Abstract
In recent years, the native woodlands of Europe, including those of Britain and Ireland, have increasingly come under threat from a range of biotic and abiotic factors, and are therefore a conservation priority demanding careful management in order to realise their inherent ecological and cultural benefits. Because the distribution of genetic variation across populations and regions is increasingly considered an important component of woodland management, we carried out a population genetic analysis on black alder (Alnus glutinosa) across Northern Ireland in order to inform “best practice” strategies. Our findings suggest that populations harbour high levels of genetic diversity, with very little differentiation between populations. Significant F IS values were observed in over half of the populations analysed, however, which could reflect inbreeding as a result of the patchy occurrence of alder in Northern Ireland, with scattered, favourable damp habitats being largely isolated from each other by extensive tracts of farmland. Although there is no genetic evidence to support the broad-scale implementation of tree seed zones along the lines of those proposed for native woodlands in Great Britain, we suggest that the localised occurrence of rare chloroplast haplotypes should be taken into account on a case-by-case basis. This, coupled with the identification of populations containing high genetic diversity and that are broadly representative of the region as a whole, will provide a sound genetic basis for woodland management, both in alder and more generally for species that exhibit low levels of genetic differentiation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In recent years, the native woodlands of Europe, including those of Britain and Ireland, have increasingly come under threat from a range of biotic and abiotic factors, including habitat loss and fragmentation, often as a result of land-use change, invasive species, emergent pests and diseases and global climate change (Rackham 2008). Less than 1.5 % of the land area in Britain is occupied by native forest (Brown 1997). Comparable data for Ireland suggest that less than 1 % of land area is native woodland, and this is continuing to decrease as a result of intensive agriculture and forestry practice (Cross 1998). Our remaining native woodland, therefore, is a conservation priority demanding careful management in order to realise its inherent ecological and cultural benefits (Thomas et al. 1997).
Black alder (Alnus glutinosa [L.] Gaertn.) is a key component of European broadleaved woodlands, which is also found in highly fragmented populations in the extreme northern reaches of northwest Africa (Claessens et al. 2010). The species thrives in damp and riparian habitats, which often results in a patchy distribution, but where it does form stands, it represents an important component of riverine systems by ameliorating erosion (Claessens 2003), as well as being one of the few tree species that fixes atmospheric nitrogen via symbiosis with bacteria of the genus Frankia (Bond et al. 1954). Its flowers are wind-pollinated catkins which are self-incompatible, and seeds are generally dispersed by wind or water, having cork appendages that can aid floatation for up to a year (McVean 1953).
The increased threats to native tree species, including alder, which has been impacted in the last few decades by a disease caused by the oomycete Phytophthora alni (Brasier et al. 1995), demands the development of “best practice” management strategies. Where restocking or replanting is required, it has been recommended that seed from the same area are used to reflect local provenance (Herbert et al. 1999). The Forestry Commission in Great Britain has consequently delineated “seed zones” to assist in the selection of appropriate material. Such areas, however, are defined primarily by climatic and broad ecological factors, and do not take into account the distribution of genetic variation across populations and regions, which is increasingly considered an important component of woodland management (Müller-Starck et al. 1992; Ennos et al. 1998). A recent study on European ash (Fraxinus excelsior) demonstrated that populations across Northern Ireland are represented by a single gene pool, and thus suggests that material for replanting need not be locally sourced (Beatty et al. 2015). In the present study, we analysed populations of alder from across the same region, since increasing deforestation and land-use change for agriculture is putting many suitable habitats at risk necessitating the development of rational management programmes.
Materials and methods
Sampling and DNA extraction
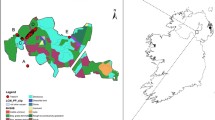
Samples were collected from 24 sites across Northern Ireland and 1 site each in the Republic of Ireland and Scotland that had been previously designated as ancient or semi-natural woodland based on data collected for the Woodland Trust Inventory of ancient and long-established woodland in Northern Ireland (www.backonthemap.org.uk), the National Survey of Native Woodlands 2003–08 in the Republic of Ireland (www.npws.ie) and the Scottish Ancient Woodland Inventory (www.snh.gov.uk; Fig. 1 and Table 1). A single leaf was collected from each of 30 trees per site and stored in silica gel, and GPS coordinates recorded for every tree sampled. DNA was extracted using the CTAB method of Doyle and Doyle (1987). Nuclear genotypes were obtained for between 14 and 29 individuals per population (Table 1; total = 632; mean = 24.308), and chloroplast haplotypes were obtained for between 12 and 30 individuals per population (Table 1; total = 673; mean = 25.885).
Locations of sites sampled in this study. Numbers correspond to those in Table 1
Genotyping
All trees were genotyped for 11 nuclear and 6 chloroplast microsatellite loci. For nuclear microsatellite genotyping, we used 11 of the 12 previously reported loci developed for alder, with the exception of locus Ag23, which could not be consistently amplified (Lepais and Bacles 2011). Forward primers included a 19 bp M13 tail (CACGACGTTGTAAAACGAC) and reverse primers included a 7 bp tail (GTGTCTT). The 11 nuclear loci were amplified in 3 separate multiplex reactions (Ag05, Ag10, Ag14, Ag30 with 6-FAM; Ag01, Ag13, Ag27, Ag35 with HEX; Ag09, Ag20, Ag25 with PET) and combined for capillary electrophoresis.
A. glutinosa chloroplast sequences in the GenBank database were searched for mononucleotide repeats of nine or more (Provan et al. 2001). Primers were designed using the Primer3 program to amplify the six loci in two multiplexes (Table S1, Supporting Information).
PCR was carried out in a total volume of 10 μl containing 100 ng genomic DNA, 5 pmol of 6-FAM-, HEX- or PET-labelled M13 primer, 0.05 pmol of each M13-tailed forward primer, 5 pmol each reverse primer, 1× PCR reaction buffer, 200 μM each dNTP, 2.5 mM MgCl2 and 0.25 U GoTaq Flexi DNA polymerase (Promega, Sunnyvale, CA, USA). PCR was carried out on a MWG Primus thermal cycler (Ebersberg, Germany) using the following conditions: initial denaturation at 94 °C for 3 min followed by 40 cycles (30 for chloroplast loci) of denaturation at 94 °C for 30 s, annealing at 58 °C for 30 s, extension at 72 °C for 30 s and a final extension at 72 °C for 5 min. Genotyping was carried out on an AB3730xl capillary genotyping system. (Applied Biosystems, Foster City, CA, USA). Allele sizes were scored using the GeneMapper software package (v4.1; Applied Biosystems) using LIZ-500 size standards, and were checked by comparison with previously sized control samples. Chromatograms were all inspected visually.
Data analysis
GenePop (V3.4; Raymond and Rousset 1995) was used to test for linkage disequilibrium between nuclear microsatellite loci. To estimate genetic diversity within the populations, levels of observed (H O ) and expected (H E ) heterozygosity, levels of allelic richness (A R ) and fixation indices (F IS) were calculated using the Fstat software package (V2.9.3.2; Goudet 2001). Significance of F IS was determined by 10,000 randomisation steps. Chloroplast microsatellite allele sizes were combined into haplotypes, and levels of genetic diversity (H) based on haplotype frequencies were calculated using the Arlequin software package (V3.5.1.2; Excoffier and Lischer 2010). To account for differences in sample sizes, levels of haplotype richness (R h ) were also calculated using Haplotype Analysis (V1.05; Eliades and Eliades 2009).
The overall level of genetic differentiation between populations was estimated using Φ ST, which gives an analogue of F ST (Weir and Cockerham 1984) calculated within the analysis of molecular variance (AMOVA) framework (Excoffier et al. 1992) using Arlequin. To further identify possible patterns of genetic structuring, the software package BAPS (V5; Corander et al. 2003) was used to identify clusters of genetically similar populations. The program uses a greedy stochastic optimization algorithm to determine K, the number of clusters. Ten replicates were run for all possible values of K up to K = 26, the number of populations sampled. Multiple independent runs always gave the same outcome.
Results
No significant evidence of consistent linkage disequilibrium (i.e. involving the same loci) was detected between any of the 11 nuclear microsatellites analysed (35 out of 1430 tests). Between 5 (Ag20) and 19 (Ag14) alleles were detected per locus, with a total of 130 (mean = 11.818 per locus; Table 1; Figs. S1–S11, Supporting Information). Within populations, levels of allelic richness (A R) averaged over loci ranged from 3.786 (Gortin Glen) to 5.056 (Rostrevor), with a mean value of 4.664 (Table 1). Thirteen private alleles (10 % of the total number of alleles) were detected, with the number per population ranging from 0 to 2 (see Figs. S1–S11, Supporting Information). The majority (11) of these were restricted to a single individual, with 1 of those remaining being found in 2 individuals and the other found in 3 individuals from the Roe Valley population (16/632 = 2.5 % of all trees studied). Levels of observed (H O) and expected (H E) heterozygosity ranged from 0.530 (Gortin Glen) to 0.680 (Ballinderry Bridge; mean = 0.612), and from 0.584 (Gortin Glen) to 0.708 (Marble Arch; mean = 0.663) respectively. The heterozygote deficit observed in all of the populations gave rise to F IS values which were significantly different from 0 in 17 of the 26 populations studied, ranging from 0.008 (Lough Beg) to 0.155 (Correl Glen; mean = 0.078).
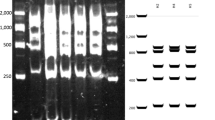
Four of the six chloroplast microsatellite loci studied were polymorphic, exhibiting either two or three alleles (Table S1, Supporting Information). Combining allele sizes across loci gave seven haplotypes. Two of these (H1 and H2) were found in the vast majority (648 out of 673) of the trees studied (Table 1; Fig. 2). Of the remainder, H3 was found in 19 individuals, 17 of which belonged to the Fardross Forest population. Levels of haplotype diversity (H) ranged from 0.069 (Castle Archdale) to 0.582 (Crom). Levels of haplotype richness (R h ) ranged from 0.414 (Castle Archdale) to 2.196 (Fardross Forest).
Levels of population differentiation based on the nuclear and chloroplast markers were Φ ST = 0.0195 and Φ ST = 0.1864 respectively (Table 2). Population-pairwise Φ ST values ranged from −0.034 (Hillborough/Ballinderry Bridge) to 0.078 (Portaferry/Lough Beg) for the nuclear microsatellites, and from −0.058 (Hillsborough/Gortin Glen) to 0.850 (Portaferry/Castle Archdale). The BAPS analysis assigned 25 of the 26 populations to the same genetic cluster, the exception being the Glenarriff population.
Discussion
The findings of the present study suggest that alder populations across Northern Ireland harbour high levels of genetic diversity, with very little differentiation between populations. It is somewhat difficult to put the levels of genetic diversity observed into any significant context, since only a single population genetic analysis of alder using microsatellites has been carried out. In it, a study on fragmented populations from Northern Africa (Lepais et al. 2013), gene diversity values (equivalent to expected heterozygosity) ranged from 0.48 to 0.59 for diploid populations, with a mean value of 0.54 (unusual tetraploid populations from Morocco exhibited higher values). These were lower than the values observed in Northern Ireland in this study, which ranged from 0.584 to 0.699, with a mean of 0.663. This value is similar to that exhibited by the sole northern population analysed in the previous study, which was from Perthshire in Scotland (0.67). We also genotyped 23 individuals from a population in Tarbet, Scotland, which had H E = 0.662, as well as 24 individuals from Coolure, Co. Westmeath (H E = 0.617). Thus, the levels of genetic diversity in Northern Irish populations of alder would appear to be comparable to those from the rest of Great Britain and Ireland. Significant F IS values were found in over half of the populations analysed, and were generally in excess of those reported in both an allozyme study on Slovakian populations (Gömöry and Paule 2002) and a microsatellite study on a single Scottish population (Lepais and Bacles 2011), but lower than that reported in an allozyme study in Poland (Mejnartowicz 2008). This could be a result of inbreeding, reflecting the patchy occurrence of alder in Northern Ireland, with scattered, favourable damp habitats being largely isolated from each other by extensive tracts of farmland. Nevertheless, this is not entirely consistent with the apparent high levels of gene flow, most likely via pollen given the low levels of genetic differentiation observed at the nuclear microsatellite loci. Alternative explanations could involve Wahlund effects, as a result of substructuring within populations, and/or unevenness in patterns of recruitment (“sweepstakes recruitment”).
Analysis of genetic structuring based on nuclear microsatellites did not reveal any obvious patterns. Overall levels of differentiation were low, with around 2 % of the total diversity being partitioned between populations (Table 2), a figure similar to that in Slovakian populations (Gömöry and Paule 2002). Consequently, all but one of the populations (including those from the Republic of Ireland and Scotland) were assigned to a single genetic cluster in the BAPS analysis. Assignment of the Glenarriff population to an alternative cluster is most likely an artefact of the BAPS algorithm, which has been shown previously to tend to overestimate the true number of clusters, particularly where levels of differentiation are low (Latch et al. 2006). An examination of the allele frequencies at each of the 11 loci (Figs. S1–S11, Supporting Information) indicates that any difference in the Glenarriff population is largely due to slight differences in the frequency of a low number of alleles at a few of the loci.
Chloroplast markers tend to reveal more genetic structuring in natural plant populations due to their lower effective population size and, in angiosperms, being maternally inherited and thus dispersed via seed (Provan et al. 2001). This was reflected in Φ ST values for chloroplast microsatellites that were an order of magnitude higher than those for nuclear microsatellites, indicating that 19 % of the genetic variation was partitioned between populations. Nevertheless, this genetic variation was not partitioned geographically on any broad scale. Thus, from a management point of view, any recommendations concerning restocking should be taken at the population or local level, particularly where the population in question contains a high proportion of genotypes not found elsewhere. An obvious example of this is the Fardross Forest population, which is dominated by an otherwise relatively rare haplotype. In the event of complete loss of trees from a woodland, for example following a catastrophic disease outbreak, it would be prudent to use some form of “genetic matching” to identify populations with broadly similar haplotype compositions. Additionally, as a general recommendation for good conservation genetic practice, particular attention should be paid to populations of a species that contain high levels of diversity and exhibit multiple haplotypes (Allendorf and Luikart 2007). In alder, for example, the Fardross Forest population contains four of the five non-unique haplotypes found in Northern Ireland, including the two haplotypes that dominate the remaining populations, with the remaining non-unique haplotype being found in a single individual from each of the extreme southeasterly populations (Portaferry and Hollymount). Such smaller-scale geographical localization of haplotypes represents a further factor that should be taken into account when assessing potential material for restocking.
The findings of the present study provide a good framework for the development of best practice management for native woodlands, particularly for species that exhibit low levels of genetic differentiation, including many tree species. Although there is no genetic evidence in alder or in ash (Beatty et al. 2015) to suggest the broad-scale implementation of seed zones along the lines of those proposed for Great Britain, there is enough evidence to suggest that the localised occurrence of rare haplotypes should be taken into account on a case-by-case basis. This, coupled with the identification of populations containing high genetic diversity and that are broadly representative of the region as a whole, will provide a sound genetic basis for woodland management.
References
Allendorf FW, Luikart G (2007) Conservation and the genetics of populations. Blackwell, Oxford
Beatty GE, Brown JA, Cassidy EM, Finlay CMV, McKendrick L, Montgomery WI, Reid N, Tosh DG, Provan J (2015) Lack of genetic structure and evidence for long-distance dispersal in ash (Fraxinus excelsior) populations under threat from an emergent fungal pathogen: implications for restorative planting. Tree Genet Genomes
Bond G, Fletcher W, Ferguson T (1954) The development and function of the root nodules of Alnus, Myrica and Hippophae. Plant Soil 5:309–323
Brasier C, Rose J, Gibbs J (1995) An unusual Phytophthora associated with widespread alder mortality in Britain. Plant Pathol 44:999–1007
Brown N (1997) Re-defining native woodland. Forestry 70:191–198
Claessens H (2003) The alder populations of Europe. For Comm Bull 126:5–14
Claessens H, Oosterbaan A, Savill P, Rondeux J (2010) A review of the characteristics of black alder (Alnus glutinosa (L.) Gaertn.) and their implications for silvicultural practices. Forestry 83:163–175
Corander J, Waldmann P, Sillanpää MJ (2003) Bayesian analysis of genetic differentiation between populations. Genetics 163:367–374
Cross JR (1998) An outline and map of the potential natural vegetation of Ireland. Appl Veget Sci 1:241–252
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:1–15
Eliades N-G, Eliades DG (2009) Haplotype Analysis: software for analysis of haplotype data (available from www.uni-goettingen.de/en/134935.html)
Ennos RA, Worrell R, Malcolm DC (1998) The genetic management of native species in Scotland. Forestry 71:1–23
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes - application to human mitochondrial DNA restriction data. Genetics 131:479–491
Gömöry D, Paule L (2002) Spatial and microgeographical genetic differentiation of plack alder (Alnus glutinosa Gaertn.) populations. For Ecol Managem 160:3–9
Goudet J (2001) Fstat, version 2.9.3, A program to estimate and test gene diversities and fixation indices. http://www2.unil.ch/popgen/softwares/fstat.htm
Herbert R, Samuel S, Pattison G (1999) Using Local Stock for Planting Native Trees and Shrubs. Forestry Commission Practice Note 8. Forestry Commission, Edinburgh
Latch EK, Dharmarajan D, Glaubitz JC, Rhodes OE (2006) Relative performance of Bayesian clustering software for inferring population substructure and individual assignment at low levels of population differentiation. Conserv Genet 7:295–302
Lepais O, Bacles CFE (2011) De novo discovery and multiplexed amplification of microsatellite markers for Black alder (Alnus glutinosa) and related species using SSR-enriched shotgun pyrosequencing. J Hered 102:627–631
Lepais O, Muller SD, Saad-Limam SB, Benslama M, Rhazi L, Belouahem-Abed D, Doud-Bouattour A, Gammar AM, Ghrabi-Gammar Z, Bacles CFE (2013) High genetic diversity and distinctiveness of rear-edge climate relicts maintained by ancient tetraploidization for Alnus glutinosa. PLoS ONE 8, e75029
McVean DN (1953) Alnus glutinosa (L.) Gaertn. J Ecol 41:447–466
Mejnartowicz L (2008) Genetic variation within and among naturally regenerating populations of alder (Alnus glutinosa). Acta Soc Bot Ploniae 77:105–110
Müller-Starck G, Baradat P, Bergmann F (1992) Genetic variation within European tree species. New For 6:23–47
Provan J, Powell W, Hollingsworth PM (2001) Chloroplast microsatellites: new tools for studies in plant ecology and systematic. Trends Ecol Evol 16:142–147
Rackham O (2008) Ancient woodlands: modern threats. New Phytol 180:571–586
Raymond M, Rousset F (1995) GenePop (version 1.2): population genetic software for exact tests and ecumenicism. J Hered 86:248–249
Thomas RC, Kirby KJ, Reid CM (1997) The conservation of fragmented ecosystem within a cultural landscape—the case of ancient woodland in England. Biol Conserv 82:243–252
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Acknowledgments
This study was funded by the Natural Heritage Research Partnership (NHRP) between the Northern Ireland Environment Agency (NIEA) and Quercus, Queen’s University Belfast (QUB). John Farren acted as NIEA Client Officer.
Data archiving statement
All data will be deposited in DRYAD on acceptance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. Beaulieu
This aticle is part of the Topical collection on Germplasm Diversity
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
(DOC 58 kb)
Table S2
(DOC 141 kb)
Figs. S1-S11
Bubble plots showing allele frequencies at each locus. Size of bubbles are proportional to allele frequency. (PPTX 1629 kb)
Rights and permissions
About this article
Cite this article
Beatty, G.E., Montgomery, W.I., Tosh, D.G. et al. Genetic provenance and best practice woodland management: a case study in native alder (Alnus glutinosa). Tree Genetics & Genomes 11, 92 (2015). https://doi.org/10.1007/s11295-015-0919-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-015-0919-1