y., Jin, L., Xiao, P., Lu, Y., & Bao, J.-S. (2009). Total phenolics, flavonoids, antioxidant capacity in rice grain and their relations to grain color, size and weight. Journal of Cereal Science, 49(1), 106–111.
Shih, F. F. (2004). Rice Proteins. In E. T. Champagne (Ed.), Rice chemistry and technology (3rd ed., pp. 143–157). St Paul, Minnesota: American Association of Cereal Chemists Inc.
Shirasawa, K., Shiokai, S., Yamaguchi, M., Kishitani, S., & Nishio, T. (2006). Dot-blot-SNP analysis for practical plant breeding and cultivar identification in rice. Theoretical and Applied Genetics, 113(1), 147–155.
Strehmel, N., Hummel, J., Erban, A., Strassburg, K., & Kopka, J. (2008). Retention index thresholds for compound matching in GC-MS metabolite profiling. Journal of Chromatography, 871, 182–190.
Tarpley, L., & Roessner, U. (2007). Metabolomics: Enabling systems-level phenotyping in rice functional genomics. In N. M. Upadhyaya (Ed.), Rice functional genomics: Challenges, progress and prospects (pp. 91–107). New York: Springer.
Tikunov, Y., Lommen, A., de Vos, C. H. R., Verhoeven, H. A., Bino, R. J., Hall, R. D., et al. (2005). A novel approach for nontargeted data analysis for metabolomics. Large-scale profiling of tomato fruit volatiles. Plant Physiology, 139(3), 1125–1137. doi:10.1104/pp.105.068130.
van Berloo, R. (2008). GGT 2.0: Versatile software for visualization and analysis of genetic data. Journal of Heredity, 99(2), 232–236. doi:10.1093/jhered/esm109.
Verhoeven, H. A., Jonker, H. H., de Vos, R. C. H., & Hall, R. D. (2011). Solid-phase micro-extraction (SPME) GC-MS analysis of natural volatile components in melon and rice. In N. G. Hardy, & R. D. Hall (Eds.), Plant metabolomics methods. Ithaca: Humana Press (in press).
Wheeler, M. L., & Pi-Sunyer, F. X. (2008). Carbohydrate issues: Type and amount. Journal of the American Dietetic Association, 108(4), S34–S39.
Wright, M., Tung, C.-W., Zhao, K., Reynolds, A., McCouch, S. R., & Bustamante, C. D. (2010). ALCHEMY: A reliable method for automated SNP genotype calling for small batch sizes and highly homozygous populations. Bioinformatics (Oxford), 26(23), 2952–2960. doi:10.1093/bioinformatics/btq533.
Yokoyama, W. H. (2004). Nutritional properties of rice and rice bran. In E. T. Champagne (Ed.), Rice chemistry and technology (pp. 595–606). Minneapolis: American Association of Cereal Chemists, Inc.
Yu, S., Nehus, Z. T., Badger, T. M., & Fang, N. (2007). Quantification of vitamin E and y-oryzanol components in rice germ and bran. Journal of Agricultural and Food Chemistry, 55, 7308–7313.
Zeisel, S. H., Da Costa, K. A., Franklin, P. D., Alexander, E. A., Lamont, J. T., Sheard, N. F., et al. (1991). Choline, an essential nutrient for humans. The FASEB Journal, 5(7), 2093–2098.
Zhao, K., Wright, M., Kimball, J., Eizenga, G., McClung, A., Kovach, M., et al. (2010). Genomic diversity and introgression in O. sativa reveal the impact of domestication and breeding on the rice genome. PLoS ONE, 5(5), e10780. doi:10.1371/journal.pone.0010780.
Acknowledgments
All the authors acknowledge funding from the EU for the project META-PHOR (FOOD-CT-2006-036220) which was part of FP VI. RDH, RCHV and RM acknowledge additional support from the Centre for BioSystems Genomics, an initiative under the auspices of the Netherlands Genomics Initiative. MAF acknowledges additional support from the International Rice Research Institute (IRRI). CB acknowledges support from IRRI, KKU and NAFRI. University of Copenhagen acknowledges additional support from HarvestPlus to the Danish Plant Research Network entitled ‘Supporting the Foundation for Iron and Zinc Biofortification in Beans, Rice and Wheat’. Bordeaux Metabolome Facility acknowledge additional support from IBiSA and INRA CNOC, and Daniel Jacob for developing and maintaining the MeRy-B database.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Calingacion, M.N., Boualaphanh, C., Daygon, V.D. et al. A genomics and multi-platform metabolomics approach to identify new traits of rice quality in traditional and improved varieties. Metabolomics 8, 771–783 (2012). https://doi.org/10.1007/s11306-011-0374-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11306-011-0374-4