Abstract
The wall-associated kinases (WAKs) are a family of transmembrane proteins involved in pathogen responses and cell elongation in Arabidopsis. They belong to the major receptor-like kinase (RLK) family in plants. Given their architecture and connection to the cell wall, WAKs are thought to perceive and propagate extracellular signals. This study reports the characterization of the WAK family in a woody species based on the v3.0 genome assembly of Populus trichocarpa. In silico analysis revealed a total of 175 PtWAK sequences classified into four groups based on protein domains. Of the PtWAKs, 91.5 % were found in tandem-duplicated clusters contributing to the expansion of the family in poplar. Microarray and EST expression data mining revealed contrasting temporal and spatial expression patterns in stress treatments for several WAK members. The WAKs in poplar form the largest WAK family encountered to date in plants. The combination of phylogenetic and transcriptional data showed that members in nonexpanded clusters were mainly expressed in developmental processes, whereas PtWAKs that had evolved independently in a species-specific way were structured in clusters and were involved in resistance responses. This paper offers an overview of WAK family structure in P. trichocarpa, which will be useful for further functional analysis of the PtWAK family.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The plant cell wall is an important dynamic structure. It mechanically connects cells in tissues and controls the shape of plant cells (Wolf et al. 2012). There is increasing new evidence that this remarkable structure is also critical for sensing external stimuli and transmitting information into the intracellular space. For instance, transmembrane proteins connecting the apoplasm and cytoplasm compartments may well be able to perceive wall status and variation in cell volume (Humphrey et al. 2007). The wall-associated kinases (WAKs) are a set of receptor-like kinases (RLKs; Shiu and Bleecker 2001) involved in signaling pathways (Kohorn et al. 2009). For example, AtWAK1 is able to interact with oligogalacturonic acids playing a key functional role in responses to pathogens (Brutus et al. 2010). Various studies have suggested that WAKs are able to bind pectins through the presence of specific protein domains located in their extracellular region (He et al. 1998; Wagner and Kohorn 2001; Decreux and Messiaen 2005). This extracellular region contains one or two epidermal growth factor (EGF)-like domains (He et al. 1999; Verica and He 2002; Zhang et al. 2005) that could be involved in protein–protein interactions (Rao et al. 1995; Stenflo et al. 2000). The intracellular region of WAKs displays a Ser/Thr kinase domain that might be involved in signal transduction. It has been shown that when oligogalacturonic acids are produced by wall disruption, AtWAK2 signaling by kinase activity modulates a MAP kinase activity (AtMAPK6; Kohorn et al. 2009; Kohorn et al. 2012).
Until now, WAKs have been described exhaustively in two herbaceous model species: Arabidopsis thaliana (He et al. 1999; Verica and He 2002) and Oryza sativa (Zhang et al. 2005). The Arabidopsis genome contains 27 WAKs: five are termed AtWAKs (He et al. 1999) and 22 AtWAK like (AtWAKLs; Verica and He 2002). This gene family is larger in the rice genome, with 125 OsWAKs classified into 67 RLKs, 28 receptor-like cytoplasmic kinases (RLCKs), 13 receptor-like proteins (RLPs), and 12 predicted short proteins and five pseudogenes (Zhang et al. 2005).
In terms of genome location, the five AtWAKs are distributed along a 30-kb cluster on chromosome I (He et al. 1999); 14 AtWAKLs are also found on chromosome I and eight are located on the other four chromosomes (Verica and He 2002). This clustering was also observed in the organization of rice WAKs.
In woody species, WAKs, as receptors, may play a decisive part in development and in response to environmental constraints. The main characteristic of trees is wood production through secondary growth and perennialism, which implies short-term and long-lasting responses in continually changing environmental conditions. In addition, environmental stresses can induce specific modifications in secondary cell wall characteristics (Jourez et al. 2001), notably G layer deposition in the tension wood, which requires cellular communications between the apoplasm and the cytoplasm. The involvement of WAKs as potential mediators of the reaction wood formation in response to gravitropic stimulus has been proposed (Telewski 2006; Tocquard et al. 2014).
The evolutionary history of plant RLK genes, including Populus genes, was presented in a previous study (Lehti-Shiu et al. 2009). However, particular emphasis was not given to the WAK gene family. In the present study, we carried out a genome-wide identification of WAK family members from Populus trichocarpa (PtWAKs), which is considered to be an important model for basic and applied research in forest biology (Yang et al. 2009). Extensive database searches and manual reannotation were made. A phylogenetic tree was constructed to evaluate the evolutionary relationships among WAK amino acid sequences between Arabidopsis, rice, and poplar and to gain insights into potential mechanisms of WAK gene family expansion in poplar. Additionally, the public microarray data and ESTs were used to monitor the expression of PtWAKs in different organs during various stages of development and under biotic and abiotic conditions, in a panel of Populus clones. Finally, we carried out a combined reading between the patterns of expansion and gene expression related to different physiological responses, in order to uncover physiological functions of chosen WAK members. This work provides a basis for a better understanding of the genomic organization and probable roles of these genes in plant growth/development and disease resistance in poplar.
Methods
WAK sequence retrieval from public databases
The DNA and protein-predicted sequences of WAKs were retrieved from poplar v3.0 genome assembly (P. trichocarpa, Torr. & Gray, Tuskan et al. 2006) in v9.0 gene annotation on the public database Phytozome (P. trichocarpa v3.0, DOE-JGI, http://www.phytozome.net/). Protein sequences AtWAK1-5 of A. thaliana were used as queries for a tBLASTx on Phytozome using default parameters. Retrieved DNA sequences of PtWAKs were then used as queries for a BLASTn on Phytozome. In order to correct predicted sequences, an EST search was conducted on the NCBI resource database (http://www.ncbi.nlm.nih.gov/) using BLASTn on both genomic and transcript PtWAK sequences. All sequences were then manually expertized to fit Phytozome-predicted open reading frames. When two predicted genes were separated by less than 5 kb, the first one coding an extracellular part of a WAK and the other a kinase part, a single sequence was created by merging the two parts and removing one stop codon. All changes are described in Online resource 1. As there was no standard WAK annotation assigned to these newly identified genes in poplar, we gave each of them an identity based on the order of their location on each poplar linkage group (LG) listed in increasing order.
Protein domains were predicted with Conserved Domain Database (CDD; Marchler-Bauer et al. 2011), a resource from the NCBI. The corresponding regions were extracted from the alignment performed with MUSCLE (Edgar 2004) in order to identify conserved amino acids between WAKs within each predicted domain. These sequences were then analyzed using the Multiple Expectation Maximization for Motif Elicitation (MEME) program (Bailey et al. 2009) with default parameters.
Alignment and phylogenetic analysis
The kinase domains of WAK protein sequences were aligned using the MUSCLE interface in Geneious Pro v5.3.6 (Biomatters, Auckland, New Zealand). The phylogenetic analysis was performed on the kinase region of the alignment after removing incomplete sequences. The phylogenetic tree was generated using the neighbor-joining (NJ), maximum parsimony (MP), and Bayesian inference (BI) methods with 1,000 bootstrap replicates using the JTT + G + I model determined with MEGA v5.1 (Tamura et al. 2011).
The congruence between the NJ, MP, and BI topologies was tested using the Kishino–Hasegawa test (Kishino and Hasegawa 1989) implemented in the PAUP*4.0b10 program (Swofford 2000), using the PaupUp graphical interface (Calendini and Martin 2005). The tree representation was constructed on the interactive Tree Of Life Web service (iTOL; Letunic and Bork 2011).
Transcriptomics data analyses
PtWAK expression patterns among tissues/conditions/genotypes
Most of the data used for transcriptomics data analysis in this study were robust multi-array average (RMA) normalized transcriptomics data obtained from Affymetrix Poplar Genome Array (Platform accession GPL4359). For the drought experiment in the apex only, data were obtained from the Agilent array with custom probe design based on v2.0 poplar gene models (Lafon-Placette et al., unpublished data). Values of gene expression were downloaded from the v2.2 PopGenIE Website (http://popgenie.org/, Sjödin et al. 2009). Analyses of WAK expression were conducted on different datasets: (i) tissue profiling (GSE13990, Wilkins et al. 2009a), (ii) light treatments (GSE13990, Wilkins et al. 2009a), (iii) pathogen infection (Melampsora medusae) (GSM412653, Levée et al. 2009; GSE9673 Azaiez et al. 2009), (iv) drought stress on roots (GSE17223, Cohen et al. 2010), (v) drought stress on leaves (GSE15242, Wilkins et al. 2009b; GSE17226, Balzergue et al., unpublished data; GSE17230, Cohen et al. 2010; and GSE21171, Hamanishi et al. 2010), (vi) drought/rewatering cycle on apex (Lafon-Placette et al., unpublished data), (vii) genotypes (GSE15242, Wilkins et al. 2009b; GSE17226, Balzergue et al., unpublished data; GSE17230, Cohen et al. 2010; and GSE21171, Hamanishi et al. 2010), (viii) aluminium stress (GSE19297, Grisel et al. 2010), and (ix) hypoxia stress by flooding (GSE13109, Hauberg et al., unpublished data). To avoid experimental bias when combining several studies (batch effect), data were normalized with the ComBat method (Johnson et al. 2007) in R v2.15.2 (R Development Core Team 2008) using “control” and “pathogen”/“drought” as covariates for the pathogen/drought effect. No covariate was defined for genotype effect, thus assuming no genotype effect to normalize data (only batch effect). Significant tissue/condition/genotype effects were tested using Welch’s t test or one-way ANOVA for two or more samples, respectively. Adjusted Bonferroni correction was used to decrease false discovery rate. Statistical tests were considered significant at *p < 0.05. Average linkage hierarchical clustering analysis was applied using Euclidean distance. These statistical analyses were performed with MeV software, v4.8.1(Saeed et al. 2003).
Entropy and random comparison
To evaluate the PtWAK global expression, 72 genes were randomly selected using the “Random List” tool from the PopGenIE Website (Sjödin et al. 2009). To define tissue/condition/genotype specificity of PtWAK expression, the Shannon entropy value was calculated for each gene (Schug et al. 2005) using expression levels from the experiments described above instead of EST counts. Shannon entropy value ranks a gene according to the degree to which its expression pattern is similar to ubiquitous uniform expression among several samples. Thus, the more similar to ubiquity (i.e., the less specific) is the expression pattern, the higher is the Shannon entropy value. To evaluate PtWAK effect on entropy distribution, a χ 2 homogeneity test was performed at *p < 0.05, **p < 0.01, or ***p < 0.001 with R v2.15.2 (R Development Core Team 2008).
Results and discussion
A genome-wide analysis of the WAK multigenic family in P. trichocarpa was initiated. The monitoring of the dynamics of this family and of the transcriptional expression patterns in different tissues and stress conditions brings some insight into the structural and functional diversities of this family.
Features of WAK gene family in Populus
Exploration on the Phytozome portal (Goodstein et al. 2012) and NCBI databases enabled us to identify 175 putative PtWAKs. This represents 85 % more than A. thaliana and 28.5 % more than O. sativa. Marked expansions of multigenic families have already been described in poplar (Lan et al. 2009; Petre et al. 2011). This is especially observed for RLKs, the largest gene family in most plant species (Kohler et al. 2008), which comprises the WAK subfamily. According to Lehti-Shiu et al. (2009), the RLK family expanded at a significantly higher rate than other kinases.
The fact that the poplar WAK family had more members than rice tempers the hypothesis of Zhang et al. (2005) whereby the expansion of WAKs arises specifically in monocot species. Although we cannot rule out the hypothesis of WAK gene loss events in Arabidopsis, the high number of WAKs in poplar and rice compared with the number of WAKs in Arabidopsis suggests, alternatively, that both poplar and rice genomes present a lineage-specific expansion of this WAK family.
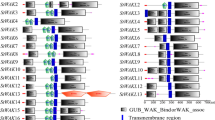
In poplar, from the 175 WAK members identified, 141 PtWAK genes were distributed unevenly among 15 of the 19 LGs corresponding to the 19 poplar chromosomes. The remaining 34 PtWAK genes were localized on eight independent scaffolds not yet grouped to the chromosomes. Among the 141 PtWAKs, 12 were isolated genes and 129 (91.5 %) were grouped into 20 clusters containing 2 to 16 genes separated from 1 to 13 kb (Fig. 1). Such a high proportion of clustered genes was unexpected because in Arabidopsis, only 60 % of members were found in clusters (Shiu and Bleecker 2003). It is widely accepted that the tandem duplications drive most of the specific lineage expansion of various gene families involved in tolerance to any environmental deviances, and represent a noteworthy way to generate possible functional diversity of paralogs (Paterson et al. 2010; Lopez et al. 2012). The expansion of the PtWAK family, as hypothesized for RLKs (Hanada et al. 2008), could be linked to the evolution of poplar to adapt to fluctuating environmental stimuli.
Representation of PtWAK types on 15 of the 19 poplar linkage groups (LGs). Black bars represent poplar LGs; their numbers are indicated on the left. Stacked symbols represent clusters. Within each cluster, the first PtWAK gene on an LG is found at the top of the pile, and the last is at the bottom. Clustered genes are noted PtWAKx-y, meaning that this cluster contains (y − x + 1) PtWAK genes. Empty squares, PtWAK-RLK; filled squares, PtWAK-RLCK; empty circles, PtWAK-RLP; and filled circles, PtWAK short genes
Putative PtWAKs were only annotated as protein kinases in the poplar genome v3.0. They were predicted to encode a variable extracellular part with different domain organization, a single transmembrane region and an intracellular kinase domain. Our classification adapted from Zhang et al. (2005) distributed PtWAKs into four groups (Table 1). A PtWAK-RLK had one or more extracellular protein domains and an intracellular kinase domain, a PtWAK-RLCK had a single intracellular kinase domain, and a PtWAK-RLP had only extracellular protein domains. WAK genes without a functional domain were classified as PtWAK short genes, but their protein sequence showed at least 70 % of similarity with other WAKs. For comparison, the deduced protein sequences of OsWAK short genes showed 40 % similarity with the longer WAK sequence.
Analysis of the protein extracellular part
Numerous domains (GubWAK, WAK, WAKassoc, EGF-Ca2+, cEGF, and EGF3) may be found in the extracellular part of the WAK proteins, which enhance its variability potential.
From the N-terminal, the majority of PtWAK proteins displayed the wall-associated receptor kinase galacturonan-binding (Gub_WAK_bind, called GubWAK) domain, which was described by Decreux et al. (2006). In PtWAKs, this domain had two different parts. The first part contained a highly conserved motif (YF)PF comprising aromatics (F and Y) and cyclic (P) amino acids. The second part of the domain comprised a C-X5–6-F-X-(VL)-X-C motif dividing the set of sequences into two groups (Fig. 2). The conserved motifs could form a possible site of adhesion to a ligand in the cell wall matrix, where AtWAKs were shown to bind cell wall pectins in the N-terminal of their extracellular region in a calcium-induced conformation (Decreux and Messiaen 2005).
Conserved motifs in poplar WAK protein domains. The conserved motifs are indicated in the order they appear in the different PtWAK domains. Arrows indicate conserved amino acids described in PFAM database. A alanine, R arginine, N asparagine, D aspartic acid, C cysteine, Q glutamine, E glutamic acid, G glycine, H histidine, I isoleucine, L leucine, K lysine, M methionine, F phenylalanine, P proline, S serine, T threonine, W tryptophan, Y tyrosine, V valine
Paradoxically, two domains were never found together in the same protein: the WAK domain, and the wall-associated receptor kinase C-terminal domain (WAK_assoc, termed WAKassoc). The role of the WAK domain remains unknown. It is described in the PFAM database as being associated with the kinase domain in plant WAK proteins. Our analysis showed three conserved parts in the WAK domain. Fifty percent of the proteins contained the first part, which was highly conserved (Fig. 2) and consisted of polar and noncharged residues (N, T, and S) plus some hydrophobic amino acids (V and L). The second and third parts were the most frequent (94 %). The second part mainly comprised hydrophobic amino acids. A highly conserved structure (CK(LV)AF(IL)V) was distinguishable in this part, composed of a charged lysine between a cysteine and a sequence of hydrophobic amino acids (Fig. 2). The third part was composed mainly of cyclic (P), aromatic (W and Y), and aliphatic (L and V) amino acids. The WAKassoc domain is described as a cysteine-rich domain, since it contained four highly conserved cysteines as described in the PFAM database (Punta et al. 2012).
The last downstream extracellular domain was represented by the EGF-like family with three EGF-like motifs. The calcium-binding EGF domain (EGF_CA, called EGF-Ca2+) was the only domain found in Arabidopsis, rice and Populus. It contained six highly conserved cysteines. We note that the second cysteine is not represented in Fig. 2 because the MEME program did not detect this second cysteine as conserved, although it was present in the dataset of sequences and the alignment (Online resource 2). This might be due to a too-broad variability of the other amino acids flanking this cysteine. The (EGF2)-like domain described in rice and Arabidopsis was not found in poplar WAKs. Instead, two other EGF-like domains were observed: the complement Clr-like EGF_like domain (cEGF) and the EGF_3 domain (EGF3). The latter presented the same conserved residues described in the PFAM database except that the cysteine at the last position was missing and replaced by an aromatic residue (Y/F), whereas the poplar cEGF domain was as expected by the PFAM database description. The substitution of the sixth cysteine of the EGF3 domain by an aromatic residue (Y/F) suggested a possible cation–π interaction between an aromatic compound and a cation (Dougherty 2012). The main cation of the cell wall with which the protein may interact is calcium. This is consistent with EGF domains studied mainly in animals (Rao et al. 1995; Morgan et al. 1999; Stenflo et al. 2000; Wouters et al. 2005), which interact in a calcium-induced conformation. EGF-like domains can also be O-glycosylated and are known to take part in domain/domain interaction, both domains in part bridged by calcium chelation (Rao et al. 1995; Stenflo et al. 2000). The presence of one or more EGF-like domains on the extracellular part of WAK proteins also suggested potential protein-protein interactions in the extracellular part of these WAKs.
It is interesting to note that all the domains described above exhibit highly conserved cysteines that could indicate that WAK proteins may be regulated via redox processes and/or via protein/protein interactions (Wrzaczek et al. 2010; Freisinger 2011). In addition, WAK proteins presented notable conserved domain architecture in the extracellular region, potentially offering a physical zoning of protein domains, which might be set in parallel with cell wall layers deposition, which is crucial in wood formation.
Phylogenetic analysis
The WAK family in Populus consists of a broad RLK subclass that differs in both intracellular kinase sequence and extracellular domain compositions. GubWAK, WAK, and WAKassoc protein domains are exclusive to the plant kingdom, and have been organized around the kinase domain during the course of plant evolution. Here, the phylogenetic analysis focused exclusively on the intracellular kinase domain, which is the single region common to most PtWAKs, and the heart of the enzymatic activity. Accordingly, this analysis concerned the PtWAK-RLK and PtWAK-RLCK groups (see PtWAK classification in the section entitled “Features of WAK gene family in Populus”). After removing incomplete sequences from the alignment, a subset of 112 sequences was used for the phylogeny. The three topologies obtained using NJ, MP, and BI phylogenetic reconstruction were congruent (p = 0.274); NJ and MP bootstrap values and BI posterior probability values are indicated at each node (Fig. 3). Two monophyletic clades were markedly differentiated (i.e. WAK Clades I and II), although the intracellular kinase domains of these WAKs were highly conserved (because of a single Ser/Thr catalytic domain and few downstream target recognition domains that must be evolutionarily constrained). In Fig. 3, the composition of the extracellular region is shown for each protein. The PtWAK proteins with WAK and EGF-like (EGF-Ca2+, cEGF, and EGF3) extracellular domains were found only in clade I, and were unexpectedly organized. Some subclades preferentially contained one type of extracellular domain combination. This result was supported by high posterior probability and bootstrap values (Fig. 3), suggesting an interesting parallel evolution between the kinase domain and the WAK/EGF-like domains of PtWAK-RLK. By contrast, the WAKassoc domain was found in two distinct subclades belonging to WAK clades I and II. When a focused phylogenetic analysis of the WAKassoc domain was performed, the 18 proteins were not split into two groups (Online resource 3). These observations suggest that the 18 proteins recognize similar extracellular signals, but may be involved in two different intracellular signaling pathways. The last domain GubWAK was spread over the entire phylogeny, highlighting its older origin.
Phylogenetic tree of PtWAKs. The conserved kinase domain was analyzed. The protein domain architecture is indicated on the right. Bar length and color shapes are proportional to the number of amino acids of the protein and to the NCBI’s CDD prediction of domain length, respectively. The tree obtained by the BI method is represented, and posterior probability and bootstrap values of >0.5 are indicated at nodes as BI/NJ/MP
Using the same approach, a phylogenetic analysis of PtWAKs, OsWAKs, and AtWAKs was also carried out. The topologies obtained using MP and BI methods were congruent (p = 0.842), but the three were not congruent with NJ topology. Although these analyses showed 18 highly conserved monophyletic groups, their organization was not rigorously conserved between the three topologies (data not shown). However, from the combined reading of the two phylogenetic analyses (Figs. 3 and 4), two divergent WAK family evolutionary threads can be drawn. A first thread (WAK group I) with several clades emerged containing members from the three species. In the second thread (WAK group II), one clade specifically addressed PtWAK. This result was supported by high bootstraps and posterior probability. This suggests that the evolution of the first WAK group had occurred in the monocot–dicot common ancestor, while the clade from the second WAK group evolved independently in poplar.
Transcriptomics data analyses
Intuitively, we might argue that such a remarkable expansion of the WAK family and its protein extra-intracellular domain organization could confer advantages on Populus processes involved in innate immunity and abiotic stress responses and/or in plant growth and development. Two main fundamental questions then arise: (i) what are the PtWAK members that show contrasting transcriptional expressions during specific physiological responses? (ii) what kind of evolutionary information could be highlighted in this panel of modulated PtWAKs given their genomic organization and their protein domain architecture?
In our study, 119 PtWAKs among the 175 predicted WAK genes retrieved in v3.0 poplar genome matched v2.2 gene models that were included in the publicly available transcriptomics analyses (Azaiez et al. 2009; Levée et al. 2009; Wilkins et al. 2009a, b; Cohen et al. 2010; Grisel et al. 2010; Hamanishi et al. 2010). In the v2.2 genome assembly, only 120 PtWAKs were identified (data not shown). To our knowledge, since the v3.0 genome assembly has been released in August 2012, no transcriptomics analyses based on these data are available yet. We were able to obtain expression values for 72 genes (Affymetrix array) (Online resource 4). For comparison, another technology (Agilent custom probe design array) found a similar number with 76 PtWAKs expressed in apex. In view of the large number of sequences identified in the poplar v3.0 genome assembly, and the redundancy of sequences in the multigenic family, our results give a good indication of PtWAK expression profile, still limited, however, by probe design in chips. The WAK family presents a high level of homology between its members, especially among members within clusters. As a consequence, one cannot exclude that a probe designed on the v2 genome assembly could hybridize transcripts from different genes. Alternatively, different probes could hybridize only one v3 gene. Keeping this limitation in mind, the observation of differential expression pattern suggests the modulation of expression of at least one WAK member.
Overall, the 72 PtWAKs exhibited a lower expression level than a randomly selected set of genes (Online resource 5a). Likewise, only 35 ESTs were retrieved from the database (Online resource 1, April 2013), representing a very small group of 17 PtWAKs. This is in agreement with the commonly found low abundance of WAK transcripts in Arabidopsis (He et al. 1996; Kohorn et al. 2006). This indicated that these genes were transcribed at a very low level, or that their mRNAs were not accessible because of a possible too-short half-life, or that these genes presented spatial and temporal expression patterns not recorded in the public libraries.
The WAK family has been shown to be involved in plant growth/development and plant-microbe interactions/abiotic stress responses in Arabidopsis (He et al. 1998; Sivaguru et al. 2003; Verica et al. 2003; Hou et al. 2005; Meier et al. 2010), and in plant–microbe interaction in rice (Li et al. 2009). An entropy calculation approach was used to evaluate the global expression of PtWAKs. This approach first indicated that the expression of PtWAKs was not significantly tissue-dependent (Online resource 5b). This contrasted with AtWAKs and AtWAKLs expressions, which were largely tissue-specific in green tissues and roots/flowers, respectively (Verica and He 2002; Verica et al. 2003). However, a detailed analysis yielded further information. Twenty-five PtWAKs showed a significant tissue-specific expression and were mainly overexpressed in mature leaves, roots, and flowers (Fig. 5). Secondly, the expression of 16 PtWAKs was significantly affected by changing environmental conditions (Online resource 5c), after pathogen attacks, light exposure or drought treatment. In line with the hypothesis that the ability to link with oligogalacturonic acids is a key factor for the pathogen attack response (Brutus et al. 2010), all the PtWAKs that were overexpressed during pathogen stress belonged to the PtWAK-RLK group, in which members contain a GubWAK domain (Fig. 5; Online resource 1). However, we noted that the PtWAK-RLCK Potri.017G117500, which has no GubWAK domain, was overexpressed during pathogen attacks, suggesting the existence of additional pathogen recognition mechanisms or a lack of structural annotation for this gene model. In addition, the expression of PtWAKs was significantly modified during light limitation (five members), and by water availability in shoot apical meristem (five members, Online resource 6), in roots (one member), and in leaves (one member, Fig. 5). These two abiotic parameters could be viewed as being two interconnected driving forces involved in the hydraulic conductivity of the plant, in which WAKs could play a sensing role (Lally et al. 2001; Kohorn et al. 2006; Ben Bâaziz et al. 2012). Lastly, neither aluminium nor flooding stresses had any significant effect on the expression of any of the 72 PtWAKs (Online resource 4).
Clustered heatmap showing expression patterns of significant tissue/condition/genotype specific expressed WAKs in poplar. PtWAK v3.0 gene model names are indicated. Only PtWAKs showing differential expression in at least one tissue/environmental condition/genotype are displayed (42 PtWAKs). For each tissue/environmental condition/genotype, mean relative expression values (three to four replicates) are shown. Scale from 0.0 to 1.0 corresponds to drought experiment in the apex; scale from 1.0 to 1.2 is for all the affymetrix array experiments. Relative expression value was calculated by dividing the absolute value (RMA normalized) by the median of absolute values of all samples, except for the apex assay. In this case, relative expression was obtained by dividing each array-normalized expression value by the median of absolute values of all samples, and was then log2-transformed. An asterisk indicates significant tissue/environmental condition/genotype effect on PtWAK expression according to Welch’s t test/one-way ANOVA for two or more samples followed by adjusted Bonferroni correction (*p < 0.05). Hierarchical clustering was performed for PtWAKs according to their expression patterns. “Carpaccio,” “DN34,” and “Soligo” are Populuscanadensis genotypes; “NM36” is P. nigra × P. maximowiczii while “AP-947,” “AP-1005,” “AP-1006,” “AP-2278,” “AP-2298,” and “AP-2300” are P. balsamifera genotypes. Blue- and red-shaded PtWAK names belong to PtWAK-RLK and PtWAK-RLCK gene groups, respectively
Analysis of these expression data together with WAK family genome organization gave some interesting results. Except for Potri.008G059900, PtWAK members showing significant transcript accumulation during pathogen attack all belonged to the largest tandem clusters (chromosome VII, IX, XII, and XVII). Intriguingly, WAK members of these chromosomes were spread preferentially over the monophyletic WAK clade II, which is specific to Populus (Fig. 4). These findings suggest duplication resulting from strong selection pressure exerted by poplar pathogens. This hypothesis has also been advanced by Lehti-Shiu et al. (2009) studying the RLK family in Arabidopsis, poplar, and rice. This may be functionally linked to phytopathogen features, which generally present narrow host spectrum specificities and display a corresponding genetic evolution (Jones and Dangl 2006). By contrast, most PtWAK members that were differentially expressed during developmental/growth processes were not integrated into an expanded tandem cluster. These WAKs were found on chromosomes III, V, VII, XIV, XV, and XVIII, and were distributed in the monophyletic WAK clade I, which is shared with rice and Arabidopsis (Fig. 4). Genomic nontandem receptors involved in development could be more conserved between plants, probably because of the common role they play. A few tandem members belonging to chromosomes IV, VII, and XVII were also modulated in development processes. They could represent potential developmental innovations that meet specific woody-species needs. Their presence in clusters could be explained by specific duplication in poplar after its divergence.
The large proportion of tandem duplication of WAK genes raises the question of the functional redundancy of duplicates (or paralogs). According to Moore and Purugganan (2005), paralogs may undergo divergent evolutionary fates during subsequent genome evolution such as subfunctionalization (partition of original functions), neofunctionalization (acquisition of novel functions), or nonfunctionalization (loss of original functions), which could be paired with divergence in expression patterns. Of the 11 main phylogenetic homologous blocks of PtWAK, nine showed near-similar expression patterns with respect to the tissues or stress conditions examined (Fig. 3; Online resource 4), suggesting that most expressed PtWAK genes have been retained by subfunctionalization. However, neofunctionalization may also have been involved, albeit on a smaller scale, with only two phylogenetic blocks: one composed of two segmental duplicates, the other containing three tandem duplicates. These five WAK sequences strongly diverged in their expression patterns. The segmental duplicate Potri.014G0386000 was predominantly expressed in roots and female catkin, whereas its counterpart Potri.002G130600 seemed specifically expressed in differentiating xylem. Furthermore, among the three tandem duplicated members Potri.002G075900, Potri.002G076000, and Potri.002G076100, only the second was differentially expressed in mature leaves.
Finally, our analysis revealed a randomly expressed PtWAKs distribution between genotypes and Populus species. 24 PtWAKs were modulated by drought stress in one or more of the ten genotypes studied here. In other words, a WAK gene may be differentially expressed in various genotypes in response to the same constraint. This suggests complex regulations of the WAK gene family that may have been set up during evolution in response to a changing environment.
Conclusions
The WAK family in poplar comprises a very large number of members. The WAK proteins contain various extracellular domains putatively playing divergent functional roles. Together, these features make WAKs appealing candidates to investigate new communication networks that may relay the wall status to the cytoplasm in the context of the CPMCW continuum described by Baluška et al. (2003). The cell wall is an essential compartment affected not only by both pathogen attacks and developmental processes but also by abiotic stresses such as drought and various mechanical constraints. Our study reports that a panel of PtWAKs was modulated at the transcriptional level in some of these physiological processes, suggesting that related encoded proteins could be involved, potentially as receptors of pathogen ligands or as sensors of hydromechanical cues. Thus this study provides new insight into the WAK gene family in Populus. Our findings may also be a starting point for a study of the physiological roles of WAKs in cellular regulatory networks in woody species.
References
Azaiez A, Boyle B, Levée V, Séguin A (2009) Transcriptome profiling in hybrid poplar following interactions with Melampsora rust fungi. Mol Plant Microbe Interact 22:190–200
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble S (2009) MEME Suite: tools for motif discovery and searching. Nucleic Acids Res 37:202–208
Baluška F, Šamaj J, Wojtaszek P, Volkmann D, Menzel D (2003) Cytoskeleton–plasma membrane–cell wall continuum in plants. Emerging links revisited. Plant Physiol 133:482–491
Ben Bâaziz K, Lopez D, Rabot A, Combes D, Gousset A, Bouzid S, Cochard H, Sakr S, Venisse JS (2012) Light-mediated Kleaf induction and contribution of both the PIP1s and PIP2s aquaporins in five tree species: walnut (Juglans regia) case study. Tree Physiol 32:423–434
Brutus A, Sicilia F, Macone A, Cervone F, De Lorenzo G (2010) A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc Natl Acad Sci U S A 107:9452–9457
Calendini F, Martin JF (2005) PaupUP v1.0.3.1, A free graphical frontend for Paup* Dos software
Cohen D, Bogeat-Triboulot MB, Tisserant E et al (2010) Comparative transcriptomics of drought responses in Populus: a meta-analysis of genome-wide expression profiling in mature leaves and root apices across two genotypes. BMC Genomics 11:630
Decreux A, Messiaen J (2005) Wall-associated kinase WAK1 interacts with cell wall pectins in a calcium-induced conformation. Plant Cell Physiol 46:268–278
Decreux A, Thomas A, Spies B, Brasseur R, Van Cutsem P, Messiaen J (2006) In vitro characterization of the homogalacturonan-binding domain of the wall-associated kinase WAK1 using site-directed mutagenesis. Phytochemistry 67:1068–1079
Dougherty DA (2012) The cation–π interaction. Acc Chem Res 46:885–893
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Freisinger E (2011) Structural features specific to plant metallothioneins. J Biol Inorg Chem 16:1035–1045
Goodstein DM, Shu S, Howson R et al (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40:1178–1186
Grisel N, Zoller S, Künzli-Gontarczyk M, Lampart T, Münsterkötter M, Brunner I, Bovet L, Métraux JP, Sperisen C (2010) Transcriptome responses to aluminum stress in roots of aspen (Populus tremula). BMC Plant Biol 10:185
Hamanishi ET, Raj S, Wilkins O, Thomas BR, Mansfield SD, Plant AL, Campbell MM (2010) Intraspecific variation in the Populus balsamifera drought transcriptome. Plant Cell Environ 33:1742–1755
Hanada K, Zou C, Lehti-Shiu MD, Shinozaki K, Shiu SH (2008) Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol 148:993–1003
He ZH, Fujiki M, Kohorn BD (1996) A cell wall-associated, receptor-like protein kinase. J Biol Chem 271:19789–19793
He ZH, He D, Kohorn BD (1998) Requirement for the induced expression of a cell wall associated receptor kinase for survival during the pathogen response. Plant J 14:55–63
He ZH, Cheeseman I, He D, Kohorn BD (1999) A cluster of five cell wall-associated receptor kinase genes, Wak1-5, are expressed in specific organs of Arabidopsis. Plant Mol Biol 39:1189–1196
Hou X, Tong H, Selby J, Dewitt J, Peng X, He ZH (2005) Involvement of a cell wall-associated kinase, WAKL4, in Arabidopsis mineral responses. Plant Physiol 139:1704–1716
Humphrey TV, Bonetta DT, Goring DR (2007) Sentinels at the wall: cell wall receptors and sensors. New Phytol 176:7–21
Johnson WE, Li C, Rabinovic A (2007) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8:118–127
Jones JDG, Dangl JL (2006) The plant immune system. Nature 444:323–329
Jourez B, Riboux A, Leclercq A (2001) Anatomical characteristics of tension wood and opposite wood in young inclined stems of poplar (Populus euramericana cv ‘ghoy’). IAWA J 22:133–157
Kishino H, Hasegawa M (1989) Evaluation of the maximum likelihood estimate of the evolutionnary tree topologies from DNA sequences data, and the branching order in Hmonoidea. J Mol Evol 29:170–179
Kohler A, Rinaldi C, Duplessis S, Baucher M, Geelen D, Duchaussoy F, Meyers BC, Boerjan W, Martin F (2008) Genome-wide identification of NBS resistance genes in Populus trichocarpa. Plant Mol Biol 66:619–636
Kohorn BD, Kobayashi M, Johansen S, Friedman HP, Fischer A, Byers N (2006) Wall-associated kinase 1 (WAK1) is crosslinked in endomembranes, and transport to the cell surface requires correct cell-wall synthesis. J Cell Sci 119:2282–2290
Kohorn BD, Johansen S, Shishido A, Todorova T, Martinez R, Defeo E, Obregon P (2009) Pectin activation of MAP kinase and gene expression is WAK2 dependent. Plant J 60:974–982
Kohorn BD, Kohorn SL, Todorova T, Baptiste G, Stansky K, McCullough M (2012) A dominant allele of Arabidopsis pectin-binding wall-associated kinase induces a stress response suppressed by MPK6 but not MPK3 mutations. Mol Plant 5:841–851
Lally D, Ingmire P, Tong HY, He ZH (2001) Antisense expression of a cell wall-associated protein kinase, WAK4, inhibits cell elongation and alters morphology. Plant Cell 13:1317–1331
Lan T, Yang ZL, Yang X, Liu YJ, Wang XR, Zeng QY (2009) Extensive functional diversification of the Populus glutathione S-transferase supergene family. Plant Cell 21:3749–3766
Lehti-Shiu MD, Zou C, Hanada K, Shiu SH (2009) Evolutionary history and stress regulation of plant Receptor-Like Kinase/Pelle genes. Plant Physiol 150:12–26
Letunic I, Bork P (2011) Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39:475–478
Levée V, Major I, Levasseur C, Tremblay L, MacKay J, Séguin A (2009) Expression profiling and functional analysis of Populus WRKY23 reveals a regulatory role in defense. New Phytol 184:48–70
Li H, Zhou SY, Zhao WS, Su SC, Peng YL (2009) A novel wall-associated receptor-like protein kinase gene, OsWAK1, plays important roles in rice blast disease resistance. Plant Mol Biol 69:337–346
Lopez D, Bronner G, Brunel N et al (2012) Insights into Populus XIP aquaporins: evolutionary expansion, protein functionality, and environmental regulation. J Exp Bot 63:2217–2230
Marchler-Bauer A, Lu S, Anderson JB et al (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39:225–229
Meier S, Ruzvidzo O, Morse M, Donaldson L, Kwezi L, Gehring C (2010) The Arabidopsis wall associated kinase-like 10 gene encodes a functional guanylyl cyclase and is co-expressed with pathogen defense related genes. PLoS One 5:e8904
Moore RC, Purugganan MD (2005) The evolutionary dynamics of plant duplicate genes. Curr Opin Plant Biol 8:122–128
Morgan WD, Birdsall B, Frenkiel TA, Gradwell MG, Burghaus PA, Syed SEH, Uthaipibull C, Holder AA, Feeney J (1999) Solution structure of an EGF module pair from the Plasmodium falciparum merozoite surface protein 1. J Mol Biol 289:113–122
Paterson AH, Freeling M, Tang H, Wang X (2010) Insights from the comparison of plant genome sequences. Annu Rev Plant Biol 61:349–372
Petre B, Major I, Rouhier N, Duplessis S (2011) Genome-wide analysis of eukaryote thaumatin-like proteins (TLPs) with an emphasis on poplar. BMC Plant Biol 11:33
Punta M, Coggill PC, Eberhardt RY et al (2012) The Pfam protein families database. Nucleic Acids Res 40:290–301
R Development Core Team (2008). R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org
Rao Z, Handford P, Mayhew M, Knott V, Brownlee GG, Stuart D (1995) The structure of a Ca2+-binding epidermal growth factor-like domain: its role in protein-protein interactions. Cell 82:131–141
Saeed A, Sharov V, White J et al (2003) TM4: A free, open-source system for microarray data management and analysis. Biotechniques 34:374–378
Schug J, Schuller WP, Kappen C, Salbaum JM, Bucan M, Stoeckert CJ (2005) Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol 6:R33
Shiu SH, Bleecker AB (2001) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci U S A 98:10763–10768
Shiu SH, Bleecker AB (2003) Expansion of the Receptor-Like Kinase/Pelle gene family and Receptor-Like Proteins in Arabidopsis. Plant Physiol 132:530–543
Sivaguru M, Ezaki B, He ZH, Tong H, Osawa H, Baluška F, Volkmann D, Matsumoto H (2003) Aluminum-induced gene expression and protein localization of a cell wall-associated receptor kinase in Arabidopsis. Plant Physiol 132:2256–2266
Sjödin A, Street NR, Sandberg G, Gustafsson P, Jansson S (2009) The Populus Genome Integrative Explorer (PopGenIE): a new resource for exploring the Populus genome. New Phytol 182:1013–1025
Stenflo J, Stenberg Y, Muranyi A (2000) Calcium-binding EGF-like modules in coagulation proteinases: function of the calcium ion in module interactions. Biochim Biophys Acta 1477:51–63
Swofford DL (2000) PAUP*: phylogenetic Analysis Using Parsimony (and Other Methods) Version 4. Sinauer Associates, Sunderland
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Telewski FW (2006) A unified hypothesis of mechanoperception in plants? Am J Bot 93:1466–1476
Tocquard K, Lopez D, Decourteix M, Thibaut B, Julien JL, Label P, Leblanc-Fournier N, Roeckel-Drevet P (2014) The molecular mechanisms of reaction wood induction. In: Gardiner B, Barnett J, Saranpää P, Gril J (eds) Biol. React. Wood, Springer, Berlin Heidelberg, pp 107–138
Tuskan GA, Difazio S, Jansson S et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313:1596–1604
Verica JA, He ZH (2002) The cell Wall-Associated Kinase (WAK) and WAK-Like kinase gene family. Plant Physiol 129:455–459
Verica JA, Chae L, Tong H, Ingmire P, He ZH (2003) Tissue-specific and developmentally regulated expression of a cluster of tandemly arrayed cell wall- associated kinase-like kinase genes in Arabidopsis. Plant Physiol 133:1732–1746
Wagner TA, Kohorn BD (2001) Wall-associated kinases are expressed throughout plant development and are required for cell expansion. Plant Cell 13:303–318
Wilkins O, Nahal H, Foong J, Provart NJ, Campbell MM (2009a) Expansion and diversification of the R2R3-MYB family of transcription factors. Plant Physiol 149:981–993
Wilkins O, Waldron L, Nahal H, Provart NJ, Campbell MM (2009b) Genotype and time of day shape the Populus drought response. Plant J 60:703–715
Wolf S, Hématy K, Höfte H (2012) Growth control and cell wall signaling in plants. Annu Rev Plant Biol 63:381–407
Wouters MA, Rigoutsos I, Chu CK, Feng LL, Sparrow DB, Dunwoodie SL (2005) Evolution of distinct EGF domains with specific functions. Protein Sci 14:1091–1103
Wrzaczek M, Brosché M, Salojärvi J, Kangasjärvi S, Idänheimo N, Mersmann S, Robatzek S, Karpiński S, Karpińska B, Kangasjärvi J (2010) Transcriptional regulation of the CRK/DUF26 group of receptor-like protein kinases by ozone and plant hormones in Arabidopsis. BMC Plant Biol 10:95
Yang X, Kalluri UC, DiFazio SP, Wullschleger SD, Tschaplinski TJ, Cheng ZM, Tuskan GA (2009) Poplar genomics: state of the science. Crit Rev Plant Sci 28:285–308
Zhang S, Chen C, Li L, Meng L, Singh J, Jiang N, Deng XW, He ZH, Lemaux P (2005) Evolutionary expansion, gene structure, and expression of the rice wall-associated kinase gene family. Plant Physiol 139:1107–1124
Acknowledgments
This work was supported by the French Ministery of Higher Education and Research (doctoral contract). Editing and proofreading were done by Auvergne Traduction Technique.
Data archiving statement
A full list of the Phytozome gene sequences used for this paper is included in Online ressource 1. Data used for transcriptomic analyses are referenced on GEO database (accession numbers: GSE13990, GSM412653, GSE9673, GSE17223, GSE15242, GSE17226, GSE17230, GSE21171, GSE19297, and GSE13109).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. L. Wegrzyn
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online resource 1
P. trichocarpa WAKs. PtWAKs were classified based on their chromosomal location. Alias names of v1 and v2 of poplar genome annotation are indicated. Domain structure, EST names and numbers per sequence, and WAK genomic, transcript, CDS, and protein sequences are given (PDF 29 kb)
Online resource 2
Alignment of EGF-Ca2+ domains (PDF 51 kb)
Online resource 3
Phylogenetic tree of PtWAKs containing the WAKassoc domain. The phylogenetic tree of PtWAKs using conserved WAKassoc domain is represented. The tree obtained by the BI method is represented, and posterior probability and bootstrap values of >50 are indicated at nodes as BI/NJ/MP (PDF 33 kb)
Online resource 4
Heatmap representation of all PtWAK expression patterns among different poplar tissues/environmental conditions/genotypes. PtWAK v3.0 gene model names are indicated. For each tissue/environmental condition/genotype, mean relative expression values (three to four replicates) are displayed. Sample and WAK, relative expression value was calculated by dividing the absolute value (RMA normalized) by the median of absolute values of all samples, except for the apex assay. In this case, relative expression was obtained by dividing each array-normalized expression value by the median of absolute values of all samples, and was then log2-transformed. An asterisk indicates significant tissue/environmental condition/genotype effect on PtWAK expression according to Welch’s t test/one-way ANOVA for two or more samples followed by adjusted Bonferroni correction (*p < 0.05). Blue-, red-, and green-shaded PtWAK names belong to PtWAK-RLK, PtWAK-RLCK, and PtWAK-RLP gene groups, respectively (PDF 144 kb)
Online resource 5
Expression pattern comparison of PtWAK and randomly selected genes;Seventy-two PtWAK genes or randomly selected genes were used for this analysis. a Box-plot representation of PtWAK and random gene expression level distribution. Expression values are RMA-normalized data from all experiments performed in this work (see “Methods”). The significant PtWAK effect on expression level was detected with Welch’s t test (***p < 0.001). For the Shannon entropy calculation, see “Methods.” b Tissue specificity of PtWAK and random genes as measured by Shannon entropy distribution. Low entropy values indicate high tissue specificity. χ2 value is given (not significant, p = 0.064). c Condition specificity of PtWAK and random genes as measured by Shannon entropy distribution. Low entropy values indicate high condition specificity. χ2 value is given (**p < 0.01). d Genotype specificity of PtWAK and random genes as measured by Shannon entropy distribution. Low entropy values indicate high genotype specificity. χ2 value is given (*p < 0.05) (PDF 55 kb)
Online resource 6
PtWAKs whose apex expression is affected depending on water availability. The mean relative expression values (three samples per condition) and their standard errors shown here were significantly affected by water availability variations according to one-way ANOVA followed by adjusted Bonferroni correction (*p < 0.05). Relative expression was obtained by dividing each array-normalized expression value by the median of absolute values of all samples for the PtWAK, and was then log2-transformed. Values for control (black charts), drought (grey charts) and rewatering after drought (white charts) are given (PDF 24 kb)
Rights and permissions
About this article
Cite this article
Tocquard, K., Lafon-Placette, C., Auguin, D. et al. In silico study of wall-associated kinase family reveals large-scale genomic expansion potentially connected with functional diversification in Populus . Tree Genetics & Genomes 10, 1135–1147 (2014). https://doi.org/10.1007/s11295-014-0748-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-014-0748-7