Abstract
Membrane-bound ectonucleoside triphosphate diphosphohydrolases (E-NTPDases) in the inner ear regulate complex extracellular purinergic type-2 (P2) receptor signalling pathways through hydrolysis of extracellular nucleoside 5′-triphosphates and diphosphates. This study investigated the distribution of NTPDase5 and NTPDase6, two intracellular members of the E-NTPDase family, and linked this to regulation of P2 receptor signalling in the adult rat cochlea. These extracellular ectonucleotidases preferentially hydrolyse nucleoside 5′-diphosphates such as UDP and GDP. Expression of both enzymes at mRNA and protein level was detected in cochlear tissues and there was in vivo release of soluble NTPDase5 and 6 into cochlear fluids. Strong NTPDase5 immunostaining was found in the spiral ganglion neurones and supporting Deiters’ cells of the organ of Corti, while NTPDase6 was confined to the inner hair cells. Upregulation of NTPDase5 after exposure to loud sound indicates a dynamic role for NTPDase5 in cochlear response to stress, whereas NTPDase6 may have more limited extracellular roles. Noise-induced upregulation of co-localised UDP-preferring P2Y6 receptors in the spiral ganglion neurons further supports the involvement of NTPDase5 in regulation of P2Y receptor signalling. Noise stress also induced P2Y14 (UDP- and UDP-glucose preferring) receptor expression in the root processes of the outer sulcus cells, but this was not associated with localization of the E-NTPDases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hearing relies on a sensitive mechanoelectrical transduction process in the cochlea of the inner ear. The cochlea contains sensory, secretory, neural and supporting cells which are all essential to the sound transduction process. A complex extracellular purinergic signalling system contributes to cochlear homeostasis, altering cochlear sensitivity and neural output via ATP-gated ion channels (P2X receptors) and G protein-coupled P2Y receptors [1].
Regulation of extracellular nucleotide levels is critical to the maintenance of cochlear homeostasis. P2 receptor signalling pathways in the cochlea are modulated by the surface-located ectonucleoside triphosphate diphosphohydrolases (E-NTPDases) [2–5]. These enzymes maintain low nanomolar concentrations of ATP in cochlear fluids under quiescent conditions. Stressors such as acoustic overstimulation and hypoxia induce ATP release from cells [6], with concomitant activation of P2X2 and P2X7 receptor subunits in cochlear epithelial and neural tissues. During exposure to loud sound, P2X2 receptors are thought to provide a K+ shunt conductance out of the endolymph to reduce the endocochlear potential [7], whilst the P2X7 receptors likely modulate the olivocochlear efferent neural feedback with consequent reduction in hearing sensitivity [8]. Noise exposure leads to upregulation of P2X2 receptors [9] and membrane-bound NTPDases (NTPDase1–3) [5, 10]. The roles of P2Y receptors in the inner ear are less clear, but they are extensively expressed in the cochlear partition and have multiple roles in the maintenance of K+ transport and cycling, micromechanics of the hair cells and supporting cells, neuromodulation and cochlear development [1]. A role for P2Y receptors in the propagation and sensing of noise damage [11, 12] is also emerging.
NTPDases can control ligand availability at P2 receptors either inside or outside the cell [13]. NTPDase1–3 and NTPDase8 are cell-surface-located enzymes with two transmembrane regions and a large extracellular domain which contains the catalytic site [13, 14]. NTPDase4-7 are intracellular enzymes that preferentially hydrolyze nucleoside diphosphates and triphosphates apart from ATP. The principal substrates of NTPDase5 and NTPDase6 are nucleoside diphosphates UDP and GDP [13, 14]. These two enzymes can be cleaved at the N terminus and released from cells in a soluble form [15–21]. They are localised to the endoplasmic reticulum and Golgi apparatus respectively, and may be involved in glycosylation reactions [18, 22]. NTPDase5, also known as proto-carcinogenic progression of hamster embryo cells (PCPH) [23], is present in normal and tumour samples from rat and human tissue [24, 25], but the evidence for release of soluble forms is currently limited to in vitro studies. NTPDase5 and NTPDase6 were first identified over a decade ago [26], but their tissue distribution and function remain both unexplored and enigmatic.
The role of uridine and guanine nucleotides such as GDP and UDP in the cochlea is unknown, but they have extracellular roles in neurotransmission, vascular tone and chloride secretion in other tissues [27, 28]. This study set out to investigate the expression and distribution of NTPDase5 and NTPDase6 in the cochlea and their putative role in regulation of P2Y receptor signalling by such nucleotides under both quiescent and stressed (noise exposure) conditions.
Materials and methods
Animals
Experiments were carried out on young adult male Wistar rats (8–10 weeks) supplied by the University of Auckland Animal Resources Unit. All procedures were approved by the University of Auckland Animal Ethics Committee and the experiments conformed to international guidelines on the ethical use of animals.
Analysis of NTPDase5 and NTPDase6 mRNA transcripts
Rats were euthanised with sodium pentobarbitone (100 mg/kg i.p.) and their cochleae removed. The cochleae were decapsulated in sterile 0.1 M phosphate-buffered saline (PBS, pH 7.4) and homogenised in lysis buffer (100 mM Tris–HCl pH 8.0, 500 mM LiCl, 10 mM EDTA, pH 8.0, 1% LiDS, 5 mM dithiothreitole) using a sterile Teflon pestle. Cochlear mRNA was isolated using a Dynabeads® mRNA kit (Dynal ASA, Oslo, Norway) as previously described [10]. First strand complementary cDNA was produced by reverse transcription, and then used as template in 50 µl PCR reactions. Rat-specific primer pairs were designed (Oligo Perfect, Invitrogen) from published NTPDase5 (GenBank accession number NM_199394) and NTPDase6 (GenBank accession number AJ277748) sequences (Table 1). Control reactions omitted reverse transcriptase (−RT) from the first strand cDNA reaction mixture to control for genomic DNA contamination. The PCR products were cloned (TA cloning kit, Invitrogen) and resulting plasmids purified (QIA filter Plasmid Midi; Qiagen). The identity of the inserts was confirmed by DNA sequencing.
Western blot analysis
Immunoblotting was used to detect NTPDase5 and NTPDase6 in cochlear tissues and to investigate post-translational modifications (e.g. protein oligomerisation). Rat liver tissues were used as positive controls [19, 29].
Antibodies used for immunoblotting and immunohistochemistry were anti-human polyclonal antibodies for NTPDase5 and NTPDase6. These antibodies were generated by immunisation of rabbits with synthetic peptides corresponding to amino acids EVAKDSIPRSHWKK (NTPDase5; [16]) and TRAAPGARWGQQAH (NTPDase6; [19]). The specificity of both antibodies has been demonstrated on COS-7 cells transiently transfected with plasmids containing the coding sequences for NTPDase5 and NTPDase6 as well as cross-reacting with rat NTPDase5 and NTPDase6 on astrocytes [30].
Rat cochleae were extracted from the auditory bulla, the bony otic capsule removed and tissue homogenised in cold extraction buffer (125 mM Tris–HCl pH 6.8, 10 mM EDTA, 50 mM NaCl, 5% SDS, 20% v/v glycerol) containing a 1% protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany). Similarly, control liver tissues were homogenised in the ice-cold buffer. All tubes were placed on ice, sonicated in brief bursts and centrifuged (16,000 rpm for 10 min at 4°C). Loading buffer (0.1% bromophenol blue; 20% v/v glycerol; 2% SDS) was added to the supernatants, followed by heat denaturation (99°C for 3 min), and centrifugation (2,000 rpm for 2 min). Typically, 20 µg of cochlear protein, and 5 µg of liver protein, in a total volume of 25 µl was loaded into a 10% polyacrylamide gel (BioRad Laboratories, CA, USA) and proteins separated in a sodium dodecyl sulphate (SDS) buffer (25 mM Tris base, 19 mM glycine, 0.1% SDS). A protein ladder was also run on each gel (Blue Ranger Prestained Protein Molecular Weight Marker, Pierce, Rockford, IL, USA). Proteins were transferred overnight to a PVDF membrane in a 4°C transfer buffer (25 mM Tris base, 190 mM glycine, 15% methanol in sterile water, pH 8.4). Transferred proteins were incubated for 2 h at room temperature in blocking solution (5% non-fat milk powder; 2% normal goat serum in Tris-buffered saline, composition: 20 mM Tris–HCl pH7.2; 137 mM NaCl; 0.1% Tween-20). This was followed by overnight incubation at 4°C with either primary antibody (NTPDase5 or NTPDase6, 1:1,000 dilution) or control pre-immune serum (1:1,000 dilution). A control where the primary antibody was omitted was also included. After washes in Tris-buffered saline, a peroxidase-labelled secondary antibody (goat anti-Rabbit IgG; 1:8,000 dilution; Amersham Biosciences, NZ) was applied for 1 h. Protein bands were visualised using an Enhanced Chemiluminescence system according to the manufacturer’s instructions (Amersham Biosciences). The membranes were directly visualised using a Fuji LAS-3000 Imager with a cooled CCD camera and analysed using Fuji Multigauge v3.0 software.
Immunohistochemistry
After transcardial perfusion with a saline solution (0.9% NaCl, 1 mg/ml NaNO2, 0.02% heparin) and fixative (4% PFA in 0.1 M PB, pH 7.4), the cochleae were removed from the auditory bullae, decalcified for 7 days (5% EDTA in 0.1 M PB), cryoprotected overnight in 30% sucrose, snap-frozen in isopentane, and stored at −80°C. The cochleae were cryosectioned either at 20 µm for immunoperoxidase histochemistry or at 50 µm for confocal immunofluorescence. Tissues were permeabilised (1% Triton-X for 1 h) and non-specific binding sites blocked (2% non-fat milk powder and 1.5% normal goat serum). Sections were incubated overnight at 4°C with one of the primary antibodies: NTPDase5 and NTPDase6 polyclonal antibodies at 1 µg/ml, or commercial (Alomone Labs Ltd., Jerusalem, Israel) P2Y6 and P2Y14 polyclonal antibodies at 1:200 dilutions. These peptide-derived antibodies have been previously characterised [30–33]. In control reactions, the primary antibody was replaced with the pre-immune serum, omitted or preabsorbed with the immunising peptide. Only the peptide block control was presented in figures as representative for all controls. Immunoperoxidase reaction was detected using a secondary biotin-conjugated goat anti-rabbit IgG, followed by reaction visualisation using an avidin–biotin–peroxidase complex (ABC kit, Vector Laboratories) and diaminobenzidine (DAB kit, Vector). Images were obtained using a Zeiss Axioskop 2 Mot-plus microscope fitted with a digital camera (Zeiss Axiocam). The images were processed with Axiovision 3.1 software as previously described [34]. Immunofluorescence was detected with Alexa 488 secondary antibody (Invitrogen) and sections examined using a laser scanning confocal microscope (TCS SP2, Leica Leisertechnik, Heidelberg, Germany). Sections were excited with a 488-nm wavelength argon–krypton laser, and a software emission filter narrowed (bandpass 519 nm) to reduce background. Image acquisition was controlled by Scanware software (Leica). A series of six to ten optical sections were collected for each specimen, and image analysis was performed on an optical section from the centre of the stack.
RP-HPLC analysis of nucleotidase activities in the cochlear perfusate
Artificial perilymph (AP) or divalent-cation-free artificial perilymph (DCF-AP) solutions were perfused through the cochlear perilymphatic spaces of control animals and perfusate collected for in vitro incubation with nucleotides and nucleosides. Samples were analysed by reverse phase HPLC (RP-HPLC) to establish hydrolysis profiles for extracellular nucleotides. The artificial perilymph solution contained 137 mM NaCl, 12 mM NaHCO3, 1 mM NaH2PO4, 5 mM KCl, 11 mM glucose, 2 mM CaCl2, 1 mM MgCl2 [35, 36]. Ca2+ and Mg2+ were omitted from the divalent-cation-free AP. pH of both solutions was adjusted to 7.4, and osmolality recorded with a freezing point osmometer (FiskeMark3, USA) as 300 mOsm/kg H2O (AP) and 297 mOsm/kg H2O (DCF-AP).
For cochlear perfusions, the rats were anaesthetized with pentobarbitone sodium (60 mg/kg i.p.), placed in a heated (37°C) head holder with the ventral side up, tracheostomized and left to breathe on their own. Body temperature was maintained at 37°C using a heated blanket. Prior to the opening of the tympanic bulla, the cisterna magna was opened to release the positive pressure that cerebrospinal fluid (CSF) exerts on the perilymph and prevent the contamination of the effluent with CSF. The bulla was opened and two small holes hand-drilled in the cochlea with an ultrafine hypodermic needle (30G, BD, Singapore). The inlet hole was in the basal turn scala tympani and the outlet hole in the basal turn scala vestibuli. Plastic tubing was placed inside the inlet and outlet holes and both tubes were connected to separate Hamilton syringes secured on a push–pull syringe pump (Harvard Apparatus, USA). The cochlea was perfused at a constant perfusion rate of 2.5 µl/min. The initial AP perfusate (∼12 µl) was discarded then AP was further collected for 45 min, followed by DCF-AP perfusion for a further 45 min. Collected perfusates (AP and DCF-AP) were centrifuged separately (4,000 rpm at 4°C for 10 min) before ultracentrifugation (100,000×g) at 4°C for 30 min. Supernatants were aliquoted (10 µl) into sterile tubes. Nucleosides or nucleotides of HPLC grade (2 mM final concentration; Sigma) were then incubated for 1 h at 37°C with AP and DCF-AP perfusates, to establish the hydrolysis profile of the putative soluble ectonucleotidases NTPDase5 and NTPDase6. To control for non-enzymatic nucleotide breakdown, the nucleotides were also incubated for 1 h at 37°C in non-perfused AP. Samples were then snap-frozen on dry ice and stored at −80°C until processing for RP-HPLC. Both AP and DCF-AP samples were batch-processed. Supernatants were transferred to HPLC injection bottles with septa lids (Biolab, NZ). Nucleotides and nucleosides were separated through an Adsorbosphere nucleoside–nucleotide column (Altech, NZ; 7 µm pore size, 250 mm length × 4.6 mm ID) and quantified using RP-HPLC (Agilent 1200 series, Agilent Technologies, Palo Alto, USA) with diode-array detector at 254 nm. All reagents and chemicals used for RP-HPLC were HPLC-grade. The aqueous mobile phase (Buffer A: 60 mM NH4H2PO4; 5 mM tetrabutylammonium phosphate, pH 5.0), and the organic mobile phase (Buffer B: 80% methanol; 5 mM TBAP) were filtered prior to use. The mobile gradient proportion of Buffer B was increased from 10% B to 50% over the first 10 min, and then decreased to 10% over the following 10 min. Peaks were identified by comparison of retention times with endogenous standards, then peak areas recorded as ratios based on the added substrate (nucleoside:nucleoside monophosphate; nucleoside monophosphate:nucleoside diphosphate; nucleoside diphosphate:nucleoside triphosphate). Nucleotide hydrolysis in AP and DCF-AP solution was determined for each nucleotide series (NTP/NDP/NMP).
Noise exposure
Rats (n = 5 per group) were exposed to octave band noise centred at 4.5 kHz presented for 24 h at 90, 100 or 110 dB sound pressure level (SPL). Noise exposures were carried out in a custom-built acoustic chamber (Shelburg Acoustics, Sydney, Australia) with internal speakers and external controls (sound generator and frequency selector). Sound intensity inside the chamber was tested using a calibrated Rion NL-40 sound level metre (Tokyo, Japan) to ensure minimal deviations of sound intensity. Control animals were housed in the Animal Resources Unit at ambient sound conditions (45–55 dB SPL, 0.5–20 kHz).
Real-time RT-PCR
The mRNA expression of NTPDase5 and 6 in the cochlea was quantified by real-time PCR using specific primers and TaqMan® MGB probes carrying a 5′ reporter FAM (6-carboxy fluorescein) and a 3′ non-fluorescent quencher (Applied Biosystems). The primers and probes are shown in Table 1.
Real-time PCR was carried out in MicroAmp Optical 384-well reaction plates using TaqMan® Universal PCR Master Mix (Applied Biosystems) and a mix of unlabelled primers and FAM-labelled TaqMan MGB probes (Custom TaqMan® Gene Expression Assays). As a template, 1 μl of sample cDNA was added to a total reaction volume of 12.5 μl. The samples were tested in duplicate and data were expressed as a mean of two replicates. Negative controls without a template or reverse transcriptase were included in every PCR run. Quantitation of a house-keeping gene, glyceraldehyde-3 phosphate dehydrogenase (GAPDH), was performed for all samples as an endogenous reference. GAPDH-specific primers and VIC-labelled probes are proprietary to Invitrogen. Amplification and fluorescence detection were carried out using the 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The thermal cycling protocol included 2 min at 50°C, 10 min at 95°C and a 40-cycle profile: 15 s at 95°C and 1 min at 60°C. The data were analysed using the Sequence Detector v2.3 software (Applied Biosystems). Gene expression levels of NTPDase5 and 6 were normalised to the reference GAPDH gene expression. The relative gene expression (fold change) of soluble NTPDases was calculated using the 2T −ΔΔC method [37].
Semi-quantitative analysis of NTPDase5 and 6 protein expression
The levels of NTPDase5 and NTPDase6 expression in cochlear tissues were determined by measuring the mean pixel intensity of fluorescence as previously described [10]. Images were analysed using identical acquisition parameters. The middle image from each series was then used in further analysis, after grayscale conversion. Image J Software tools were used to demarcate the boundaries of areas of interest, and for NTPDase5 this was the spiral ganglion cytoplasm. Cells to be measured were chosen by placing a 6 × 6 grid pattern diagonally over the image and selecting the first ten ganglion neurons that crossed the intersect points. Background intensity was measured by selection of an equal pixel area in the image not labelled by NTPDase5 antibody. Mean pixel intensity, with background subtraction, was recorded for ten spiral ganglion neurones from each animal (n = 5 per group). All specimens were processed in a double-blind manner.
Data analysis
Results are presented as the mean ± SEM, and statistical analysis performed using Student’s t test or one-way ANOVA followed by post-hoc Games–Howell test. The α level was set at P = 0.05.
Results
NTPDase5 and NTPDase6 mRNA are expressed in rat cochlea
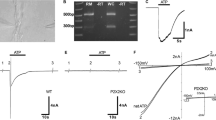
The expression of NTPDase5 and NTPDase6 in the rat cochlea was analysed by RT-PCR, and mRNA transcripts were detected for both enzymes (Fig. 1a, c). Generated PCR products corresponded to the predicted sizes of DNA fragments (541 bp for NTPDase5 and 489 bp for NTPDase6). Control reactions omitting reverse transcriptase (−RT) or cDNA template (NTC) gave no reaction product. Subsequent sequencing and restriction enzyme analysis confirmed the identity of the NTPDase5 and NTPDase6 amplicons.
NTPDase5 and NTPDase6 expression in rat cochlear tissues. RT-PCR generated amplicons (SYBR Safe™ agarose gel) are of expected size for both NTPDase5 at 541 bp (a) and NTPDase6 at 489 bp (c), compared with a 100 bp DNA ladder. No bands are observed in control lanes, −RT reverse transcriptase omitted) and NTC (no template control). DNA sequencing verified the identity of amplified cDNAs. Immunoblots with non-reduced total cochlear protein and liver tissues show major bands around 95 and 47 kDa for NTPDase5 (b). Lower molecular weight band is consistent with the monomeric NTPDase5 isoform, whereas bands at higher molecular weight probably represent dimerized form of NTPDase5. These bands were absent from the pre-immune (PI) and no primary antibody controls. NTPDase6 antibody demonstrated a double-band pattern (46–49 kDa) in the cochlea (d)
Immunoblotting of NTPDase5 and NTPDase6
A principal immunoreactive band around 95 kDa and a smaller band around 47 kDa were detected in cochlear tissues with NTPDase5 antibody under non-reducing conditions (Fig. 1b). This is consistent with NTPDase5 protein bands from rat brain astrocytes identified at approximately 50 kDa (monomer) and 100 kDa (dimer) weights [30]. Neither of these two bands was seen in the pre-immune serum control or in the absence of the primary antibody. Liver tissue used as positive control also shows the two main bands, however the monomeric form is dominant (Fig. 1b). Western blot analyses of normal and tumour samples from rat mammary tissue, human laryngeal, breast and testicular tissues have also demonstrated the presence of oligomeric forms of NTPDase5 [24, 25, 29, 38].
NTPDase6 immunoblots (Fig. 1d) demonstrated two close faint bands around 46–49 kDa consistent with limited NTPDase6 expression in the cochlea (see Fig. 4). The pre-immune serum control and the no primary antibody control were clear. The same NTPDase6 antibody was reported as labelling only one band at 49 kDa on cultured newborn rat brain astrocytes protein [30]. The double band in cochlear tissue may represent different glycosylation of the NTPDase6 protein, as the molecular weight difference appears to be only 2–3 kDa. Recombinant NTPDase6 expressed in COS-7 cells has also been shown to exhibit heterogeneous post-translational modifications [19].
NTPDase5 localisation in rat cochlea
NTPDase5-specific immunofluorescence was detected in the spiral ganglion afferent neurons and the organ of Corti (Fig. 2a). The same distribution was observed with immunoperoxidase histochemistry (Fig. 2b, c), whilst the control sections remained unlabelled (insets). While the majority of spiral ganglion neurons and their central processes were strongly immunolabelled, the efferent fibres in the intraganglionic spiral bundle (igsb) were unlabelled (Fig. 2b). No NTPDase5 labelling was seen in the lateral wall tissues, including the spiral ligament and the stria vascularis. Faint staining in the outer sulcus cells was also observed in the control sections and was therefore rendered non-specific. At high magnification, the spiral ganglion showed a typical intracellular pattern of immunostaining with marked labelling of the central neuronal processes (Fig. 2d). NTPDase5 immunolabelling in the organ of Corti was limited to supporting cells such as Deiters’ and inner border cells (Fig. 3a). Light staining observed in the headplates and footplates of the inner and outer pillar cells was also apparent in the control (inset). Detail of cytoplasmic NTPDase5 localisation in type I spiral ganglion neurons is shown in Fig. 3b. Some neurons appear unstained, suggesting a possibility that NTPDase5 is not expressed in type II neurones, usually identified by their smaller size and grouping in the vicinity of the intraganglionic spiral bundle [39]. This, however, needs to be confirmed by immunolabelling specific for type II neurones. Even though the images show only the middle cochlear turn, the same staining pattern was observed in all cochlear turns.
NTPDase5 distribution in adult rat cochlea. a NTPDase5 immunofluorescence was observed in the spiral ganglion (sgn) and organ of Corti. Images were taken at the mid-cochlear level. b Immunoperoxidase histochemistry demonstrated strong NTPDase5 immunolabelling in the cell bodies of the spiral ganglion neurons (sgn), whilst the efferent neural processes located in the intraganglionic spiral bundle (igsb) were not stained. c In the organ of Corti, NTPDase5 immunoreactivity was observed in the supporting Deiters’ cells (Dc) but not in the sensory inner hair cells (ihc) and outer hair cells (ohc). The inner border cells, medial to the inner hair cells, were immunopositive. d Stereo image (reconstructed from a stack of confocal images) showing labelling of central neuronal processes (cp) of the spiral ganglion neurons. No labelling was found in control tissues in which the antibody was absorbed with the corresponding peptide (insets). sV scala vestibuli, sM scala media, sT scala tympani. Scale bar 100 µm (a) and 20 µm (b–d)
Detail of NTPDase5 immunolocalisation in adult rat cochlea. a NTPDase5-specific antibody showed strong supranuclear cytoplasmic immunoperoxidase labelling of Deiters’ cells (Dc). Weaker labelling was observed in the inner border cells (IBC), but the adjacent inner hair cell (ihc) was unlabelled. Residual labelling in head and footplate of outer pillar cells (oPC) was observed in control tissues after peptide block (inset). b Type I spiral ganglion neurons (based on their size and distribution), demonstrated strong NTPDase5-specific labelling in the cytoplasm, which was not observed in the surrounding satellite cells. Peptide block controls showed no immunolabelling in ganglion cell perikarya (inset). Scale bar 10 µm. Dc Deiters’ cell, iPC inner pillar cell, N nucleus, ohc outer hair cell, oPC outer pillar cell
NTPDase6 localisation to the inner hair cells of adult rat cochlea
Immunostaining with the NTPDase6 antibody in adult rat cochlea produced a very distinctive pattern. Even at low power, NTPDase-specific immunolabelling was exclusively observed in the inner hair cell area (Fig. 4a, b). The inner hair cell labelling was present in all cochlear turns (Fig. 4a). No other cell type in the organ of Corti, spiral ganglion or lateral wall was stained. High-power imaging demonstrated that NTPDase6 was scattered throughout the cytoplasm of the inner hair cells (Fig. 4d). There was no immunolabelling in the inner hair cell area in the control cochlea (Fig. 4e).
NTPDase6 distribution in adult rat cochlea. a Even at a low magnification, NTPDase6 immunoperoxidase labelling was evident in the inner hair cells (ihc) in each turn of the cochlea. b NTPDase6-specific inner hair cell labelling was also observed with confocal immunofluorescence; it was absent from the outer hair cells (ohc), stria vascularis (sv), spiral ligament (sl), or spiral limbus (s limbus). Detail of cytoplasmic NTPDase6 immunoreactivity was evident in the inner hair cells using both immunoperoxidase (c) and confocal immunofluorescence (d). e Inner hair cells are unlabelled in peptide block controls. Abbreviations: Bm basilar membrane, Cc Claudius cells, sM scala media, sT scala tympani, sV scala vestibule, Tm tectorial membrane. Scale bar 50 µm (a, b); 20 µm (c–e)
Evidence for soluble NTPDase5 and NTPDase6 in cochlear fluids of control rats
The hydrolytic profile of nucleoside monophosphates, diphosphates and triphosphates incubated in AP or DCF-AP perfused through the cochlear perilymphatic compartment is shown in Fig. 5. Prior to these experiments, all commercial nucleoside triphosphates and diphosphates (Sigma) were analysed by HPLC to evaluate the level of contamination with nucleoside diphosphates and monophosphates. The contributions of GDP, UDP and ADP in the corresponding nucleoside triphosphate samples were 8%, 7.5% and 4% respectively, and the contributions of GMP, UMP, and AMP in the corresponding nucleoside diphosphate samples were 3%, 10%, and 4.5% respectively. These values were subtracted from nucleotide hydrolysis in cochlear perfusate.
Nucleotide hydrolysis profile in cochlear perfusate. Hydrolysis of extracellular nucleotides in artificial perilymph (AP) or divalent-cation-free artificial perilymph (DCF-AP) perfused through the cochlea and subsequently incubated with NTP/NDP/NMP substrates. a Nucleoside 5′-monophosphates AMP and UMP were not hydrolysed to their corresponding nucleosides, in contrast to GMP which was similarly dephosphorylated in both AP and DCF-AP. b Strong preference for UDP and GDP hydrolysis was divalent-cation-dependent, whilst ADP was not the substrate for the enzymes present in cochlear perfusates. c Nucleoside 5′-triphosphate hydrolysis was marginal when ATP was used as a substrate and slightly higher with UTP and GTP. This hydrolysis was not divalent-cation-dependent. Data presented as mean ± SEM (n = 5) of the HPLC peak area ratio (product/substrate), after correction for non-enzymatic hydrolysis. *P < 0.05, two-tail paired t test
Our results show that AMP and UMP were not hydrolysed in AP or DCF-AP perfused through the cochlea, whilst 4% of GMP was hydrolysed to guanosine in divalent-cation-independent manner (Fig. 5a). Minimal hydrolysis of nucleoside triphosphates (1–4%) was observed for all triphosphates tested (ATP, UTP, GTP), and this hydrolysis was also divalent-cation-independent (Fig. 5c). By contrast, considerable hydrolysis of UDP to UMP and GDP to GMP (but not ADP to AMP) was observed in cochlear perfusate in the presence of divalent cations (AP; 2 mM Ca2+ and 1 mM Mg2+; Fig. 5b). Comparisons between UDP and GDP hydrolysis in AP and DCF-AP (n = 5) demonstrated strong divalent-cation dependence (P = 0.014 for GDP and P = 0.026 for UDP, paired two-tailed t test). This is consistent with soluble NTPDase5 and NTPDase6 activities in the cochlear perfusate which require divalent cations.
Changes in NTPDase5 and NTPDase6 expression with noise exposure
NTPDase5 and NTPDase6 mRNA transcript levels were analysed in the noise-exposed cochleae using quantitative RT-PCR. NTPDase5 was upregulated 2.2-fold (P < 0.05; one-way ANOVA) in the cochleae exposed to 100 dB SPL (Fig. 6a), whilst at 90 and 110 dB SPL relative expression levels of NTPDase5 were not altered significantly. No statistically significant changes in NTPDase6 transcript levels were found in the cochleae exposed to noise (Fig. 6a).
Changes in NTPDase5 and NTPDase6 mRNA and protein expression levels in cochleae exposed to loud sound. a Transcript levels of NTPDase5 and NTPDase6 in noise-exposed cochleae relative to non-noise-exposed control animals. NTPDase5 was upregulated (2.2-fold increase) in the cochlea exposed to 100 dB SPL for 24 h, whilst transcript levels of NTPDase6 were not altered with noise exposure. Data presented as mean ± SEM (n = 5). *P < 0.05, one-way ANOVA with post-hoc Games–Howell test. b The intensity of NTPDase5 immunofluorescence in the spiral ganglion neurons was increased in the cochlea exposed to 100 dB SPL for 24 h (B2) compared to non-noise-exposed cochlea (B1). NTPDase6 immunolabelling was confined to the inner hair cell (ihc) in control and noise-exposed cochleae (B3, B4) and the average pixel intensity of immunostaining was comparable. No labelling in the inner hair cells was observed in peptide block controls (inset). Weak P2Y6 immunolabelling was observed in the spiral ganglion (sgn) of non-noise-exposed animals (B5), but in the cochlea exposed to 100 dB SPL the intensity of immunostaining was substantially increased (B6). P2Y14 was not expressed in normal adult cochlea (B7), but after noise exposure at 100 dB SPL, P2Y14 expression was detected in the root processes (rp) of the outer sulcus (os) cells in the spiral ligament (sl; B8). Immunostaining was absent in the peptide block controls (inset). Scale bars 20 µm (B1, B2, B5–B8); 10 µm (B3, B4). c Semi-quantitative analysis of NTPDase5 immunolabelling in the spiral ganglion neurons after exposure to different sound pressure levels. Abbreviations: igsb intraganglionic spiral bundle, sv stria vascularis. Number of animals: n = 5 per group. *P < 0.05; one-way ANOVA
Noise exposure increases NTPDase5 immunostaining in the spiral ganglion
The intensity and distribution of NTPDase5 and NTPDase6 immunostaining was investigated in the cochlea exposed to loud sound at different sound pressure levels. Confocal immunofluorescence demonstrated increased intensity of NTPDase5 immunolabelling in the spiral ganglion perikarya in the cochleae exposed to 100 dB SPL for 24 h (Fig. 6b2). Semi-quantitative analysis of NTPDase5 immunolabelling (Fig. 6c) demonstrated significant difference between the control and the 100-dB-noise-exposed groups (P < 0.05; one-way ANOVA), consistent with the upregulation of NTPDase5 expression. There was no significant difference between the control and the 90- or 110-dB groups. Other cell types that showed NTPDase5 expression (e.g. Deiters’ cells), were not sufficiently preserved after noise exposure to be able to reliably quantify any change in expression.
No change in NTPDase6 distribution or intensity of immunostaining was observed in the noise-exposed cochleae. NTPDase6-specific immunoreactivity was confined to the inner hair cells in both control and noise-exposed cochleae (Fig. 6b3, b4). Semi-quantitative analysis of immunostaining intensity within the inner hair cells did not show significant differences between noise-exposed and control cochleae (data not shown).
Noise exposure and P2Y6 and P2Y14 receptor expression in the cochlea
P2Y receptors can be activated by extracellular pyrimidine nucleotides such as UTP and UDP. P2Y receptors are thus activated by ligands that are also potential hydrolysis substrates for NTPDase5 and NTPDase6. Therefore, the distribution of the UDP-preferring P2Y6 and the UDP and UDP-glucose-preferring P2Y14 receptors were investigated in the noise-exposed cochlea. P2Y6 receptor immunoreactivity in the spiral ganglion neurones in the non-noise-exposed cochlea (Fig. 6b5) appeared to be upregulated after exposure to 100 dB SPL for 24 h (Fig. 6b6). No P2Y14 receptor immunolabelling was observed in the normal rat cochlea (Fig. 6b7); however, exposure to 100 dB SPL induced P2Y14 receptor-specific immunoreactivity in the outer sulcus root cell region of the spiral ligament (Fig. 6b8). Peptide block controls were unlabelled (insets, Fig. 6b).
Discussion
This study demonstrated the expression and specific distribution of intracellular nucleotidases NTPDase5 and NTPDase6 in the adult rat cochlea and the release of these enzymes into cochlear fluids. NTPDase5 is likely involved in the regulation of P2Y receptor signalling, whilst NTPDase6 may be more preferentially involved in intracellular processes vital for the inner hair cell metabolic function.
NTPDase5 was localised to the supporting Deiters’ cells and spiral ganglion neurons, whilst NTPDase6 was very specifically confined to the inner hair cells. These findings suggest differential functional roles for these two enzymes. Cytoplasmic NTPDase5 localisation is consistent with the intracellular localisation of PCPH, the same entity as NTPDase5, reported in human tumour and normal cells [38, 40]. Cell bodies of the type I neurons are known to contain abundant rough endoplasmic reticulum (ER) and the NTPDase5 intracellular distribution in the present study was also comparable to calreticulin distribution in murine SGN [41]. Other studies have suggested that the subcellular localisation of NTPDase5 to the ER and preference for UDP as substrate imply that this enzyme is involved in the regulation of intracellular glycosylation [22]. NTPDase5 may also have an extracellular enzymatic function, as supported by the release of soluble enzymes into cochlear fluids.
Other members of the NTPDase family (NTPDase1–3) have been differentially localised to the nearby OHC synaptic region where they likely regulate responses of P2X (P2X2, P2X7) and P2Y (P2Y4) receptors [1, 4, 5]. Hydrolysis of extracellular UTP by the plasma membrane-located NTPDases regulates P2Y4 receptor signalling and generates significant UDP accumulation [42]. Release of soluble NTPDase5 from adjacent Deiters’ cells into the extracellular space may be responsible for the removal of UDP and generation of UMP, which is further hydrolysed to uridine by ecto-5′-nucleotidase. NTPDase5 may thus contribute to salvage and recycling of uridine following an uptake via nucleoside transporters.
The localisation of membrane-bound NTPDase1 and NTPDase3 to SGN cell bodies is proposed to be involved in the regulation of the P2X2 and P2X7 receptors signalling, although the impact of this regulation on neural output is unknown. Consideration of the nature of the P2Y receptors on these cells could give some indication of the functional role of NTPDase5 in SGN. In cultured rat sympathetic ganglion neurons expressing P2Y4 and P2Y6 receptors, activation by UTP or UDP inhibits preactivated G protein-coupled inward rectifier K+ (GIRK) channels [43]. Activation of P2Y6 receptors in vivo could therefore modulate neuronal excitability. P2Y4 and P2Y6 receptor signalling responses in SGN (see also Huang et al. 2010—this issue) are likely controlled by NTPDase5 in conjunction with membrane-bound NTPDase1 and NTPDase3.
Evidence for the presence of soluble forms of NTPDase5 and NTPDase6 in the cochlear perfusate is supported by the preferential hydrolysis of nucleoside diphosphates (GDP and UDP > > ADP) and the divalent-cation dependence of hydrolysis. Recombinant NTPDase5 and NTPDase6 have a marked preference for GDP and UDP over ADP, with Km for GDP in the micromolar range and ADP hydrolysis either too low to measure, or with an activity rate of 5–10% of the GDP hydrolysis rate [18, 21]. The absence of divalent cations dramatically decreases activity for recombinant soluble NTPDase5 and NTPDase6. Other possible candidates for this nucleotidase enzymatic activity include soluble NPP2 (autotaxin) and Ca2+-activated nucleotidase (CAN). While NPP2 does require divalent cations for enzymatic activity, it also hydrolyses nucleoside mono-, di- and triphosphates [44]. In contrast, the enzymatic activity of CAN has been shown to also have a marked preference for nucleoside diphosphates UDP and GDP but not ADP, and has a strict requirement for Ca2+ for activity [45]. While a soluble recombinant human form (SCAN) is expressed in cell culture medium, no corresponding rat soluble recombinant homologue is produced. Therefore, NTPDase5 and/or NTPDase6 remain the most likely candidates for this enzymatic action. This conclusion is based on the comparison of hydrolysis profiles in the presence and absence of divalent cations, but there remains the possibility that a rundown in release of NTPDases during the sample collection period could also account for the differences in nucleotide hydrolysis between these samples. A follow-up study should reverse the collection order to control for this possibility.
Release of NTPDase5 and 6 into cochlear fluids observed in this study may be induced by stress due to temporal bone surgery. In other cells, factors that trigger the release of extracellular nucleotides can result in co-release of soluble ectonucleotidases involved in the regulation of their concentration [46]. Hence, loud sound and hypoxia are likely triggers for the release of soluble nucleotidases in the cochlea [7]. However, this does not preclude sustained release of NTPDase5 and NTPDase6 under quiescent conditions, which would help maintain low extracellular nucleoside diphosphate levels and hence suppress P2Y receptor activation when hearing sensitivity is greatest.
This study showed evidence for NTPDase5 upregulation after noise exposure (4.5 kHz octave band noise presented at 100 dB SPL for 24 h). Interestingly, the lower (90 dB SPL) and the higher (110 dB SPL) sound levels did not change the expression of NTPDase5. Exposure to mild noise (90 dB SPL) may not be sufficient to change expression levels of NTPDase5, whilst cell loss after exposure to 110 dB SPL may obscure changes in NTPDase5 transcript levels. A significant increase in NTPDase5 immunolabelling was observed in the spiral ganglion neurons after exposure to 100 dB SPL for 24 h. In addition, UDP-preferring P2Y6 receptor appeared to be upregulated in SGN in response to 100 dB SPL. Increased NTPDase5 and P2Y6 expression in the SGN is consistent with an upregulation of other components of the purinergic signalling system with noise [5, 9, 10]. In addition, loud sound induces an inflammatory response in the cochlea with the infiltration of inflammatory cells and an increase in the expression of a variety of genes involved in inflammatory responses [47, 48]. UDP is a pro-inflammatory signal [49], and its release from damaged neurons may promote the macrophage migration into the cochlea. Upregulation of NTPDase5 could thus reflect an anti-inflammatory signal in the noise-exposed cochlea. In contrast, noise-induced expression of P2Y14 receptor in the outer sulcus coincides only with the expression of NTPDase8 (unpublished data), whilst NTPDase5 and NTPDase6 are not expressed in that region of the cochlea.
NTPDase6 was found exclusively in the cytoplasm of the inner hair cells. In the cochlea, both outer and inner hair cells are sensory cells but they play fundamentally different roles. Activation of stereocilia bundles on inner hair cells results in subsequent release of neurotransmitter from the synaptic region and firing of auditory nerve fibres conducting the encoded signal to the brain. An active process of cochlear mechanics centred on the electromotile outer hair cells enhances hearing sensitivity and frequency selectivity by modulating inner hair cell output. Polarised inner hair cells are therefore the main sensory cells involved in hearing transduction and exhibit specialised intracellular and biochemical organisation.
The NTPDase6 labelling pattern seen in the inner hair cells was similar to that reported in cultured astrocytes with a punctate and granular distribution throughout the cytoplasm and around the nucleus [30]. P2Y6 receptor expression occurs in the cuticular plate region of the adult rat inner hair cells (Huang et al. 2010—this issue), suggesting that NTPDase6 in the cochlea may also be associated with regulation of P2Y receptor signalling.
In conclusion, this study presents intriguing initial evidence of the expression, distribution and release of NTPDase5 and 6 in the cochlea. Soluble NTPDases likely have intra- and extracellular roles in cochlear function and response to stress.
References
Housley GD, Bringmann A, Reichenbach A (2009) Purinergic signaling in special senses. Trends Neurosci 32(3):128–141
Vlajkovic SM, Thorne PR, Housley GD et al (1998) Ecto-nucleotidases terminate purinergic signaling in the cochlear endolymphatic compartment. NeuroReport 9(7):1559–1565
Vlajkovic SM, Thorne PR, Housley GD et al (1998) The pharmacology and kinetics of ecto-nucleotidases in the perilymphatic compartment of the guinea-pig cochlea. Hear Res 117(1–2):71–80
Vlajkovic SM, Thorne PR, Sévigny J et al (2002) Distribution of ectonucleoside triphosphate diphosphohydrolases 1 and 2 in rat cochlea. Hear Res 170(1–2):127–138
Vlajkovic SM, Vinayagamoorthy A, Thorne PR et al (2006) Noise-induced up-regulation of NTPDase3 expression in the rat cochlea: implications for auditory transmission and cochlear protection. Brain Res 1104(1):55–63
Muñoz DJ, Kendrick IS, Rassam M et al (2001) Vesicular storage of adenosine triphosphate in the guinea-pig cochlear lateral wall and concentrations of ATP in the endolymph during sound exposure and hypoxia. Acta Otolaryngol 121(1):10–15
Thorne PR, Muñoz DJB, Housley GD (2004) Purinergic modulation of cochlear partition resistance and its effect on the endocochlear potential in the guinea pig. JARO 5(1):58–65
Nikolic P, Housley GD, Thorne PR (2003) Expression of the P2X7 rceptor subunit of the adenosine 5′-triphosphate-gated ion channel in the developing and adult rat cochlea. Audiol Neurootol 8(1):28–37
Wang JC, Raybould NP, Lin L et al (2003) Noise induces up-regulation of P2X2 receptor subunit of ATP-gated ion channels in the rat cochlea. NeuroReport 14(6):817–823
Vlajkovic SM, Housley GD, Muñoz DJB et al (2004) Noise exposure induces up-regulation of ecto-nucleoside triphosphate diphosphohydrolases 1 and 2 in rat cochlea. Neurosci 126(3):763–773
Gale JE, Piazza V, Ciubotaru CD et al (2004) A mechanism for sensing noise damage in the inner ear. Curr Biol 14(6):526–529
Piazza V, Ciubotaru CD, Gale JE et al (2007) Purinergic signalling and intercellular Ca2+ wave propagation in the organ of Corti. Cell Calcium 41(1):77–86
Robson SC, Sévigny J, Zimmermann H (2006) The E-NTPDase family of ectonucleotidases: structure function relationships and pathophysiological significance. Purinergic Signalling 2(2):325–441
Zimmermann H (2001) Ectonucleotidases: some recent developments and a note on nomenclature. Drug Dev Res 52(1–2):44–56
Mulero JJ, Yeung G, Nelke ST et al (1999) CD39-L4 is a secreted human apyrase, specific for the hydrolysis of nucleoside diphosphates. J Biol Chem 274(29):20064–20067
Mulero JJ, Yeung G, Nelken ST et al (2000) Biochemical characterisation of CD39L4. Biochem 39(42):12924–12928
Hicks-Berger CA, Chadwick BP, Frischauf A-M et al (2000) Expression and characterisation of soluble and membrane-bound human nucleoside triphosphate diphosphohydrolase 6 (CD39L2). J Biol Chem 275(44):34041–34045
Braun N, Fengler S, Ebeling C et al (2000) Sequencing, functional expression and characterization of rat NTPDase6; a nucleoside diphosphatase and novel member of the ecto-nucleoside triphosphate diphosphohydrolase family. Biochem J 351(3):639–647
Yeung G, Mulero JJ, McGowan DW et al (2000) CD39L2, a gene encoding a human nucleoside diphosphatase, predominantly expressed in the heart. Biochem 39(42):12916–12923
Ivanenkov VV, Murphy-Piedmonte DM, Kirley TL (2003) Bacterial expression, characterization, and disulfide bond determination of soluble human NTPDase6 (CD39L2) nucleotidase: implications for structure and function. Biochem 42(40):11726–11735
Murphy-Piedmonte DM, Crawford PA, Kirley TL (2005) Bacterial expression, folding, purification and characterization of soluble NTPDase5 (CD39L4) ecto-nucleotidase. Biochim Biophys Acta 1747(2):251–259
Trombetta ES, Helenius A (1999) Glycoprotein reglucosylation and nucleotide sugar utilization in the secretory pathway: identification of a nucleoside diphosphatase in the endoplasmic reticulum. EMBO J 18(12):3282–3292
Paez JG, Recio JA, Rouzaut A et al (2001) Identity between the PCPH proto-oncogene and the CD39L4 (ENTPD5) ectonucleoside triphosphate diphosphohydrolase gene. Int J Oncol 19(6):1249–1254
Blánquez MJ, Regadera J, Mariño J et al (2002) Gradual deregulation and loss of PCPH expression in the progression of human laryngeal neoplasia. Mol Carcinog 35(4):186–195
Regadera J, Blanquez MJ, Gonzalez-Peramato P et al (2006) PCPH expression is an early event in the development of testicular germ cell tumors. Int J Oncol 28(3):595–604
Chadwick BP, Frischauf AM (1998) The CD39-like gene family: identification of three new human members (CD39L2, CD39L3, and CD39L4), their murine homologues, and a member of the gene family from Drosophila melanogaster. Genomics 50(3):357–367
Communi D, Boeynaems JM (1997) Receptors responsive to extracellular pyrimidine nucleotides. Trends Pharmacol Sci 18(3):83–86
Santos TG, Souzab DO, Tascaa CI (2006) GTP uptake into rat brain synaptic vesicles. Brain Res 1070(1):71–76
Solanas M, Escrich E, Rouzaut A et al (2002) Deregulated expression of the PCPH proto-oncogene in rat mammary tumors induced with 7, 12-dimethylbenz[a]anthracene. Mol Carcinog 33(4):219–227
Wink MR, Braganhol E, Tamajusuku ASK et al (2006) Nucleoside triphosphate diphosphohydrolase-2 (NTPDase2/CD39L1) is the dominant ectonucleotidase expressed by rat astrocytes. Neurosci 138(2):421–432
Xiang Z, Burnstock G (2006) Distribution of P2Y6 and P2Y12 receptor: their colocalization with calbindin, calretinin and nitric oxide synthase in the guinea pig enteric nervous system. Histochem Cell Biol 125(4):327–336
Kudirka JC, Panupinthu N, Tesseyman MA et al (2007) P2Y nucleotide receptor signaling through MAPK/ERK is regulated by extracellular matrix: involvement of β3 integrins. J Cell Physiol 213(1):54–64
Amadio S, Montilli C, Picconi B et al (2007) Mapping P2X and P2Y receptor proteins in striatum and substantia nigra: an immunohistological study. Purinergic Signalling 3(4):389–398
Vlajkovic SM, Abi S, Wang CJH, Housley GD, Thorne PR (2007) Differential distribution of adenosine receptors in the rat cochlea. Cell Tissue Res 328(3):461–471
Zhang SY, Robertson D, Yates G et al (1999) Role of L-Type Ca2+ channels in transmitter release from mammalian inner hair cells I. Gross sound-evoked potentials. J Neurophysiol 82(6):3307–3315
Robertson D, Paki B (2002) Role of L-type Ca2+ channels in transmitter release from mammalian inner hair cells. II. Single-neuron activity. J Neurophysiol 87(6):2734–2740
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆CT method. Methods 25(4):402–408
Blánquez MJ, Arenas MI, Conde I et al (2004) Deregulated expression of the PCPH proto-oncogene in human breast cancers. Int J Oncol 5(4):821–830
Hafidi A (1998) Peripherin-like immunoreactivity in type II spiral ganglion cell body and projections. Brain Res 805:181–190
Recio JA, Zambrano N, Pena L et al (2000) The human PCPH proto-oncogene: cDNA identification, primary structure, chromosomal mapping, and expression in normal and tumor cells. Mol Carcinog 27(3):229–236
Cryns K, Thys S, Van Laer L et al (2003) The WFS1 gene, responsible for low frequency sensorineural hearing loss and Wolfram syndrome, is expressed in a variety of inner ear cells. Histochem Cell Biol 119(3):247–256
Kukulski F, Levesque SA, Lavoie EG et al (2005) Comparative hydrolysis of P2 receptor agonists by NTPDases 1, 2, 3 and 8. Purinergic Signalling 1(2):193–204
Filippov AK, Fernandez-Fernandez JM, Marsh SJ et al (2004) Activation and inhibition of neuronal G protein-gated inwardly rectifying K+ channels by P2Y nucleotide receptors. Mol Pharmacol 66(3):468–477
Lee J, Jung ID, Nam SW et al (2001) Enzymatic activation of autotaxin by divalent cations without EF-hand loop region involvement. Biochem Pharmacol 62(2):219–224
Failer BU, Braun N, Zimmermann H (2002) Cloning, expression, and functional characterization of a Ca2+-dependent endoplasmic reticulum nucleoside diphosphatase. J Biol Chem 277(40):36978–36986
Yegutkin G, Bodin P, Burnstock G (2000) Effect of shear stress on the release of soluble ecto-enzymes ATPase and 5'-nucleotidase along with endogenous ATP from vascular endothelial cells. Br J Pharmacol 129(5):921–926
Hirose K, Discolo CM, Keasler JR et al (2005) Mononuclear phagocytes migrate into the murine cochlea after acoustic trauma. J Comp Neurol 489(2):180–194
Kirkegaard M, Murai N, Risling M et al (2006) Differential gene expression in the rat cochlea after exposure to impulse noise. Neurosci 142(2):425–435
Inoue K, Koizumi S, Tsuda M (2007) The role of nucleotides in the neuron-glia communication responsible for the brain functions. J Neurochem 102(5):1447–1458
Acknowledgements
This study was supported by the Auckland Medical Research Foundation. MOK was also supported by the University of Auckland Doctoral Scholarship.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
O’Keeffe, M.G., Thorne, P.R., Housley, G.D. et al. Distribution of NTPDase5 and NTPDase6 and the regulation of P2Y receptor signalling in the rat cochlea. Purinergic Signalling 6, 249–261 (2010). https://doi.org/10.1007/s11302-010-9190-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11302-010-9190-y