Abstract
While walnuts are grown worldwide, two species are predominantly cultivated in China, Juglans regia and Juglans sigillata. J. regia is a widespread species, while J. sigillata is an indigenous species mainly distributed in southwestern China. In Tibet, unique plateau climatic conditions and relatively low-intensity agriculture aid walnuts to preserve a great variety of genetic resources. Knowing the genetic diversity and genetic structure of walnut populations in Tibet is essential when planning genetic conservation and screening for superior germplasm resources. The objectives of this study were to reveal the genetic diversity and genetic structure of walnut populations in Tibet and to infer the relationship between two species, J. regia and J. sigillata, using 12 molecular markers. The results based on screening 209 walnut trees from nine populations showed that the level of polymorphism is moderately high. However, the number of rare alleles (allele frequency <0.05) was higher than that reported in previous studies. An analysis of molecular variance revealed that significant genetic variation existed both among populations of J. regia (10.25 %, p < 0.0001) and J. sigillata (11.07 %, p < 0.0001) and between the two species (7.91 %, p < 0.0001). A Bayesian approach divided the 209 walnut samples into two clusters and presented the differentiation pattern of these two species. The assignment analysis revealed the presence of J. regia × J. sigillata hybrids among the sampled individuals. The results suggest unique germplasm preservation among the walnut populations in Tibet and that introgression between J. regia and J. sigillata may account for the convoluted boundary between the two species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Juglans includes approximately 20 species (Gleeson 1982) distributed over a wide geographical range, including Southern Europe, East Asia, and the Americas. The species within this genus display differentiation in morphology, particularly in nut characteristics (Krüssmann et al. 1985). In China, the two predominantly cultivated species are Juglans regia and Juglans sigillata. J. regia is grown locally in the Balkans, Iran, Turkey, Central Asia, the Himalayas, and China, while J. sigillata is indigenous to China, distributed mainly in southwestern China. Within the genus Juglans, J. sigillata exhibits the closest relationship with J. regia (Kuang 1997). They are readily distinguishable from each other based on leaflet number (9–11 in J. sigillata and 5–9 in J. regia) and nut morphology (J. sigillata has deep pits and seal-like depressions on the surface of the nuts, whereas J. regia has a wrinkled nut surface). There is a long-lasting disagreement whether J. sigillata and J. regia are completely different species or whether they simply represent different ecological types. Manning (1978) grouped J. regia and J. sigillata into the same species based on morphological similarities. Previous molecular marker-based results from chloroplast sequences supported a sister-group relationship between J. regia and J. sigillata (Aradhya et al. 2004). Furthermore, research based on microsatellite markers was not able to distinguish the two species (Wang et al. 2008; Gunn et al. 2010). Although most studies have considered J. sigillata and J. regia to be different ecological types of the same species, one important factor that could affect the accuracy of these results is noteworthy: the spontaneous hybridization of the two species. Crossing between J. regia × J. sigillata revealed that the two species could produce progeny (Fan et al. 2005). There is a risk that if too many potential hybrids were sampled in a study, the potential hybrids may be mistaken for one of their parents. In Tibet, J. regia is mainly distributed in the southeast along the Brahmaputra river, while J. sigillata is often distributed further south and east. The two species are sympatric only in a small region. Such distributions should reduce the possibility of inaccurate sampling and enable the detection of the extent of introgression between the two species.
Tibet comprises a large part of the Tibetan Plateau, which possesses a great range of climates and landforms. Walnut trees are mainly seed-propagated across the Tibetan Plateau due to the relatively primitive cultivation technologies available. Furthermore, for religious reasons, the cutting of walnut trees is forbidden in Tibet, especially large old trees (Duan 1984). Currently, genotypes of significant commercial importance are generally propagated vegetatively and grown in large orchards. Some important characteristics that are not readily visible may have already been lost in the process of cultivation (e.g., resistances to diseases and extremely cold weather), while local landraces grown in Tibet under low-intensity agricultural systems may preserve abundant genetic diversity. Recent studies have shown that traditionally managed populations may serve as reservoirs for genetic variation within the crop species (Miller and Schaal 2006; Jarvis et al. 2008). Therefore, evaluation of the genetic diversity and genetic structure of Tibetan walnut may prove highly important for the future conservation and breeding programs of walnut.
Several techniques to estimate the genetic diversity in walnut species have been applied, including isozymes (Fornari et al. 2001), restriction fragment length polymorphism (RFLP; Fjellstrom and Parfitt 1994), randomly amplified polymorphic DNA (RAPD; Fatahi et al. 2010; Erturk and Dalkilic 2011), inter-simple sequence repeat markers (ISSR; Christopoulos et al. 2010), amplified fragment length polymorphism (AFLP; Bayazit et al. 2007; Ma et al. 2011), and single nucleotide polymorphisms (SNPs; Ciarmiello et al. 2013). Simple sequence repeat (SSR; Pollegioni et al. 2009) markers exhibit hypervariability and codominance and are therefore highly informative. SSRs are also mostly neutral markers that are maintained even when evolutionary forces act upon in natural populations. Currently, hundreds of specific SSR primer pairs are available to Juglans, and they have been demonstrated to be useful in many kinds of genetic analyses on walnuts (Wang et al. 2008; Karimi et al. 2010; Ebrahimi et al. 2011; Pollegioni et al. 2011, 2012).
Although there may be abundant walnut genetic resource in Tibet, broad-scale research on the genetic diversity and structure of Tibet’s walnut populations at the DNA level is still lacking. The objectives of the present study were to reveal the genetic diversity and genetic structure of walnut germplasm in Tibet and to investigate whether molecular markers can distinguish the introgression of sympatric J. regia and J. sigillata in Tibet.
Materials and methods
Plant materials
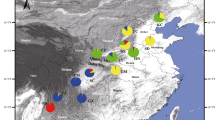
Based on knowledge obtained from field investigations by other researchers and on information supplied by local forestry agencies, five representative populations of J. regia (N = 100) and four representative populations of J. sigillata (N = 109) were sampled within the main distribution range of walnut in Tibet. The populations are named after the county, where they are located (JC—Jacha, LX—Langxian, BY—Bayi, BM—Bomi, LZ—Luozha, JL—Jilong), except for ML and DJ population (both located in Linzhi county), which are named after the village of their location (DJ—Dongjiu, ML—Milin). Five J. regia populations located along the Brahmaputra river are comparatively close, four of them located in Langxian, Linzhi, Bayi, and Bomi counties, which all belong to the Nyingchi Prefecture. JC population is located in the Shannan Prefecture. Locations of the four J. sigillata populations are scattered: JL population is located in the Jilong county of the Xigaze Region which is far away from the other populations. LZ population is located in the Luozha county of the Shannan Prefecture. DJ and BM populations are located in Linzhi and Bomi counties, respectively, which both belong to the Nyingchi Prefecture (Fig. 1, Table 1). All populations were autochthonous and their sites were unmanaged. All sampled trees were estimated to be older than 100 years based on their diameter at breast height (DBH). The individuals of J. regia and J. sigillata were distinguished based on leaflet numbers (9–11 in J. sigillata, 5–9 in J. regia) and nut morphology (J. sigillata has deep pits and seal-like depressions on the nut surface, whereas J. regia has a wrinkled nut surface) (Xi and Zhang 1996). The J. regia populations were labeled with an “r” suffix, and the J. sigillata populations with an “s” suffix.
The average distance between a pair of populations within the same species was approximately 50 km. The sympatric J. regia and J. sigillata populations were at the sample site BM, and they were labeled as BM-r and BM-s. Among the nine walnut populations, only the DJ-s population was a completely natural population, while the other populations were all landraces, which had undergone human-mediated selection and grew closely associated with human habitats (Fig. 1). Sampled trees within a population were separated by a distance of more than 50 m to avoid close relationships among them. The leaf material collected from each tree was preserved in silica gel and taken to the laboratory for storage at −80 °C until DNA extraction and molecular marker analyses. Herbarium vouchers for each walnut tree were collected and deposited in the Chinese Academy of Forestry (CAF) for future reference.
DNA extraction
Genomic DNA was isolated from leaves dried in silica gel using Foregene Plant DNA Isolation Kit according to the manufacturer’s instructions (http://www.foregene.com/).
Microsatellite amplification
DNA was amplified using 12 microsatellite primer pairs selected from a previous study on J. regia and Juglans nigra (Dangl et al. 2005; Woeste et al. 2002), namely WGA1, WGA4, WGA42, WGA70, WGA71, WGA72, WGA76, WGA89, WGA225, WGA276, WGA321, and WGA376. SSR reactions were conducted according to the protocol of Victory et al. (2006) with some modifications concerning reaction volumes and numbers of cycles. Amplification reactions were performed in a 15-μL volume containing 1× reaction buffer (TaKaRa Biotechnology, Dalian, China), 30 ng of genomic DNA, 0.2 mM of each dNTP (Promega, Beijing, China), 0.4 mM of each primer, and 0.9 unit of Taq DNA polymerase (TaKaRa). For DNA amplifications, a GeneAmp PCR System (9700; PerkinElmer, Waltham, MA) was programmed as follows: an initial 3-min incubation at 94 °C; followed by 30 cycles of 45 s at 94 °C, 30 s at the annealing temperature (Dangl et al. 2005; Woeste et al. 2002), 45 s at 72 °C; and a final incubation at 72 °C for 5 min. Amplified DNA fragments were separated using capillary electrophoresis (ABI 377 genetic analyzer). Data were collected with the associated data collection software (PE Applied Biosystems). The fragment analyses were performed with GeneScan 3.1, and data were assembled as multilocus microsatellite genotypes across the 12 loci. Microsatellite data were reformatted from an excel file to an Arlequin (.arp) file using the program CONVERT (Glaubitz 2004). In all cases, PCR reactions were performed at least twice to ensure that absence of bands was not due to a failed reaction.
Data analysis
Estimates of genetic variation (observed numbers of alleles per locus and effective numbers of alleles) as well as heterozygosity statistics (expected homozygosity and heterozygosity) and estimated null allele frequencies were calculated at each locus and over all loci using the program Popgene version 1.32 (Yen et al. 1997) and Micro-Checker (Oosterhout et al. 2004). Departures from Hardy–Weinberg expectations at each locus were tested by the likelihood ratio (G test) procedure provided by Popgene version 1.32. The unbiased estimators of Wright’s F-statistics (Weir and Cockerham 1984), within-population inbreeding coefficient (F IS) and among-population genetic differentiation coefficient (F ST), were computed for each locus across all populations. Statistical significances of F IS and F ST were tested using a nonparametric approach with 1000 permutations described in Excoffier et al. (1992). An analysis of molecular variance (AMOVA) was conducted separately for each species, within species among populations, and within populations among individuals using the program Arlequin version 3.5 (Excoffier and Lischer 2010). The Bayesian model-based clustering algorithm implemented in the program STRUCTURE (Pritchard et al. 2000) was used to determine whether individuals from the same species grouped together. This method attempts to assign individuals to several genetic groups in order to minimize within-group linkage disequilibrium and deviation from the Hardy–Weinberg equilibrium. As suggested by Falush et al. (2007), STRUCTURE analysis was performed using the admixture model on the whole dataset with no previous population information and the correlated allele frequencies between populations option. In this study, the range of possible number of clusters (K) tested ranged from 1 to 12. Based on the initial results, a series of six independent runs were performed for each K value with a burn-in period of 10,000 steps followed by 105 MCMC replicates. Furthermore, the ad hoc statistic ΔK defined by Evanno et al. (2005) was used to detect the most likely number of populations. The ΔK-statistics is based on the second-order rate of change of L (K) (the posterior probability of the data among given K) between successive K values over six replicates. As demonstrated by Evanno et al. (2005), it is possible to identify the number of clusters corresponding to the uppermost hierarchical level of genetic partitioning between populations. Therefore, the groups inferred by the first STRUCTURE analysis were subsequently processed separately in order to identify possible substructure. Any individual with ≤70 % probability of assignment to a cluster was considered as a possible hybrid. A neighbor-joining analysis of relationships based on Nei’s genetic distances among all populations of J. regia and J. sigillata was performed. An unweighted pair-group method with arithmetic mean (UPGMA) tree was constructed based on Nei’s (1972) genetic distances. Bootstrap support for this tree was determined by resampling loci 1000 times using the POP TREE2 software (Takezaki et al. 2010) to visualize genetic relationships. The number of migrants between species per generation, (1/F ST − 1)/4 was estimated using the program GenAlEx software 6.3 (Peakall and Smouse 2006). Genetic differentiation between population pairs was measured by Wright’s F ST coefficient (Weir and Cockerham 1984) using the Arlequin software. F ST was used for this study because it does not assume a stepwise mutation model and appeared to be a more sensitive measure of intra-specific variation compared to R ST (Balloux and Lugon-Moulin 2002). A nonparametric pairwise correlation test between the matrices of F ST/(1 − F ST) and the natural logarithm of geographic distances was applied to test for isolation by distance (Rousset 1997). The p value for the Z score of the Mantel association parameter was inferred using 1000 permutations. These calculations were performed using the GenAlEx software 6.3.
Results
Microsatellite polymorphism
The microsatellite variation pattern of the Tibetan Juglans populations is summarized in Tables 2 and 3 and Table S1. Highly significant (p < 0.01) departures from Hardy–Weinberg expectations (deficiencies or excesses) across J. regia populations were found for WGA72, WGA76, WAG89, and WGA4, while across J. sigillata populations, deviations were found for WGA01, WGA72, WAG76, WGA89, WGA4, WGA376, WGA321, and WGA225 (G test) (Table 2). The within-population inbreeding coefficient (F IS) was significantly negative for WGA42, WGA70, WGA321, and WGA276 and significantly positive for WGA1, WGA76, WGA376, and WGA225 in J. regia, but significantly positive for all loci in J. sigillata. The F IS value being significantly greater than zero indicated heterozygote deficiency. The null allele frequency (F nul) estimated for J. regia ranged from 0.0035 for WGA70 to 0.1589 for WGA76, while in J. sigillata, the value ranged from 0.0186 for WGA71 to 0.2477 for WGA376 (Table 2). All 12 microsatellite loci were polymorphic. In all, 107 alleles were detected in the 100 J. regia trees and 145 alleles were detected in the 109 J. sigillata trees. The number of alleles per locus ranged from 7 at locus WGA70, WGA71, and WGA225 to 22 at locus WGA276, with an average of 13.5. The genetic diversity greatly varied across the SSR loci. The average observed (H o) and expected (H e) heterozygosities were 0.512 (SD = 0.249) and 0.586 (SD = 0.243) in J. regia and 0.556 (SD = 0.164) and 0.706 (SD = 0.164) in J. sigillata, respectively (Table 3).
Genetic variation and genetic structure within and among Tibetan walnut populations
The results indicated that all J. sigillata populations possess higher genetic diversities than do J. regia populations. Among J. sigillata populations, JL-s had the highest estimates of genetic variation (H e = 0.693, Nei = 0.678) and LZ-s had the lowest estimates (H e = 0.593, Nei = 0.583). Among J. regia populations, BM-r had the highest diversity (H e = 0.589, Nei = 0.572) and JC-r had the lowest estimates (H e = 0.478, Nei = 0.463) (Table 3). The overall F IS (inbreeding level within population) varied from 0.123 (BM-r) to −0.047 (ML-r) in J. regia and from 0.247 (JL-s) to −0.069 (LZ-s) in J. sigillata. F IS values were large and significantly positive in BM-r, DJ-s, JL-s, and BM-s, indicating heterozygote deficiencies in these populations (Table 3). AMOVA showed that the percentage of variation between the two species was 7.91 % (p < 0.0001) (Table 4), which was much higher than that found in previous research [0.07 % in Gunn et al. (2010)]. Our result indicated that the difference between the two species was notable, which meant that it was necessary to analyze the genetic structures of J. regia and J. sigillata separately. Among population, proportion of variance was significant for both species based on AMOVA (J. regia 10.25 %, p < 0.0001; J. sigillata 11.07 %, p < 0.0001). These data suggested that a higher amount of genetic variance was distributed among populations for both J. regia and J. sigillata compared to previous research on Juglans (Victory et al. 2006; Wang et al. 2008; Gunn et al. 2010) (Table 5).
Cluster analysis of J. regia and J. sigillata populations
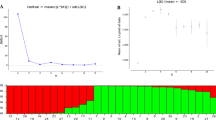
The genetic structures of nine Tibetan walnut populations were evaluated by Bayesian cluster analysis using STRUCTURE (Pritchard et al. 2000). Loci WGA-1, WGA-76, WGA-376, and WGA-225 showed significant positive F IS values in both species and frequencies of null alleles were not negligible (Table 2). Therefore, we removed those loci from the STRUCTURE analysis to avoid the negative influence of them on the level of clustering. When using the admixture and correlated frequency model, the log-likelihood value (L[K]) determined as a function of K (number of clusters) averaged over six replicates increased almost linearly from K = 1 (Ln P[D] = −5536.8) up to K = 2 (Ln P[D] = −4630.4) and increased slowly from K = 3 to K = 10 (Ln P[D] = −3978.5) (Fig. 2a). The approach of Evanno et al. (2005) strongly supported K = 2 as the most likely number of clusters, because the highest second order of change of the log-likelihood of the data (ΔK), as a function of K, was detected at K = 2 (Fig. 2b). The estimated overall number of hybrids was 15.8 % (33/209) (Fig. 3). A neighbor-joining tree revealed that the J. regia and J. sigillata populations clustered essentially according to their presumed species, except for the BM-s population, which showed a closer relationship with J. regia populations (Fig. 4).
Inference of K, the most probable number of clusters, using STRUCTURE software, based on microsatellite analysis of 209 total walnut samples. a Log-likelihood value of data L (K) as a function of K averaged over 10 replicates. b Second order of change of the log-likelihood of the data (ΔK) as a function of K, calculated over 10 replicates
Population structure inference for 209 Tibetan walnut samples by Bayesian assignment using STRUCTURE software (K = 2 clusters). Each individual is represented by a vertical line and populations are separated by a vertical black line. Different colors in the same line indicate the individual’s estimated membership percentage in K clusters (admixture proportion or Q value): red = cluster 1, green = cluster 2. For details of locations abbreviations and locations, see Table 1 and Fig. 1
Genetic variation and geography
Mantel correlations between the pairwise linearized genetic differentiation values F ST/(1 − F ST) and the natural logarithm of geographic distances between the sampling sites were significant (r = 0.5243, p = 0.0321). This indicates isolation by distance among the sampled walnut populations.
Discussion
Detection of genetic diversity and variation in geographic patterns among walnut populations in Tibet
This study was the first large-scale analysis of the Tibetan walnut germplasm, which has survived under relatively unusual ecological conditions and primitive agricultural management for generations. This means that the populations have been subjected to a unique evolutionary process. Artificial selection for superior characteristics and exchange of germplasm through human activity affected the genetic diversity and structure of the Tibetan walnut. Our study was based on 12 nuclear microsatellite markers developed originally for J. nigra and J. regia (Woeste et al. 2002; Dangl et al. 2005). In this study, the mean number of alleles per locus (12.08 in J. sigillata and 8.92 in J. regia) and the effective number of alleles (4.43 in J. sigillata and 3.43 in J. regia) were much higher than the levels of variability previously detected in 48 J. regia cultivars (Dangl et al. 2005), 9 walnut populations from central and southwestern China (Wang et al. 2008), 6 village landrace populations from the Yunnan Province of China (Gunn et al. 2010), and 29 populations from Italy (Pollegioni et al. 2011). However, the genetic diversity parameters (H e, and Nei’s H e) detected in this study were comparable to those reported in these previous studies. The diversity results were also similar to that found for other domesticated tree species, including Cupressus sempervirens (Bagnoli et al. 2009) and Castanea sativa (Martin et al. 2010). The main reason for this result is that many alleles in this study were rare alleles. Among the nine populations, 29.01 % (47/162) of the allele frequencies were between 0.05 and 0.01 and 33.95 % (55/162) of the alleles had a frequency lower than 0.01. Thus, the Tibetan walnut germplasm shows the preservation of unique genotypes. Microsatellite markers mostly reveal neutral variation that is subjected to evolutionary processes, such as drift and variation in population size, but which have little association with adaptive variation (Mitchell-Olds et al. 2007). In Tibet, many old variants of walnut germplasm have been largely preserved as a result of primitive cultivation techniques and some ethnic customs (Duan 1984). Thus, Tibet could be considered as a hot spot for the preservation of genetic variation in walnut.
All J. sigillata populations showed greater genetic diversity than did J. regia populations. BM-r, which possessed the greatest genetic diversity, was sympatric with a J. sigillata population. This may be a reflection of the different origins of the two taxa in Tibet. The reported natural range for J. regia includes Central Asia, the Xinjiang Province (western China), Kazakhstan, Uzbekistan, Kirghizia, the Himalayas, Iran, Azerbaijan, Armenia, Georgia, and eastern Turkey, whereas J. sigillata is native to Yunnan, Sichuan, southeastern Tibet, Guizhou, Sikkim, and Bhutan. It appears that J. regia may have immigrated to Tibet from other places. If so, in Tibet, the genetic basis of J. regia would be narrower than that of J. sigillata populations because of the founder effect (Ladizinsky 1985). As shown in Table 2, many loci exhibited significant deviations from Hardy–Weinberg expectations and high F IS values in both species. These factors indicated that inbreeding was occurring in Tibetan walnut population. As explained by Cornuet and Luikart (1996), a population that has subsequently expanded for several generations without immigration is characterized by a heterozygote deficiency across all loci. The walnut populations in Tibet are isolated by geographic barriers, and trees with a superior yield and nut quality have probably been favored and selected by the Tibetans. Such phenomena could lead to genetic drift, which can cause allelic losses and other alleles to become fixed, with following effects on the null allele frequency estimates (Garza and Williamson 2001). Similar results were found in walnut populations in southwestern China and Italy (Wang et al. 2008; Pollegioni et al. 2011). In this study, a significant heterozygote deficiency was detected in all J. sigallata populations, but only in BM-r among J. regia populations, which indicated that isolation might have had a major impact on the number of heterozygotes.
An analysis of molecular variance (AMOVA) of the walnut populations in Tibet showed that the proportion of total genetic diversity was higher within populations (89.7 % in J. regia and 88.9 % in J. sigillata) than among populations for both species (Table 5). Domesticated plant species may show low levels of molecular differentiation due to human selection and human-mediated dispersal of selected genotypes. In Tibet, walnuts are prized for their high-quality nuts, medicinal properties, and ritual uses. They are also a common commodity and have long been exchanged in markets. In addition, local people often bring walnuts as a gift when they visit their relatives and friends. Therefore, human-mediated genotypic dispersal is likely primarily responsible for the present genetic structure of walnut populations in Tibet, rather than long-distance zygotic or gametic gene dispersal. However, it should be noted that the “low differentiation” detected herein might also be the result of comparing a domesticated species with a wild one. Indeed, due to the isolation by distance, the detected differentiation between Tibetan walnut populations is higher than that between other domesticated Juglans populations (Gunn et al. 2010; Pollegioni et al. 2011).
To the best of our knowledge, most of the populations sampled in this study were landraces, all of which have experienced some degree of human-mediated selection. Although wild populations of J. sigillata have been reported in Tibet, they were not sampled in this study, with the exception of the DJ-s population. The wild populations are usually located in primary forests, which are far away from human habitats and very difficult to reach. Our results suggested that the DJ-s population possesses a moderate level of genetic diversity compared to the others. This is in sharp contrast to the wild populations of J. regia (in Xijing Province), in which the genetic diversity was much lower than in J. regia landrace populations (Wang et al. 2008). This situation indicated that a relatively strong gene exchange might have occurred between wild J. sigillata populations and other landrace populations in Tibet, and there was a good chance for wild populations to preserve unique genotypes.
Introgression and differentiation between sympatric J. regia and J. sigillata in Tibet
Our results indicated that accurate discrimination between J. regia and J. sigillata could be revealed using 12 microsatellite loci. Previous studies based on five chloroplast intergenic spacers demonstrated only marginal differences between J. regia and J. sigillata (Aradhya et al. 2004). Several recently published studies have indicated that J. regia and J. sigillata from central and southwestern China (Wang et al. 2008) and from six villages located in the foothills of Khawa Karpo, in the northwest of Yunnan Province, China (Gunn et al. 2010), could not be distinguished using SSR markers. A possible reason to explain the difference in results was that the previous research had not considered the possibility of introgression between the two species, and they all sampled sympatric J. regia and J. sigillata populations for their research. Therefore, some hybrids may be mistaken for one of the species, and the results could be biased.
The results of the structure analysis indicated that only a few individuals (7/100) in J. regia populations had a strong possibility of a complex genetic background, while quite many individuals (26/109) in J. sigillata populations did so (especially in the BM-s population that is sympatric with J. regia). The data presented here demonstrated that introgression has occurred between the two species, and the direction of gene flow has been mainly from J. regia to J. sigillata. The natural outcrossing mechanisms of the two species may determine the direction of gene flow. Although the filial generation of J. regia × J. sigillata is progenitive, the success ratio of backcrossing with the parents is still unknown. The two species might possess a different success ratio of backcrossing due to phenological and genetic incompatibilities. Based on our results, it may be easier for the filial generation to backcross with J. sigillata. A similar situation has also been reported in other species of Juglans (Pollegioni et al. 2009).
Introgression could have a two-sided effect on the genetic diversity of populations. Its effect is influenced by many factors, including the direction of introgression and the effective population size. At present, it appears that introgression might have had some positive effects on the genetic diversity of J. sigillata populations, although this view was based on results from only one sympatric location (BM), and genetic analyses on additional sympatric locations are needed to further elucidate these issues. Yet, it is notable that some rare genetic traits present in J. sigillata populations might become lost in the process of introgression.
Conclusions
The Tibetan Plateau is a hot spot for germplasm preservation. Detailed knowledge regarding the diversity and structure of existing genetic resources of walnut in Tibet may provide important information for the conservation of genetic resources and for the screening of superior germplasm. Data revealed by SSR markers indicated that Tibetan walnut populations possess average levels of genetic diversity and numerous rare alleles. These results demonstrated that the walnut populations in Tibet have not experienced a recent sharp population decline and probably contain some unique germplasm. Geographical isolation is the main factor that has shaped the genetic structure of walnut populations in Tibet, while the human-mediated transfer of walnut genotypes has reduced this isolation to some extent. Introgression between the two species was detected, although additional sympatric populations should be investigated to confirm this result and elucidate the mechanism of introgression between the two species.
References
Aradhya M, Potter D, Simon C (2004) Origin, evolution, and biogeography of Juglans: a phylogenetic perspective. V Int Walnut Symp 705:85–94
Bagnoli F, Vendramin G, Buonamici A, Doulis A, Gonzalez-Martinez SC, La Porta N, Magri D, Raddi P, Sebastiani F, Fineschi S (2009) Is Cupressus sempervirens native in Italy? An answer from genetic and palaeobotanical data. Mol Ecol 18:2276–2286
Balloux F, Lugon-Moulin N (2002) The estimation of population differentiation with microsatellite markers. Mol Ecol 11:155–165
Bayazit S, Kazan K, Gulbitti S, Cevik V, Ayanoglu H, Ergul A (2007) AFLP analysis of genetic diversity in low chill requiring walnut (Juglans regia L.) genotypes from Hatay, Turkey. Sci Hortic 111:394–398
Christopoulos MV, Rouskas D, Tsantili E, Bebeli PJ (2010) Germplasm diversity and genetic relationships among walnut (Juglans regia L.) cultivars and Greek local selections revealed by Inter-Simple Sequence Repeat (ISSR) markers. Sci Hortic 125:584–592. doi:10.1016/j.scienta.2010.05.006
Ciarmiello LF, Pontecorvo G, Piccirillo P, Luca A, Carillo P, Kafantaris I, Woodrow P (2013) Use of nuclear and mitochondrial single nucleotide polymorphisms to characterize english walnut (Juglans regia L.) genotypes. Plant Mol Biol Rep 31:1116–1130. doi:10.1007/s11105-013 -0575-2
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Dangl GS, Woeste K, Aradhya MK, Koehmstedt A, Simon C, Potter D, Leslie CA, McGranahan G (2005) Characterization of 14 microsatellite markers for genetic analysis and cultivar identification of walnut. J Am Soc Hortic Scie 130:348–354
Duan S (1984) The investigation of walnut originating in Tibet. Acta Horticult Sinica 11:231–234 (in Chinese)
Ebrahimi A, Fatahi R, Zamani Z (2011) Analysis of genetic diversity among some Persian walnut genotypes (Juglans regia L.) using morphological traits and SSRs markers. Sci Hortic 130:146–151. doi:10.1016/j.scienta.2011.06.028
Erturk U, Dalkilic Z (2011) Determination of genetic relationship among some walnut (Juglans regia L.) genotypes and their early-bearing progenies using RAPD markers. Rom Biotechnol Lett 16:5944–5952
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. doi:10.1111/j.1365-294
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:574–578
Fan ZY, Fang WL, Dong RQ (2005) Comparison and selection of pollinated Biyangpao walnut varieties. South China Fruits 34:58–59 (in Chinese)
Fatahi R, Ebrahimi A, Zamani Z (2010) Characterization of some Iranians and foreign walnut genotypes using morphological traits and RAPD markers. Hortic Environ Biotechnol 51:51–60
Fjellstrom RG, Parfitt DE (1994) Walnut (Juglans spp.) genetic diversity determined by restriction fragment length polymorphisms. Genome 37:690–700
Fornari B, Malvolti ME, Taurchini D (2001) Isozyme and organellar DNA analysis of genetic diversity in natural/naturalised European and Asiatic walnut (Juglans regia L.) populations. Acta Horticult Sci 544:167–178
Garza J, Williamson E (2001) Detection of reduction in population size using data from microsatellite loci. Mol Ecol 10:305–318
Glaubitz JC (2004) CONVERT: a user-friendly program to reformat diploid genotypic data for commonly used population genetic software packages. Mol Ecol Notes 4:309–310
Gleeson SK (1982) Heterodichogamy in walnuts: inheritance and stable ratios. Evolution 36:892–902
Gunn BF, Aradhya M, Salick JM, Miller AJ, Yongping Y, Lin L, Xian H (2010) Genetic variation in walnuts (Juglans regia and J. sigillata; Juglandaceae): species distinctions, human impacts, and the conservation of agrobiodiversity in Yunnan, China. Am J Bot 97:660–671. doi:10.3732/ajb.0900114
Jarvis DI, Brown AHD, Cuong PH, Collado-Panduro L, Latournerie-Moreno L, Gyawali S, Tanto T, Sawadogo M, Mar I, Sadiki M, Hue NTN, Arias-Reyes L, Balma D, Bajracharya J, Castillo F, Rijal D, Belqadi L, Rana R, Saidi S, Ouedraogo J, Zangre R, Rhrib K, Chavez JL, Schoen D, Sthapit B, De Santis P, Fadda C, Hodgkin T (2008) A global perspective of the richness and evenness of traditional crop-variety diversity maintained by farming communities. Proc Natl Acad Sci U S A 105:5326–5331. doi:10.1073/pnas.0803431105
Karimi R, Ershadi A, Vahdati K, Woeste K (2010) Molecular characterization of Persian walnut populations in Iran with microsatellite markers. Hortscience 45:1403–1406
Kimura M, Crow JF (1964) The number of alleles that can be maintained in a finite population. Genetics 49:725–738
Krüssmann G, Epp ME, Daniels GS (1985) Manual of cultivated broad-leaved trees & shrubs, Volume II, E-PRO. Timber Press/American Horticultural Society
Kuang KZ, Lu AM (1997) Flora of China, vol 21. Science Press, Beijing, China:33–36 (in Chinese)
Ladizinsky G (1985) Founder effect in crop-plant evolution. Econ Bot 39:191–199
Levene H (1949) On a matching problem arising in genetics. Ann Math Stat 20:91–94
Ma QG, Zhang JP, Pei D (2011) Genetic analysis of walnut cultivars in China using fluorescent amplified fragment length polymorphism. J Am Soc Hortic Sci 136:422–428
Manning WE (1978) The classification within the Juglandaceae. Ann Mo Bot Gard 65:1058–1087
Martin MA, Mattioni C, Cherubini M, Taurchini D, Villani F (2010) Genetic diversity in European chestnut populations by means of genomic and genic microsatellite markers. Tree Genetics Genomes 6:735–744
Miller AJ, Schaal BA (2006) Domestication and the distribution of genetic variation in wild and cultivated populations of the Mesoamerican fruit tree Spondias purpurea L. (Anacardiaceae). Mol Ecol 15:1467–1480. doi:10.1111/j.1365-294×.2006.02834.×
Mitchell-Olds T, Willis JH, Goldstein DB (2007) Which evolutionary processes influence natural genetic variation for phenotypic traits? Nat Rev Genet 8:845–856
Nei M (1972) Genetic distance between populations. Am Nat 106:283–292
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci U S A 70:3321–3323
Oosterhout CV, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Pollegioni P, Woeste K, Mugnozza GS, Malvolti ME (2009) Retrospective identification of hybridogenic walnut plants by SSR fingerprinting and parentage analysis. Mol Breed 24:321–335. doi:10.1007/s11032-009-9294-7
Pollegioni P, Woeste K, Olimpieri I, Marandola D, Cannata F, Malvolti ME (2011) Long-term human impacts on genetic structure of Italian walnut inferred by SSR markers. Tree Genetics Genomes 7:707–723. doi:10.1007/s11295-011-0368-4
Pollegioni P, Olimpieri I, Woeste KE, Simoni G, Gras M, Malvolti ME (2012) Barriers to interspecific hybridization between Juglans nigra L. and J. regia L species. Tree Genetics Genomes 9:291–305. doi:10.1007/s11295-012-0555-y
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145:1219–1228
Takezaki N, Nei M, Tamura K (2010) POPTREE2: software for constructing population trees from allele frequency data and computing other population statistics with Windows interface. Mol Biol Evol 27:747–752
Victory ER, Glaubitz JC, Rhodes OE, Woeste KE (2006) Genetic homogeneity in Juglans nigra (Juglandaceae) at nuclear microsatellites. Am J Bot 93:118–126
Wang H, Pei D, Gu RS, Wang BQ (2008) Genetic diversity and structure of walnut populations in central and southwestern China revealed by microsatellite markers. J Am Soc Hortic Sci 133:197–203
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Woeste K, Burns R, Rhodes O, Michler C (2002) Thirty polymorphic nuclear microsatellite loci from black walnut. J Hered 93:58–60
Xi RT, Zhang YP (1996) Fruit trees of China. Walnut Chinese Forestry Press, Beijing (in Chinese)
Yen F, Yang R, Mao J, Ye Z, Boyle T (1997) POPGENE, the Microsoft Windows-based user-friendly software for population genetic analysis of co-dominant and dominant markers and quantitative traits. Dept Renewable Resources, University of Alberta, Edmonton
Acknowledgments
The authors thank Zheng Weilie (Tibet Institute of Agriculture and Animal Husbandry, Linzhi) for logistical help and Dr. Ma Heping and Bianbaduoji (Tibet Institute of Agriculture and Animal Husbandry, Linzhi) for assistance with sampling. We warmly thank the crew from the local District Forestry Bureau for providing information of local walnut resources and assisting with sampling, and the local Tibetan people for their hospitality and help in sample collecting. We also thank Zeng Yanfei (the Chinese Academy of Forestry, Beijing) for her support in data analysis. Financial support was received from the National Science & Technology Pillar Program (2013BAD14B01) and from the National Natural Science Foundation of China (31400558).
Data Archiving Statement
The raw data of this research was submitted to the TreeGenes Database. The accession numbers of genotypes and environmental descriptors of Juglans sigillata and J. regia are under TGDR029 and TGDR030, respectively.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Kremer
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 18 kb)
Rights and permissions
About this article
Cite this article
Wang, H., Pan, G., Ma, Q. et al. The genetic diversity and introgression of Juglans regia and Juglans sigillata in Tibet as revealed by SSR markers. Tree Genetics & Genomes 11, 1 (2015). https://doi.org/10.1007/s11295-014-0804-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-014-0804-3