Abstract
Metabolomic analysis of the urinary organic acids from 39 selected children with defined respiratory chain deficiencies (RCDs) was performed using untargeted gas chromatography–mass spectrometry, revealing the presence of 255 endogenous and 46 exogenous substances. Variable reduction identified 92 variables from the endogenous substances, which could be analysed by univariate and multivariate statistical methods. Using these methods, no characteristic organic acid biomarker profile could be defined of practical value for diagnostic purposes for complex I (CI), complex III (CIII) and multiple complex (CM) deficiencies. The statistical procedures used did, however, disclose 24 metabolites that were practical highly (d > 0.75) and statistically (P < 0.05) significant for the combined and clinically closely related group of RCDs. Several of these metabolites occur in single enzyme inherited metabolic diseases, but most were not previously reported to be linked to the metabolic perturbations that are due to RCDs. Ultimately, we constructed a global metabolic profile of carbohydrate, amino acid and fatty acid catabolism, illuminating the diverse and complex biochemical consequences of these disorders. This metabolomics investigation disclosed a metabolite profile that has the potential to define an extended and characteristic biosignature for RCDs and the development of a non-invasive screening procedure for these disorders.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Impaired energy metabolism, due to genetically based dysfunction of one or more components of the mitochondrial oxidative phosphorylation (OXPHOS) system, underlies the pathology of what are generally referred to as mitochondrial disorders (MD) in humans. The OXPHOS system consists of several multimeric and heterologous enzyme complexes, designated complex I to IV (respiratory chain, RC) and complex V (F-type ATP synthase, ATPase), which are encoded by nuclear as well as mitochondrial DNA (reviewed by Anderson et al. 1981). Although the RC primarily supports the transport of electrons from reducing cofactors (e.g. NADH2 and FADH2,), metabolic consequences of RC deficiencies (RCDs) include increased concentrations of organic acids (Munnich et al. 1992; Esteitie et al. 2005) and perturbed amino acid profiles (Rabier et al. 1998) in blood and cerebrospinal fluid. These result in a diverse profile of organic acids and amino acids excreted in the urine. Unlike many other inherited metabolic disorders due to single enzyme deficiencies, however, there are no known specific diagnostic biochemical biomarkers for disorders involving RC deficiencies (Suomalainen 2010; Haas et al. 2008; Mancuso et al. 2009). As with all inherited MDs there is no cure for these deficiencies, and the variety of treatments proposed over time for different RCDs may or may not be effective (Kurlemann et al. 1995). Several scoring systems, which include some of the RCD-associated biochemical markers in body fluids as minor criteria, have been developed to assist in the diagnosis of these disorders (Bernier et al. 2002; Wolf and Smeitink 2002; Morava et al. 2006). However, the diagnostic process is still complicated and tedious. The organic acid profile from suspected cases for a MD provides only information for a tentative indication of RCDs. A complete and reliable diagnosis of RCDs requires costly and invasive procedures and relies almost exclusively on enzyme analyses on muscle biopsy material and/or molecular genetics analyses. The initial diagnosis of patients with suspected MDs would thus greatly benefit from the identification of biochemical biomarkers in an easily obtainable specimen as a useful decision-guiding instrument.
Barshop (2004) attempted to refine the metabolic markers for MDs by means of a correlation study of a retrospective analysis of 3646 randomly selected urine samples referred to a laboratory for diagnostic purposes. The metabolomic profile from this investigation confirmed that abnormalities in the organic acid profiles are a feature of most MDs and provided further quantitative and qualitative insights into these disorders. Although somewhat limited for diagnostic purposes, elevated fumaric and malic acids were found to be the most useful in distinguishing patients with a mitochondrial deficiency from those with the organic acidaemias. Barshop, however, noted that further refinements were required to develop a characteristic metabolomic profile and may improve the use of quantitative organic acid analysis in MDs. More recently, Mancuso et al. (2009) remarked that the ideal biomarker for mitochondrial disorders should improve on the timing and accuracy of the diagnosis. Furthermore, it should contribute to limiting the number of invasive diagnostic procedures and ultimately be useful for monitoring the effect of drug treatment and disease progression (Mancuso et al. 2009). In this regard, the use of urine samples provides minimal invasiveness and several practical advantages, aspects which are of great importance where improved screening using metabolic data in the diagnostic process is an imperative. The availability of urine samples from a relatively large group of clinically and biochemically well-defined South African patients with an enzymatically confirmed respiratory chain disorder (Smuts et al. 2010) presented a unique opportunity to address these issues through a metabolomics investigation.
For this investigation we asked the question whether a metabolomics approach, using an organic acid profile of urine, could expand the existing information on the metabolites elevated due to an RCD, contribute to the better understanding of the perturbations due to the RCD, and provide a critical first step towards the future development of biomarkers for RCDs. Urine was selected as the source for data generation as it is a primary option in clinical metabolic investigations and because variations in several urinary organic acids in RCDs have been reported. A significant aspect of the cohort of RCD patients we investigated was the availability of enzyme activity data of the RC complexes I–IV, measured in muscle biopsy material. This opened up the possibility of determining if the origin of these elevated metabolites could be explained in light of the well-defined, but wide-ranging, biochemical disturbances associated with RCDs. To explore these questions, the organic acid-containing section of the metabolome from RCD patients and controls was investigated, and the organic acid profile was determined by untargeted gas chromatographic–mass spectrometric (GC–MS) analysis. Metabolic profiling depends on a clear understanding of the metabolic pathways of the subsection of the metabolome in question and the analytical methods and technologies used for the analyses (Dunn and Ellis 2005). As reported here, with the metabolomics approach followed, we succeeded in producing the first comprehensive list of statistically and practically significant elevated levels of several metabolites that have been previously described for RCDs, as well as increased levels of several metabolites not previously associated with these disorders. Our procedure also enabled the construction of a model of biochemical pathways indicating the putative origin of this complex array of metabolites from various, but related, catabolic pathways. This also paved the way for the development of possible biosignature(s) for RCDs.
2 Materials and methods
2.1 Sample selection
Ethical approval for the study was obtained from the relevant Ethics Committees of the University of Pretoria (No. 91/98 and amendments) and North-West University (No. 02M02). Informed consent was obtained from the parents of patients and controls for the use of the urine samples and biopsy material (where applicable) of their children for research purposes.
The urinary samples were obtained from patients referred to the Paediatric Neurology Unit at the Steve Biko Academic Hospital, Pretoria, South Africa. The original experimental group consisted of 101 clinically selected patients, including the cohort of South African patients as described by Smuts et al. (2010). The mean age of the original patient group was 5 years, ranging from 1 to 25 years of age. To improve a case–control comparison, age as a first criterion for exclusion was used. Only data for patients of 13 years or younger, i.e. by definition a paediatric group, were initially selected for this investigation. Of these patients, 48 were identified with an RC enzyme deficiency (see Sect. 2.3). These patients constituted the group used for the metabolomics investigation (see Fig. 1) and were not further sub-divided on clinical criteria, like hypotonia or central nervous system presentations. The 92 controls were selected from a large group of 139 children referred to the clinic, but for whom no prevailing disorder could be detected. This known complexity of obtaining comparative control groups in investigations of inherited metabolic disorders is generally recognized (Barshop 2004). No match on race and gender could be achieved between the controls and patient groups, but there are no reports that indicate race or gender as confounding factors for rather comparable age groups as selected by us. The urinary organic acid profile does seem to vary with age, although little is known about age-specific changes or circadian rhythms of urinary organic acids reflecting metabolic changes and/or maturation of tubular function and how this might influence diagnosis of disorders of organic acids (Hoffman and Feyh 2005). The reference values given by Hoffman and Feyh (2005), never the less classify them according to four age groups: (1) Term newborns (>36 weeks), (2) children (<5 years), (3) children (>5 years) and (4) adults. A common practice followed in the Human Metabolomic Data Base is to classify urinary organic acids according to (1) newborns, (2) children (1–13 years) and (3) adults. From our experience with the present and other studies, the age difference between the control (mean = 4.6; s.d. = 2.7) and patient groups (mean = 8.9; s.d. = 2.9) as used here, does not produce a confounding factor.
2.2 Enzyme analyses and sample collection in the patient group
The enzyme analyses were performed on muscle biopsies from the vastus lateralis muscle of all patients complying with the Mitochondrial Disease Criteria (Wolf and Smeitink 2002). The activity of the following enzymes was determined to identify possible RC deficiencies: complex I to IV (EC 1.6.5.3, EC 1.3.5.1, EC 1.9.2.2 and EC 1.9.3.1, respectively) and citrate synthase (CS, EC 2.3.3.1), as a marker for mitochondrial content, using standard operating procedures based on existing methods (Shepherd and Garland 1969; Rahman et al. 1996; Janssen et al. 2007). Pyruvate dehydrogenase complex (PDHc, EC 1.2.4.1) was measured using the pyruvate dehydrogenase (PDH) Enzyme Activity Dipstick Assay Kit (MitoSciences, Eugene, OR). PDHc activity was measured to determine if any of the selected cases had this related deficiency. The criteria used for identification of an enzyme deficiency in this case cohort were somewhat more stringent than originally described by Smuts et al. (2010). Here two criteria were invoked to decide if a respiratory chain enzyme deficiency was present: the one was that the activity value should be lower than the lowest control value when expressed on CS; the other was that the activity also had to be less than at least the 5th percentile of the control group when expressed against either CII or CIV, if not deficient (if both were deficient only the first criterion was applied). As summarized in Table 1, using these criteria, 39 of the 48 paediatric patients complied with a muscle deficiency of complex I (CI), complex III (CIII) or a multiple deficiency of more than one RC enzyme (CM).
The urine specimen for metabolomics analysis was obtained at the time when the muscle biopsy was performed; the patients did not receive any specific treatment or supplements often given to patients with MD. Use of anti-convulsants was not stopped.
2.3 Acquisition of metabolite data
The organic substances were isolated from the urine, and consisted mostly of organic acids, derivatized and separated by gas chromatography according to a standardized technique described below, which we have refined since our first description of a South African organic aciduria case (Erasmus et al. 1985). This is also the procedure that we use in our participation in the quality assurance programme of the European research network for evaluation and improvement of screening, diagnosis and treatment of inherited disorders of metabolism (ERNDIM, www.erndim.unibas.ch). The semi-quantitative identification of the organic acids was conducted according to Chen et al. (2009). All organic acids identified above the detection limit of the equipment used were expressed as mmol per mol creatinine.
2.3.1 Extraction and derivatization of organic acids
Urine samples of approximately 20 ml were obtained from patients and controls. The samples were frozen at −80°C upon delivery and transported to the laboratory on dry ice. The samples were thawed at room temperature and an aliquot was prepared for creatinine and organic acid analysis, respectively. The volume of urine used for organic acid analysis was based on urinary creatinine values, according to the following guidelines:
-
Creatinine values higher than 8.8 mmol/l: 0.5 ml of urine.
-
Creatinine values lower than 8.8 mmol/l and higher than 0.44 mmol/l: 1 ml of urine.
-
Creatinine values lower than 0.44 mmol/l and higher than 0.18 mmol/l: 2 ml of urine.
-
Creatinine values lower than 0.18 mmol/l: 3 ml of urine.
Urine samples were transferred to silanized glass tubes (Kimax) and the internal standard (3-phenylbutyric acid, Sigma Chemical Company) was added to a final concentration of 180 mmol/mol creatinine. This ensured a fairly constant ratio between the urinary organic acids and internal standard, which contributes to more constant extraction efficiency of the organic acids, including the internal standard. 3-Phenylbutyric acid was used as internal standard due to its absence in normal urine and in known pathological conditions and because it elutes almost in the middle of the organic acid profile and theoretically co-elutes with very few, if any, other organic acids.
Urine samples were acidified with 5 N HCl (approximately 5 drops) and the pH was monitored to ensure it was less than 2. Six ml of ethylacetate (Merck Chemicals) was added to each sample and the mixture shaken on a rotary wheel for 20 min. The mixture was centrifuged for 2 min at 1300×g and the upper ethylacetate phase transferred to a clean glass tube. Three ml of diethylether (Merck Chemicals) was added to the water phase and shaken for a further 10 min. After centrifugation (1300×g, 10 min), the upper phase was removed and added to the ethylacetate. A small amount of sodium sulphate (BDH) was added to the ethylacetate/diethylether mixture to remove any residual water. After a subsequent centrifugation step, the organic phase was transferred to a clean glass tube. The organic solvents were evaporated to dryness under nitrogen at 37°C.
O-bis(trimethylsilyl)trifluoroacetamide (BSTFA): trimethylchlorosilane (TMCS): piridine (5:1:1, and volume added based on the creatinine values) was used for derivatization. The volume of urine used gave a creatinine concentration equivalent to 21 μmol/ml derivatization reagent. The samples were derivatized at 85°C for 45 min in a sand bath. The derivatized mixture was transferred to a 1.5 ml vial for GC–MS analysis. This approach ensured a fairly constant concentration of organic acids in the derivatization mixture, which improved the repeatability of the analysis.
2.3.2 GC–MS analysis
The Agilent GC–MS system used in this study consisted of a model 7890A gas chromatograph, a model 5975C mass selective detector, an HP 5970C MS and Agilent Chemstation (Revision E.02.00). A fused-silica capillary column (DB-1MS UI, 30 m, 2.50 μm i.d., 0.25 μm film thickness) was used for the fractionation. The initial GC temperature was 60°C, kept for 2 min. It was then increased to 120°C at a rate of 5°C/min, to 295°C at a rate of 7°C/min, and held at a final temperature of 295°C for 2 min. Helium (1 ml/min) was used as carrier gas at a constant flow rate. The mass spectra of all GC peaks were generated by a mass spectrometer operated at 70 eV in the electron impact mode with SCAN (50–600 amu) positive ion monitoring. The MS source and quadrupole temperatures were 230 and 150°C respectively.
2.3.3 Deconvolution, peak identification and quantification
Deconvolution and data analyses were conducted using AMDIS software (Version 2.66) linked to NIST Mass Spectral Search Program for the NIST/EPA/NIH Mass Spectral Library (Version 2.0F, built Oct. 8, 2008). An AMDIS library file (more than 800 mass spectra) was generated by transferring organic acid (TMS derivatives) mass spectra from the NIST/EPA/NIH Mass Spectral Library, as well as from the Wiley Registry™ of Mass Spectral Data (8th Edition). In addition to the mass spectra of commercial compounds and those synthesized in our laboratory, we obtained mass spectra in the 1980s from Prof. S.K. Wadman of the Wilhelmina Kinderziekenhuis, University of Utrecht, The Netherlands (Erasmus et al. 1985).
Since January 2001, more than 30 000 organic acid analyses have been performed in our laboratory for diagnostic purposes. Mass spectra of unknown compounds present in relatively high concentrations were added to the target file, using the retention times as part of the identification. The identity of some of these spectra was resolved using the software ACD/MS Fragmenter (Release 10.00). Where possible all mass spectra were confirmed by analysing authentic standards. During this period, the mass spectra of compounds that were never detected were deleted from the library, leaving the spectra of more than 800 compounds in the reference version of the library. Where authentic standards were available, their respective responds factors were used, and for those compounds where no authentic standards were available, a response factor of 1 was assumed (See the note in this regard in the legend to Table 2). The analytical setting of the AMDIS software was as follows: minimum factor = 60% and type of analysis = “Use of an internal standard for RI”. The deconvolution settings were: component width = 20; adjacent peak subtraction = 1; resolution = low; sensitivity = very low; and shape requirements = low.
The first hit of identified compounds and integrated area of the peaks were exported to Microsoft Excel©. Some peaks with uncertain identification were indicated by the AMDIS software with a question-mark against the name of the most likely organic acid in the library. These peaks were manually inspected and compared to mass spectra and retention times of the pure compound for identification purposes. Some compounds may produce multiple peaks, so that the SUMIF function of Excel© was used to calculate the sums of the areas of these peaks. A relative concentration, expressed as mmol/mol creatinine, was calculated using the formula: Concentration = Area (compound)/Area (IS) × concentration (IS) (Chen et al. 2009). A data matrix was created by aligning all the metabolites against the samples using MATLAB software and the statistical R-program.
2.4 Statistical analysis
The data pretreatment used in this investigation consisted of zero replacement, data transformation and centering. The zero values (representing the detection limit of the analytical equipment) were replaced by a random sample of values from a Beta (0.1, 1) distribution. After comparison with pareto-, auto- and non-parametric-scaling methods, a shifted log transformation was selected and applied to these data, so that the scales of the various metabolite concentrations were more comparable. The transformed data were then centred prior to multivariate analysis.
The data analysis used in this investigation consisted of principal component analysis (PCA) as an unsupervised pattern recognition method (Johnson and Wichern 1998), and a partial least squares discriminant analysis (PLS-DA) as a supervised method (Baker and Rayens 2003). From these multivariate analyses, the coefficients for all variables important in projection (VIP) were calculated, and ranked from high to low.
Effect size (Ellis and Steyn 2003) was used as a univariate analysis to ascertain the importance of a variable. The effect size d is defined as \( d = \left| {\bar{X}_{1} - \bar{X}_{2} } \right|/S_{\max } \), where \( \bar{X}_{1} \) and \( \bar{X}_{2} \) are the group means and S max is the maximum standard deviation of the two groups. An effect size of d > 0.5 can be considered as being of medium practical importance, whereas an effect size of d > 0.75 can be considered as highly practically significant. In addition, standard t-tests and the non-parametric Mann–Whitney tests were used as univariate analyses to test the equality of group means and group medians, respectively.
3 Results and discussion
3.1 Data reduction
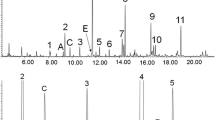
Metabolic fingerprinting of GC–MS data requires pre-processing to generate a data matrix of variables and cases of an operational size, to be followed by multivariate analysis to identify only the relevant analytical information. The work-flow that we followed is shown schematically in Fig. 1. Our selected experimental group consisted of 92 controls and 48 patients having an experimentally detected deficiency in one or more complexes of the RC. The total number of organic acids identified by GC–MS analysis of urine samples from this experimental group consisted of 301 components. From information in the Human Metabolomics Database, other published material on the origin of organic acids, and from knowledge of organic acids acquired over decades in our Laboratory for Metabolic Disorders, we could classify these 301 components into two major groups. Group 1 consisted of 255 endogenous substances, categorized as Class 1–7, as shown in Table 2. These substances originated from the metabolism of carbohydrates (Class 1, 9%); the Krebs cycle (Class 2, 3%); fatty acid metabolism (Class 3, 19%); amino acid metabolism (Class 4, 20%); neurological, stress or impaired organ function (Class 5, 11%); or from the diet or gut flora (Class 6, 15%). This group also contained substances of known metabolic pathways (e.g. purine and pyrimidine metabolism), as well as metabolites which could not be classified unambiguously (Class 7, 23%). Group 2 originated from medication or from other xenobiotic origin (Class 8). All substances from Group 2 were eliminated from the data matrix.
A scatterplot with drop-lines of the 255 organic acids for the 92 controls and the 48 patients which all complied to the two criteria based on the enzyme analyses, is shown in Fig. 2a. The organic acids are ranked from low to high retention times, as actually observed in the GC separation. The controls and patients were ranked by group, from those with the lowest total excretion of the urinary organic acids to those with the highest values. A visual inspection of this representation clearly indicates (1) a greater abundance of organic acids in the patient group than in the controls, (2) an extensive number of substances with very low or zero values, and (3) an overlap between patients with low and controls with a high excretion of urinary organic acids. The low excretion of total organic acids in some patients could be related to subjects in a near-normal clinical state when the urine sample was taken. The high values in the control group may be due to the clinical condition of individuals, which could not be discounted as a mitochondrial disorder or another metabolic condition. These observations required further selection of cases as the next step in the process of data reduction. For this, the total organic acids excreted were first calculated. Controls with total organic acid values greater than the corresponding lower quartile for patients were eliminated from the group. Patients with total organic acid concentrations less than the upper quartile of the corresponding acids excreted by the controls were similarly eliminated. From this, the final experimental group of 71 controls and 39 patients were selected, for whom the outcome is shown in Fig. 2b. Relevant meta data of these 39 patients are summarized in Table 1.
Drop-line representation of variables present in the control and patient groups. a Data for all initial 140 cases and 255 selected variables of endogenous origin. b Data for the reduced cases (110, including 71 controls and 39 patients) and the 255 variables. c Representation of the information included in the final data matrix, consisting of the 110 cases as in b and the 92 variables, selected after applying the 80% rule
The next data reduction step involved the variables. All variables which had 80% or more zeros in both the patient and the control groups were then eliminated (compare also the approach by Bijlsma et al. 2006). This yielded a final list of 92 variables, the outcome of which is shown in Fig. 2c. A comparison of the various results shown in Fig. 2 indicates that the essential characteristics of the data set were still retained after reduction of the disease cases and variables. The selected data consisted of (1) a reduced but still comprehensive set of endogenous variables, with the mean concentration of total organic acids excreted by the controls and the patients amounting to 459 (s.d. = 161) and 1822 (s.d. = 1323) mmol per mol creatinine, respectively, but (2) with the elimination of the overlap between the controls and the patients.
3.2 Selection of important variables
The subsequent data pre-treatment focused only on the variables selected after data reduction as discussed in Sect. 3.1 and shown in Fig. 2c. Mean values, as well as the standard deviation of all variables for the controls and patients, were determined on the un-scaled data, followed by t-test and Mann–Whitney analyses. The effect size analyses were subsequently conducted on the un-scaled as well as on the scaled data. The most powerful discrimination between the RCD patients and the controls was obtained from the effect size analysis of the data. For the 92 variables, d > 0.5 for 45 and 59 for the un-scaled and log-scaled data, respectively. Variables from the log-scaled data, for which d > 0.75 (practical highly significant) and were statistically significant, were selected for further analysis of the consequences of RCD on the cellular metabolism. This selection produced 44 variables, which were subsequently compared with the 44 VIPs identified by multivariate analyses. It should be noted that this list include metabolites that are not organic acids per definition, but which are co-extracted by the procedures used. They are glycerol, uric acid (a purine) and uracil (a pyrimidine).
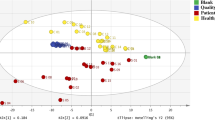
Two traditional methods of multivariate analysis (PCA and PLS-DA) proved to be valuable for variable selection and were subsequently applied to four sets of data: the three groups (CI-, CIII- and CM-deficiencies) of RCD taken separately, and also as one combined group. All 71 controls were included in every analysis. The outcome of the PCA, shown in three-dimensional score plots (PC1, PC2 and PC3) for the four data sets, is shown in Fig. 3. For all three sub-groups of RCD (Fig. 3a–c), the patient groups were distinguished from the controls, indicating that the metabolic profiles of these three groups were altered by the perturbation induced by the respective CI, CIII or CM deficiencies. These observations hold the potential to identify biomarkers that could distinguish these three RCDs. However, this was excluded by the results obtained from the PCA of the three combined groups (Fig. 3d–f). In a score plot of the PCA of the three groups taken together, but excluding the controls, they could not be distinguished from one another (Fig. 3d). The same holds true for a PCA in the presence of the controls, although the three groups could still be distinguished from the control group (Fig. 3e, f). Following these results, the search for possible biomarkers was continued with the individual (CI, CIII or CM) as well as the combined groups of patients.
Score plots of PCA shown in 3-dimensional plots of identical rotation in all six subsections of patients. The upper section of the figure depicts a model of the association of complexes CI, CII, CIII, CIV and the ATPase with the inner mitochondrial membrane. Complexes CI, CIII and CI, CII, CIII and CIV (designated together as CM) are coloured red, green and yellow, respectively. This indicates the sites of the RCD for the PCA analysis conducted for each of the three patient subgroups (CI, CIII and CM). The respective PCA analysis is shown directly below each presentation (a–c). The same controls were used for the PCAs shown in a–c, e and f, and are indicated in brown as in Fig. 2. The patient groups used in these six PCA analyses were CI (a), CIII (b), CM (c), and CI, CIII and CM together in d–f. The cumulative proportions of PC1, PC2 and PC3 for the data shown in f (blue as in Fig. 2) were 40.0, 46.6 and 51.0%. The lists of variables important in projection (VIPs) were generated for the data shown in a–c (Fig. 4a) and for f (Fig. 4b; Table 2)
In this investigation the important variables for each of the three sub-groups were identified by using three statistical methods of analysis. The three methods were distinctly different, being a univariate approach (effect size), unsupervised multivariate analysis (PCA), and a supervised multivariate analysis (PLS-DA). All variables with d > 0.75 were first identified from the VIP lists of the PCA and PLS-DA analyses for the three sub-groups. From these lists, variables originating from the diet, gut flora or of unknown source were eliminated, and were found to be four for the CI group, one in the CIII group, and zero in the CM group. The remaining important metabolites for which d > 0.75 were found to be 12 for CI, 24 for CIII and 25 for CM.
In order to compare the three lists of metabolites, we developed a Venn diagram shown in Fig. 4a, indicating the number of variables which define the three groups, as well as those which are common to all of them. Eleven of the 12 metabolites of the CI group were shared with the CIII and CM groups. These were adipic acid, fumaric acid, lactic acid, suberic acid, succinic acid, 2-hydroxyglutaric acid, 3-hydroxy-3-methylglutaric acid, 3-hydroxyisobutyric acid, homovanillic acid, 3-methylglutaric acid and 4-hydroxyphenyllactic acid. The remaining metabolite, not present among the important variables of the CIII and CM groups, was vanillylmandelic acid. Eleven of the remaining metabolites were also common to the CIII and CM groups. They were ethylhydracrylic acid, glutaric acid, glycerol, malic acid, methylsuccinic acid, phenylacetylglutamine, uracil, 2-hydroxysebacic acid, 2-methyl-3-hydroxybutyric acid, 3-hydroxyisovaleric acid and 3-methyladipic acid. The remaining metabolites not shared by any of the other groups were glyceric acid and 3-hydroxysuberic acid for the CIII, and pyroglutamic acid, 2-keto-octanoic acid and 3-hydroxyadipic acid for the CM groups, respectively. This comparison clearly indicates great overlap of the important metabolites that define the three sub-groups and substantiates the observation that the CI, CIII and CM patient groups were not distinguishable, as indicated by the PCAs shown in Fig. 3d, e. We therefore concluded that no characteristic organic acid biomarker profile could be defined for the CI, CIII and CM RCDs.
Venn diagrams highlighting the common variables observed in the four patient groups. a Diagram for subgroups CI, CIII and CM. b Diagram for the variables identified through the effect size (ES), PCA and PLS-DA for the combined patient group. In all cases all controls were used and the effect size of the log-scaled values has been used as the basic benchmark for the selection of variables. The colour scheme used in a and b is the same as in Fig. 3
Secondly, the variables from the effect size, PCA and PLS-DA lists were used to construct a Venn diagram to identify the important metabolites in the combined patient group. The outcome is shown Fig. 4b. In the case of the PCA, 8 variables, and for the PLS-DA 7 variables from their respective lists of the first 44 VIPs had d < 0.75, and were discarded, not being practically significant. In the effect size list, two of the variables with d > 0.75 were not among the 44 variables in either the PCA or the PLS-DA list. Of the remaining 11 variables with d > 0.75, five and six, respectively, were not common to either multivariate analysis. This comparison thus produced 31 variables (33.7% from the 92 variables in the final matrix, and 12.1% of the original endogenous substances) which were common to all three statistical methods of analysis. All 31 variables were subsequently evaluated to explore which metabolic pathways were affected in patients with RCD, as compared to the control group. This comparison is summarized in Table 2, and can be considered to be the most important outcome of our metabolomics investigation of RCDs.
3.3 Comparative analysis of the important metabolites
Table 2 presents the information on the 31 variables, classified according to the criteria which we applied to the initial data set of 301 variables, followed by their grouping in terms of their most important functions in intermediate metabolism. Twenty-three of the 31 variables are clearly primary or secondary metabolites associated with the metabolism of carbohydrates, fatty acids, amino acids or the Krebs cycle. Two further variables could be related to neurological or stress conditions that are known to prevail in RCD, while the remaining six variables probably originate from sources not directly related to RCD. For these 25 metabolites, the d-value for the effect size of the log-scaled data was greater than 0.75, indicating that all are highly significant from a practical point of view. Apart from one (uracil), all 25 had a P-value smaller than 0.05 in the t-tests. The Mann–Whitney test showed that the group medians differed significantly for all 25 variables at the 5% level. Fifteen of the 25 metabolites were previously reported for deficiencies in one or more complexes of the RC. Of the remaining 10 metabolites not previously mentioned in investigations on RC, most are known to be normal metabolites, or metabolites that become elevated due to an inherited metabolic defect other than RCD. The presence of 2-keto-octanoic acid was rather unexpected. It might originate from fatty acid degradation in the peroxisomes, but then 2-hydroxyoctanoic acid would be the more likely metabolite (Foulon et al. 2005). This aspect should thus need further investigations. The biological significance and potential diagnostic value of the other 24 metabolites will be discussed in sect. 3.4 below.
Table 2 also lists all other metabolites (15) that were described in other studies on RCDs. With the exception of glutaconic acid, all were included among the 92 variables in our final data matrix. Of these, acotinic acid is noteworthy for its high effect sizes (d = 1.13 and 0.94 for the log-scaled and un-scaled data, respectively) and its statistically significant (P = 0.0001) higher value for the patients than the controls. The mean value for the patient group was, however, lower than its reported normal maximum value. Three of the remaining metabolites had low d values and the other 10 were excluded by the 80% test, indicating that they were present in only a very few cases. It, therefore, appears that these 15 metabolites are not common to patients with RCDs in general, but observed in individual cases with such a deficiency.
3.4 A global metabolite profile associated with organic aciduria due to RCD
The metabolite profile as discussed here refers to changes in the cellular profile of the organic acids and a few other metabolites in cases of RCD, as derived from the information disclosed by metabolomics analysis of urine samples. We interpret the variation in this profile, compared to controls, as a result of the immediate and downstream consequences of RCDs, which include allosteric regulation of metabolic processes, as well as transcriptional/translational responses to the deficiencies. As RC is essentially involved in the cellular reduction/oxidation (redox) status and energy (ATP) production, deficiencies inevitably affect a plethora of metabolic and other processes (Reinecke et al. 2009), of which only a selective group of metabolites is unravelled here. We derived a comprehensive set of metabolic pathways shown in Fig. 5, to illustrate the complex metabolic perturbations as revealed by this investigation. Although we investigated three subsets of RCD (CI, CIII and CM), the proposed metabolic pathways affected by RCDs are based on the metabolites of the combined patient group, implicating a mutual relationship between the individual components of the RC, assuming that all complexes affected redox status and ATP production as key final consequences of the disease. Such a mutual relationship supports the prevailing view on the structure, functioning and regulation of the OXPHOS system (Boekema and Braun 2007; Dudkina et al. 2008; Reder 1988; Rossignol et al. 2003), implying that the control of mitochondrial respiration is shared among all of the OXPHOS complexes. Other investigations demonstrated the existence of a biochemical threshold effect for the expression of a specific RCD and mitochondrial energy production (Mazat et al. 2001), which accounts for the severity of the pathogenic mutation and for extrinsic factors, such as the tissue affected. Finally, kinetic studies indicated the modulation of the activity of RC complexes by variations in the concentrations of intermediate metabolites, designated as network attenuation (Heinrich and Rapoport 1974; Rossignol et al. 2003). These concepts thus support the notion of a global, perturbed metabolite profile in RCD, as presented here, rather than a linear relationship between one or a few metabolites as biomarkers for a specific complex deficiency, as seen in other inherited metabolic mitochondrial disorders like isovaleric acidaemia and propionic acidaemia.
A representation of global metabolic perturbations associated with organic aciduria due to RCD. The 25 metabolites indicated in Table 2, as being statistically and practically significant, are indicated in blue. Others frequently observed in individual cases of RCD, also included in Table 2, are shown in green. Names or structural formulas of metabolites printed in red are normal intermediates in the respective metabolic pathways. The six amino acids, from which the secondary metabolites and markers of RCD originate, are shown in red. a The extramitochondrial consequences of RCD, and its link to the intramitochondrial metabolism. Stage 1: Decreased ATP production and the decreased [ATP]/[ADP] ratio is compensated for by the action of various neurotransmitters, e.g. dopamine, and/or hormones like vasopressin, glucagon and adrenaline. Stage 2: Increased catabolism of glycogen, proteins, triacylglycerols and purines and pyrimidines. Stage 3: Intramitochondrial disarray of the Krebs cycle and production of secondary metabolites due to impaired catabolism of fatty acids and some amino acids. Production of pyroglutamic acid is probably a consequence of oxidative stress (alternatively: a degradation product of glutamine). b Proposed origin of secondary metabolites observed in RCD and formed from amino acid catabolism. The consequences for phenylalanine and tyrosine are shown on the left of the figure. Phenylacetylglutamine may be an endogenous product, as indicated, or, alternatively a bacterial metabolite from the gut. Degradation of lysine and the branched-chain amino acids resemble aspects of glutaric aciduria Type II (GLU II). Formation of 3-methylglutaric acid in RCD might be due to the mevalonate shunt; the link to the Krebs cycle and the clinical characteristic of ketosis in RCD is indicated where applicable. c Proposed origin of secondary metabolites observed in RCD and formed from fatty acid catabolism due to impaired β-oxidation, resembling glutaric aciduria Type II (GLU II)
Figure 5a summarizes the regulation of carbohydrate, protein and triacylglycerol metabolism in the extra-mitochondrial compartment and its link to RCD metabolic alterations, with a focus on the Krebs cycle, amino acid metabolism (Fig. 5b) and fatty acid metabolism (Fig. 5c). Figure 5a shows how the catabolic degradation products of carbohydrates, fatty acids and amino acids assemble in the final stages of cellular oxidation, in which the energy generated by the RC process drives the synthesis of ATP, generally providing the bulk of cellular ATP. In RCD’s a decrease in cellular ATP occurs, corresponding to a smaller ATP/ADP ratio, which is normally very high, as the ATP-ADP system is then virtually fully phosphorylated. Reduced mitochondrial production of ATP can be directly compensated for by an equivalent increase in glycolysis (Hofhaus et al. 1996), as the [ATP][ADP]/[Pi] ratio functions as the primary cytosolic sensor of altered energy production, initially via glycolysis (Pfeiffer et al. 2001). With these regulatory processes in mind, a theoretical model has been proposed (Korzeniewski 2001), indicating how compensation of OXPHOS defects could occur at the cellular level through the stimulation of ATP production by the action of various neurotransmitters, such as dopamine and/or hormones like vasopressin, glucagon and adrenaline. This is supported by the increased levels of homovanillic acid (HVA, 3-methoxy-4-hydroxyphenylacetic acid), which we found in all RCD groups, and by vanillylmandelic acid (VMA, 4-hydroxy-3-methoxyphenyl-glycolic acid) in the CI subgroup. These metabolites have not been described before for RCDs, although increased urinary HVA concentrations are known to arise from neurological stress conditions (Frankenhaeuser et al. 1985; Rauste-von Wright and Frankenhaeuser 1989). The elevated neurotransmitter–hormone model and catabolism of carbohydrates (glycogen → glycolysis) similarly apply to catabolism of proteins (→amino acid catabolism) and triacylglycerols (→fatty acid oxidation and the production of glycerol).
As indicated before, a key consequence of RCD is increased production of NADH (NADH/NAD+ ratio) and FADH2 due to the dysfunctional RC at any site within the chain. Although several enzymes are regulated by this change in redox state, extramitochondrially it can result in commonly observed (as in this study) elevated lactic acid, an increased lactate/pyruvate ratio, elevated alanine (not reported here due to our metabolome selection), as well as raised uric acid (Kuwertz-Bröking et al. 2000). Moreover, it may result in elevated TCA intermediates of which several were observed in this study, and which are included as one of the disease criteria for MD (Morava et al. 2006). Barshop (2004) highlighted increased urinary fumaric acid and malic acid but in our study the order of significance of elevated TCA metabolites was succinate >fumarate >malate. We anticipate that this order may be a result of irreversible conversion of 2-ketoglutaric acid to succinic acid, and a consequent reverse TCA cycle towards succinic acid and possibly an impaired malate–aspartate shuttle which is known to be physically closely linked to consecutive enzymes of the Krebs cycle (Beeckman and Kanarek 1981).
Accumulation of succinic acid contributes to a decreased conversion of 2-ketoglutaric acid to succinic acid, resulting in the statistically significant increase in aconitinic acid in the first half of the TCA cycle (Table 2). Although 2-ketoglutaric acid is present in the list of PCA VIPs, the expected increase in its concentration was not observed above the level of the normal maximum in any of the RC deficiencies studied here. However, 2-hydroxyglutaric acid, which has not been described in terms of RC deficiencies before, was statistically significantly increased in all three of our subgroups. Although some methane-producing prokaryotes utilize 2-hydroxyglutaric acid in a metabolic pathway (Van Beelen et al. 1984), it is not an intermediate in any known metabolic pathway in man or other eukaryotes. A first case of l-2-hydroxyglutaric aciduria was nonetheless identified in 1980 (Duran et al. 1980) and later confirmed by mutational analysis (Topçu et al. 2004; Rzem et al. 2004). Our proposal for the accumulation of 2-hydroxyglutaric acid seen in our patient groups favours the NADH-dependent conversion of 2-ketoglutaric acid to 2-hydroxyglutaric acid by mitochondrial l-malate dehydrogenase. This proposal is based on (1) the known redox potential for the 2-hydroxuglutaric acid/2-ketoglutaric acid couple (Buckel and Miller 1987), (2) the anticipated high 2-ketoglutaric content in the mitochondria due to the reversible operation of the latter part of the TCA cycle, and (3) the relative specificity of mitochondrial l-malate dehydrogenase (Banaszak and Bradshaw 1975). This mechanism was recently proposed as operating in l-2-hydroxyglutaric aciduria (Van Schaftingen et al. 2009).
The interrelated involvement of the NAD redox state (increased NADH2), formation of reactive oxygen species (ROS) and the glutathione redox state in RCDs can explain the observed elevated urinary pyroglutamic acid as also reported by Topaloğlu et al. (2008). We interpret the increase in pyroglutamic acid (Fig. 5a) as an increase in the flux through glutathione rather than a change in the thiol redox state, as the mitochondrial and cytosolic thiol redox states are not inevitably affected, as shown in studies using complex I-deficient fibroblasts (Verkaart et al. 2007).
The consequences of RCD on amino acid degradation are shown in Fig. 5b. Similarities with known inherited single enzyme deficiencies of lysine, leucine, valine and isoleucine catabolism are again conspicuous. Glutaric acid, 3-hydroxyisobutyric acid, 3-hydroxyisovaleric acid, 2-methyl-3-hydroxybutyric acid, 2-ethylhydracrylic acid (Table 2) and the glycine conjugates of isobutyric acid, isovaleric acid, and 2-methylbutyric acid are all secondary metabolites seen in glutaric aciduria type II [electrotransfer flavoprotein (ETF), ETF-ubiquinone oxidoreductase deficiency, and ETF-dehydrogenase deficiency, reviewed by Gordon (2006)]. This implies that the defective flow of electrons in RCDs impairs the functionality of the ETF-dependent enzymes, followed by an increase in their respective substrates from which the secondary metabolites, disclosed by the metabolomics analysis of urine samples from RCD patients, arises. The origin of 3-methylglutaconic acid and 3-hydroxy-3-methylglutaric acid, which we also observed in our RCD cohort (Table 2), is controversial (Walsh et al. 1999; Wortmann et al. 2005). To account for their presence in RCDs, the proposed origin of these catabolites from leucine catabolism (Duran et al. 1982) and the mevalonate shunt (Kelley and Kratz 1995) is indicated in Fig. 5b. Comprehensive genetic mapping will be required, however, to establish unequivocally the aetiology of the heterogeneous profile associated with 3-methylglutaconic acid. Finally, increased tyrosine and phenylalanine catabolism, which occurs exclusively in the liver, can account for 4-hydroxyphenyllactic acid and phenylacetylglutamine, respectively. This relates to the perturbed NADH/NAD+ ratio, and may also be an indication of liver damage in RCDs.
Finally, the consequences of RCDs for fatty acid degradation are shown in Fig. 5c. Decreased electron transfer flavoprotein (ETF) and increased electron transfer flavoprotein: ubiquinone oxidoreductase (ETF-QO) activity due to a deficient flow of electrons past CoQ10 may give rise to adipic, 3-methyladipic acid and 3-hydroxyadipic acid, suberic and 3-hydroxysuberic acid, as well as 3-hydroxysebacic acid. These are formed by ω-oxidation from their respective short- and medium-chain fatty acid-CoA precursors, comparable with the observation of impaired mitochondrial fatty acid oxidation (reviewed by Duran 2005) of short-chain (SCAD), medium-chain (MCAD) and very long-chain acyl-CoA dehydrogenase deficiencies (VLCAD). The elevated levels of two of these ω-oxidation products, 3-hydroxysebacic acid and 3-hydroxyadipic acid, again illustrate the involvement of NAD redox state. Furthermore, elevated butanoyl-CoA due to reduced SCAD activity is proposed to give rise through an acetyl-CoA- and propionyl-CoA-carboxylase catalysed production of ethylhydracrylic and methylsuccinic acid, respectively (Sweetman and Williams 2001).
3.5 Potential clinical applications
The clinical and genetic heterogeneity of RCDs are the most important aspects complicating diagnosis in these disorders. The number of known mutations is rapidly growing; around 743 different mutations had been reported on MITOMAP (http://www.mitomap.org) by October 2010. The final confirmation of a diagnosis is further complicated by the logistic, invasive and expensive nature of the current biochemical approach including enzyme analyses on muscle biopsies. Using a metabolomic technique to simplify this complex diagnostic challenge can be anticipated a priori to be a multi-faceted process, because large and complex data sets are created, requiring special skills to derive meaningful information from them.
In a metabolomics experiment the biological sample provides a profile of the metabolome at that time. In clinical studies on the diagnosis of RCDs, the samples most commonly used for biochemical analysis come from urine collected in clinics from more or less critically ill patients (Smeitink et al. 2006). Rigorous requirements for sample collection to comply with the minimum standards advocated for metabolomics investigations (Dunn and Ellis 2005; Goodacre et al. 2007) are thus not fully feasible in a metabolomics study of RCDs and many other inherited metabolic diseases. We could overcome this handicap, however, with samples from a large cohort of clinically well-described patients (Smuts et al. 2010) and from an even larger number of controls, and by specifically focusing on the organic aciduria profiles of the patients. The comparative approach of using the effect size, PCA and PLS-DA unequivocally established that no single organic acid biomarker, or combination of such biomarkers, can be defined for sub-classes of RC complex deficiencies. However, the insight into the perturbations caused by RCDs gained by the metabolomics approach has significance for possible clinical applications, and strengthens expectations for a new frontier in pediatrics research (Carraro et al. 2009).
The criteria defined for new novel biomarkers for cardiovascular disorders (Morrow and de Lemos 2007) are equally well applicable in a search for biomarkers for inherited metabolic disorders. For the present, our hypothesis is that a biosignature for RCDs, derived from a metabolomics investigation of different parts of the metabolome, rather than a search for a few highly specific biomarkers, has the potential to define a practical diagnostic instrument to detect the presence of an RCD. Ultimately, enzyme analysis of the complexes of the RC in skeletal muscle biopsy material will probably remain the golden diagnostic standard when an RCD is suspected (Smeitink et al. 2006). However, a global metabolic effect as shown here can be exploited in screening for an RCD by analysis of biofluids, such as urine, obtainable by non-invasive procedures. From our experience, the adoption of such an approach for diagnosis and in the follow-up of disease progression or treatment of patients is in the offing in the health-care systems of a developing country such as South Africa, where this investigation was conducted. The general inclusion of a biosignature for RCDs in routine clinical practice will require, however, assessment of larger, prospective and validated studies, as was recently emphasized by Mamas et al. (2011). Meanwhile, our recommended preference in clinical practice on RCDs would be to shift the emphasis from an approach “to scrape the barrel” (DiMauro and Andreu 2000; Smeitink 2003), in search of a definitive answer to a highly variable and evasive disease, to a focus on caring, affordability and personalized medicine.
4 Concluding remarks
This study demonstrates that pattern recognition of urinary organic acids in metabolomics analysis can inform on the complex differences that can be observed in the global metabolite profiles of controls and RCD patients. It rules out the possibility of identifying one unique and distinctive organic acid biomarker. Significantly, however, the global metabolite profile has the potential to define an extended biosignature and the development of a non-invasive screening procedure for RCDs. We suggest that such a procedure for diagnosis and monitoring of disease progression or treatment will add considerable value to the current clinical practice followed for RCDs.
References
Anderson, S., Bankier, A. T., & Barrell, B. G. (1981). Sequence and organization of the human mitochondrial genome. Nature, 290, 457–465.
Baker, M., & Rayens, W. (2003). Partial least squares for discrimination. Journal of Chemometrics, 17, 166–173.
Banaszak, L. J., & Bradshaw, R. A. (1975). Malate dehydrogenases. In P. D. Boyer (Ed.), The enzymes, 11 oxidation-reduction part A (pp. 369–396). New York, San Francisco, London: Academic Press.
Barshop, B. A. (2004). Metabolomic approaches to mitochondrial disease: Correlation of urine organic acids. Mitochondrion, 4, 521–527.
Barshop, B. A., Nyhan, W. L., Naviaux, R. K., et al. (2000). Kearns-Sayre syndrome presenting as 2-oxoadipic aciduria. Molecular Genetics and Metabolism, 69, 64–68.
Beeckman, S., & Kanarek, L. (1981). Demonstration of physical interactions between consecutive enzymes of the citric acid cycle and of the aspartate-malate shuttle. European Journal of Biochemistry, 117, 527–535.
Bennett, M. J., Sherwood, W. G., Gibson, K. M., & Burlina, A. B. (1993). Secondary inhibition of multiple NAD-requiring dehydrogenases in respiratory chain complex I deficiency: Possible metabolic markers for the primary defect. Journal of Inherited Metabolic Disease, 16, 560–562.
Bennett, M. J., Weinberger, M. J., Sherwood, W. G., & Burlina, A. B. (1994). Secondary 3-hydroxydicarboxylic aciduria mimicking long-chain 3-hydroxyacyl-CoA dehydrogenase deficiency. Journal of Inherited Metabolic Disease, 17, 283–286.
Bernier, F. P., Boneh, A., Dennett, X., et al. (2002). Diagnostic criteria for respiratory chain disorders in adults and children. Neurology, 59, 1406–1411.
Bijlsma, S., Bobeldijk, I., Verheij, E. R., et al. (2006). Large-scale human metabolomics studies: A strategy for data (pre-) processing and validation. Analytical Chemistry, 78, 567–574.
Boekema, E. J., & Braun, H.-P. (2007). Supramolecular structure of the mitochondrial oxidative phosphorylated system. Journal of Biological Chemistry, 282, 1–4.
Buckel, W., & Miller, S. L. (1987). Equilibrium constants of several reactions involved in the fermentation of glutamate. European Journal of Biochemistry, 164, 565–569.
Carraro, S., Giordano, G., Reniero, F., Perilongo, G., & Baraldi, E. (2009). Metabolomics: A new frontier for research in pediatrics. Journal of Pediatrics, 154, 638–644.
Chen, J., Meng, C.-K., Narayan, S. B., Luan, W., & Bennett, M. J. (2009). The use of deconvolution reporting software© and backflush improves the speed and accuracy of data processing for urinary organic acid analysis. Clinica Chimica Acta, 405, 53–59.
Christensen, E., Brandt, N. J., Schmalbruch, H., et al. (1993). Muscle cytochrome c oxidase deficiency accompanied by a urinary organic acid pattern mimicking multiple acyl-CoA dehydrogenase deficiency. Journal of Inherited Metabolic Disease, 16, 553–556.
DiMauro, S., & Andreu, A. L. (2000). Mutations in mtDNA: Are we scraping the bottom of the barrel? Brain Pathology, 10, 431–441.
Dudkina, N. V., Sunderhaus, S., Boekema, E. J., & Braun, H.-P. (2008). The higher level of organization of the oxidative phosphorylation system: Mitochondrial supercomplexes. Journal of Bioenergetics and Biomembranes, 40, 419–424.
Dunn, W. B., & Ellis, D. I. (2005). Metabolomics: Current analytical platforms and methodologies. Trends in Analytical Chemistry, 24, 285–294.
Duran, M. (2005). Disorders of mitochondrial fatty acid oxidation. In N. Blau, M. Duran, & M. E. Blaskovics (Eds.), Physician’s guide to the laboratory diagnosis of metabolic diseases (Revised 2nd ed., pp. 309–334). Heidelberg: Springer.
Duran, M., Beemer, F. A., Tibosch, A. S., et al. (1982). Inherited 3-methylglutaconic aciduria in two brothers—another defect of leucine metabolism. Journal of Pediatrics, 101, 551–554.
Duran, M., Kamerling, J. P., Bakker, H. D., Van Gennip, A. H., & Wadman, S. K. (1980). l-2-hydroxyglutaric aciduria: An inborn error of metabolism? Journal of Inherited Disease, 3, 109–112.
Ellis, S. M., & Steyn, H. S. (2003). Practical significance (effect sizes) versus or in combination with statistical significance (p-values). Management Dynamics, 12, 51–53.
Enns, G. M., Bennett, M. J., Hoppel, C. L., et al. (2000). Mitochondrial respiratory chain complex I deficiency with clinical and biochemical features of long-chain 3-hydroxyacyl-coenzyme A dehydrogenase deficiency. Journal of Pediatrics, 136, 251–254.
Erasmus, C., Mienie, L. J., Reinecke, C. J., & Wadman, S. K. (1985). Organic aciduria in late-onset biotin-responsive multiple carboxylase deficiency. Journal of Inherited Metabolic Disease, 8, 105–106.
Esteitie, N., Hinttala, R., Wibom, R., et al. (2005). Secondary metabolic effects in complex I deficiency. Annals of Neurology, 58, 544–552.
Foulon, V., Sniekers, M., Huysmans, E., et al. (2005). Breakdown of 2-hydroxylated straight chain fatty acids via peroxisomal 2-hydroxyphytanol-CoA lyase. Journal of Biological Chemistry, 280, 9802–9812.
Frankenhaeuser, M., Lundberg, U., Rauste von Wright, M., von Wright, J., & Sedvall, G. (1985). Urinary monoamine metabolites as indices of mental stress in healthy male and females. Pharmacology, Biochemistry and Behaviour, 24, 1521–1525.
Goodacre, R., Broadhurst, D., Smilde, A. K., et al. (2007). Proposed minimum reporting standards for data analysis in metabolomics. Metabolomics, 3, 231–241.
Gordon, N. (2006). Glutaric aciduria types I and II. Brain and Development, 28, 136–140.
Haas, R. H., Parikh, S., Falk, M. J., et al. (2008). The in-depth evaluation of suspected mitochondrial disease. Molecular Genetics and Metabolism, 94, 16–37.
Heinrich, R., & Rapoport, T. A. (1974). A linear steady-state treatment of enzymatic chains. European Journal of Biochemistry, 42, 89–95.
HMDB. (2010). Human metabolome database, Version 2.5. Retrieved Oct 23, 2010 from http://www.hmdb.ca.
Hoffman, G. F., & Feyh, P. (2005). Organic acid analysis. In N. Blau, M. Duran, & M. E. Blaskovics (Eds.), Physician’s guide to the laboratory diagnosis of metabolic diseases (Revised 2 ed., pp. 27–44). Heidelberg: Springer.
Hofhaus, G., Johns, D. R., Hurko, O., Attardi, G., & Chomyn, A. (1996). Respiration and growth defects in transmitochondrial cell lines carrying the 11778 mutation associated with Leber’s hereditary optic neuropathy. Journal of Biological chemistry, 271, 13155–13161.
Janssen, A. J. M., Trijbels, F. J. M., Sengers, R. C. A., et al. (2007). Spectrophotometric assay for complex I of the respiratory chain in tissue samples and cultured fibroblasts. Clinical Chemistry, 53, 729–734.
Johnson, R. A., & Wichern, D. W. (1998). Applied multivariate statistical analysis (4th ed.). Englewood Cliffs: Prentice-Hall, Inc.
Kelley, R. I., & Kratz, L. (1995). 3-methylglutaconic acidaemia in Smith-Lemli-Opitz syndrome. Pediatric Research, 37, 671–674.
Korzeniewski, B. (2001). Theoretical studies on the regulation of oxidative phosphorylation in intact tissues. Biochimica et Biophysica Acta, 1504, 31–45.
Kumps, A., Duez, P., & Mardens, Y. (2002). Metabolic, nutritional, latrogenic, and artifactual sources of urinary organic acids: A comprehensive table. Clinical Chemistry, 48, 708–717.
Kurlemann, G., Paetzke, I., Möller, H., et al. (1995). Therapy of complex I deficiency: Peripheral neuropathy during dichloroacetate theraby. European Journal of Pediatrics, 154, 928–932.
Kuwertz-Bröking, E., Koch, H. G., Marquardt, T., et al. (2000). Renal Fanconi syndrome: First sign of partial respiratory chain complex IV deficiency. Pediatric Nephrology, 14, 495–498.
Lalani, S. R., Vladutiu, G. D., Plunkett, K., et al. (2005). Isolated mitochondrial myopathy associated with muscle coenzyme Q10 deficiency. Archives of Neurology, 62, 317–320.
Mamas, M., Dunn, W. B., Neyses, L., & Goodacre, R. (2011). The role of metabolites and metabolomics in clinically applicable biomarkers of disease. Archives of Toxicology, 85, 5–17.
Mancuso, M., Orsucci, D., Coppedè, F., et al. (2009). Diagnostic approach to mitochondrial disorders: The need for a reliable biomarker. Current Molecular Medicine, 9, 1095–1107.
Mazat, J.-P., Rossignol, R., Malgat, M., et al. (2001). What do mitochondrial diseases teach us about normal mitochondrial functions… that we already knew: Threshold expression of mitochondrial defects. Biochimica et Biophysica Acta, 1504, 20–30.
Mochel, F., Slama, A., Touati, G., et al. (2005). Respiratory chain defects may present only with hypoglycemia. Journal of Clinical Endocrinology and Metabolism, 90, 3780–3785.
Morava, E., van den Heuvel, L., Hol, F., et al. (2006). Mitochondrial disease criteria: Diagnostic application in children. Neurology, 26, 1823–1826.
Morrow, D. A., & de Lemos, J. A. (2007). Benchmarks for the assessment of novel cardiovascular biomarkers. Circulation, 115, 949–952.
Munnich, A., Rustin, P., Rötig, A., et al. (1992). Clinical aspects of mitochondrial disorders. Journal of Inherited Metabolic Disorders, 15, 448–455.
Pfeiffer, T., Schuster, S., & Bonhoeffer, S. (2001). Cooperation and competition in the evolution of ATP-producing pathways. Science, 292, 504–507.
Rabier, D., Diry, C., Rotig, A., et al. (1998). Persistent hypocitrullinaemia as a marker for mtDNA NARP T 8993 G mutation? Journal of Inherited Metabolic Disorder, 21, 216–219.
Rahman, S., Blok, R. B., Dahl, H. H., et al. (1996). Leigh syndrome: Clinical features and biochemical and DNA abnormalities. Annals of Neurology, 39, 343–351.
Rauste-von Wright, M., & Frankenhaeuser, M. (1989). Females’ emotionality as reflected in the excretion of the dopamine metabolite HVA during mental stress. Psychological Reports, 64, 856–858.
Reder, C. (1988). Metabolic control: A structural approach. Journal of Theoretical Biology, 135, 175–201.
Reinecke, F., Smeitink, J. A. M., & van der Westhuizen, F. H. (2009). OXPHOS gene expression and control in mitochondrial disorders. Biochimica et Biophysica Acta—Molecular Basis of Disease, 1792, 1113–1121.
Rossignol, R., Faustin, B., Rocher, C., et al. (2003). Mitochondrial threshold effects. Biochemical Journal, 370, 751–762.
Rzem, R., Veiga-da-Cunha, M., Noël, G., et al. (2004). A gene encoding a putative FAD-dependent l-2-hydroxygluterate dehydrogenase is mutated in l-2-hydroxyglutaric aciduria. Proceedings of the National Academy of Sciences USA, 101, 16849–16854.
Shah, N. S., Mitchell, W. G., & Boles, R. G. (2002). Mitochondrial disorders: A potentially under-recognized etiology of infantile spasms. Journal of Child Neurology, 17, 369–372.
Shepherd, D., & Garland, P. B. (1969). The kinetic properties of citrate synthase from rat liver mitochondria. Biochemistry Journal, 114, 597–610.
Smeitink, J. (2003). Mitochondrial disorders: Clinical presentation and diagnostic dilemmas. Journal of Inherited Metabolic Disease, 26, 199–207.
Smeitink, J., Zeviani, M., Turnbull, D. M., & Jacobs, H. (2006). Mitochondrial medicine: A metabolic perspective on the pathology of oxidative phosphoryation disorders. Cellular Metabolism, 3, 9–13.
Smuts, I., Louw, R., du Toit, H., Klopper, B., Mienie, L. J., & van der Westhuizen, F. H. (2010). An overview of a cohort of South African patients with mitochondrial disorders. Journal of Inherited Metabolic Disease. doi:10.1007/s10545-009-9032-8.
Sperl, W., Jesina, P., Zeman, J., et al. (2006). Deficiency of mitochondrial ATP synthase of nuclear genetic origin. Neuromuscular Disorders, 16, 821–829.
Suomalainen, A. (2010). Biomarkers for mitochondrial respiratory chain disorders. Journal of Inherited Metabolic Disease. doi:10.1007/s10545-010-9222-3.
Sweetman, L., & Williams, J. C. (2001). Branched chain organic acidurias. In C. R. Scriver, A. L. Beaudet, W. S. Sly, & D. Valle (Eds.), The metabolic and molecular bases of inherited disease (2nd ed., Vol. 8, pp. 2125–2163). New York: The McGraw-Hill Companies.
Topaloğlu, R., Lebre, A. S., Demirkaya, E., et al. (2008). Two new cases with Pearson syndrome and review of Hacettepe experience. Turkish Journal of Pediatrics, 50, 572–576.
Topçu, M., Jobard, F., Halliez, S., et al. (2004). l-2-Hydroxyglutaric aciduria: Identification of a mutant gene Ci4orf160, localized on chromosome 14q22.1. Human Molecular Genetics, 13, 2803–2811.
Van Beelen, P., Labro, J. F., Keltjens, J. T., et al. (1984). Derivatives of methanopterin, a coenzyme involved in methanogenesis. European Journal of Biochemistry, 139, 359–365.
Van Schaftingen, E., Rzem, R., & Veiga-da-Cunha, M. (2009). l-2-hydroxyglutaric aciduria, a disorder of mitochondrial repair. Journal of Inherited Metabolic Disease, 32, 135–142.
Verkaart, S., Koopman, W. J. H., Sjenet, E., et al. (2007). Superoxide production is inversely related to complex I activity in inherited complex I deficiency. Biochimica et Biophysica Acta, 1772, 373–381.
Walsh, R., Conway, H., Roche, G., & Mayne, P. D. (1999). What is the origin of 3-methylglutaconic acid? Journal of Inherited Metabolic Disease, 22, 252–255.
Wolf, N. I., & Smeitink, J. A. M. (2002). Mitochondrial disorders: A proposal for consensus diagnostic criteria in infants and children. Neurology, 59, 1402–1405.
Wortmann, S., Rodenburg, R. J., Huizing, M., et al. (2005). Association of 3-methylglutaconic aciduria with sensori-neural deafness, encephalopathy, and Leigh-like syndrome (MEGDEL association) in four patients with a disorder of the oxidative phosphorylation. Molecular Genetics and Metabolism, 88, 47–52.
Acknowledgement
We would like to thank Dr. M. Duran from the Laboratory for Genetic and Metabolic Diseases, Amsterdam Medical Centre (AMC), The Netherlands, for valuable comments on the original manuscript. This study formed part of BioPAD Project BPP007, funded by the South African Department of Science and Technology. Additional financial support from North-West University is likewise acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Reinecke, C.J., Koekemoer, G., van der Westhuizen, F.H. et al. Metabolomics of urinary organic acids in respiratory chain deficiencies in children. Metabolomics 8, 264–283 (2012). https://doi.org/10.1007/s11306-011-0309-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11306-011-0309-0