Abstract
The western Himalayan yew (Taxus contorta Griffith, synonym Taxus fuana Nan Li & R.R. Mill), one of the highly prized tree species for its anticancer properties, is experiencing severe reduction in populations across its range in Pakistan. We examined a chloroplast DNA region (trnL-F) and ten nuclear microsatellite loci variations among seven populations to investigate the levels of genetic diversity and reconstruct the demographic history of T. contorta in Pakistan. A low haplotype diversity (H T = 0.182), moderate level of microsatellite diversity (H E = 0.541 ± 0.034), significant population differentiation (F ST = 0.107) and high level of inbreeding (F IS = 0.219–0.418) characterized this species. Results of an approximate Bayesian computation (ABC), mismatch distribution analysis, neutrality tests and data on glacial chronologies of the western Himalaya indicated a historic demographic expansion of T. contorta populations from small ancestral populations. This expansion is estimated to have occurred at least 37.5 thousand years ago (kya) during the late Pleistocene. An unprecedented level of habitat disturbance over the last few decades, coupled with the highly variable climate may have played an important role in shaping the genetic structure of the extant T. contorta populations. We have discussed a range of sustainable management measures for the ecological restoration of remnant populations as well as for a sustainable exploitation of this plant of high economic potential.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The levels of genetic diversity and population structure of plants are largely determined by their evolutionary histories, shaped by impacts of stochastic factors such as demographic expansion, environmental variation and natural catastrophes, that acted upon species over time (Spielman et al. 2004; Ruan et al. 2013; Liu et al. 2013a). Genetic variation also plays a role in a wide range of ecosystem processes (Hughes et al. 2008). In addition, the pattern of plant distribution and their genetic diversity has been affected by human-induced habitat fragmentation since the beginning of the Holocene (Jump and Peñuelas 2006; Miehe et al. 2009). The genetic consequences of demographic and environmental stochasticities and catastrophes in plants with restricted distribution ranges are generally a decrease in genetic variability and an increase in inbreeding which consequently can lead to a reduced reproductive fitness (Frankham 2005). Thus, the timing of conservation measures is crucial to conserve genetic diversity and potential adaptive values for the long-term persistence of a species (Jump et al. 2009; Hoffmann and Sgro 2011).
The western Himalaya, characterized by a complex mountain system, has undergone periodic climatic changes over several million years until the mid-Holocene (Yin and Harrison 2000; Owen et al. 2008; Kamp and Owen 2011). This region has also witnessed an increased human population that led to an unprecedented level of disturbance in the natural vegetation and extensive forest destruction, particularly over the last few decades (Ali et al. 2005; Pandit et al. 2007; Miehe et al. 2009; Haq 2012). Taxus contorta Griffith (synonym Taxus fuana Nan Li & R.R. Mill) is a dioecious, evergreen, understorey tree species with a distribution in subtropical to temperate regions from eastern Afghanistan to central Nepal along the western Himalaya (Möller et al. 2007; Shah et al. 2008b; Poudel et al. 2012). This species, commonly known as the western Himalayan yew, is one of the only three yew species in the Himalayas, the others being Taxus wallichiana Zucc. and Taxus mairei (Lemée & H. Léveillé) S.Y. Hu ex T.S. Liu (Poudel et al. 2012). Taxus contorta is facing severe pressure from habitat encroachment from local farmers as well as an unsustainable harvesting for domestic uses and commercial production of ‘Taxol’, an anticancer compound (Poudel et al. 2013). The species has already lost several populations from its limited distribution range in northern Pakistan, leading to further fragmentation of the remaining populations which are sometimes reduced in size to less than fifty to a few individuals (Haq 2012).
Most of the natural forests in Pakistan occur in the northern part and cover about 4.8 % of the total land area of the country. This forest area is declining at an annual rate of 4–6 % due to pressures from a rapidly growing population and agriculture land expansion, illegal commercial harvesting, and over-exploitation for local consumptive uses (Ali et al. 2005; Haq 2012). Forest destruction and fragmentation have serious impacts on less common understorey tree species by altering the species composition and the surrounding ecosystem (Benítez-Malvido and Martínez-Ramos 2003; Barbier et al. 2008). Taxus contorta has been listed as endangered in the IUCN red data book and is regulated by CITES. However, increased local uses, habitat destruction and degradation are major challenges in the conservation of yew in Pakistan (Shah et al. 2008a). An increased understanding of the historic and contemporary events and their consequences on the genetic diversity and differentiation among populations of an endangered species like T. contorta will help to design appropriate long-term conservation strategies (Liu et al. 2013b), especially for this species with its high economic potential (Poudel et al. 2013).
The genetic diversity and population structure of T. contorta in Pakistan have been characterized by Shah et al. (2008a) using random amplified polymorphic DNAs (RAPDs). A high level of genetic differentiation among populations and a low level of genetic diversity within populations were detected in this study. By contrast, RAPD and amplified fragment length polymorphism (AFLPs) markers showed a high within population genetic variation in nine studied populations of Taxus from the Himachal state of northwest India (as T. wallichiana in Mohapatra et al. 2009 according to Shah et al. (2008b) and Poudel et al. (2012), yews from this area of northwest India should be regarded as T. contorta). Comparing the results of Mohapatra et al. (2009) and Shah et al. (2008a), who used the same genetic marker, RAPDs, the genetic differentiation among populations (NW India, G ST = 0.214, ϕ ST = 0.100; Pakistan, G ST = 0.584, ϕ ST = 0.568) was higher but the genetic diversity (NW India, H E = 0.307, Pakistan, H E = 0.116) was lower within populations from Pakistan. Molecular markers such as RAPDs can sample a relatively unbiased portion of the genome, but this marker has a few technical and/or analytical drawbacks: their dominant inheritance, thus the exact level of heterozygotes cannot be determined; they can be scored only as absence or presence of bands; therefore, their homology cannot be ascertained fully; they are prone to effects of subjective user selection criteria, and RAPD assays are sometimes not sufficiently reproducible (Nybom and Bartish 2000). Although, the selection of markers depends on the practicality and objectives of a study, nuclear microsatellites or simple sequence repeats (nSSRs) have gained tremendous popularity in conservation genetic studies because of their co-dominant inheritance, multiallelic nature, high reproducibility and high throughput in genotyping for precise analyses (Sunnucks 2000). Nowadays, biparentally inherited microsatellites are often combined with uniparental organelle markers for phylogeographic studies (Qi et al. 2012; Liu et al. 2013a). In Taxus, chloroplast DNA (cpDNA) is paternally inherited and is dispersed by pollen and seeds (Collins et al. 2003). Despite the relatively slow rate of evolution in cpDNA, high levels of genetic variation have been reported in some noncoding spacers within and among species (Meng and Zhang 2013), including Taxus (Gao et al. 2007; Liu et al. 2013a). Chloroplast DNA is not subjected to recombination and has a lower effective population size and thus may provide complementary and more exact information on population differentiation, level of genetic diversity and demographic history of a species (Bai et al. 2010; Jadwiszczak et al. 2012).
In this study, we used a combination of nuclear microsatellite markers and chloroplast sequence data to investigate the genetic structure and demographic history of T. contorta in Pakistan. Specifically we aimed to (1) determine the levels of nuclear microsatellite and cpDNA diversity within and among populations, (2) estimate the degree of population differentiation and inbreeding in the context of the complex geography and habitat fragmentation in Pakistan, (3) investigate the demographic history of the species in Pakistan, whether it had been affected by past climate fluctuations, and finally, (4) discussed the implications of our finding to devise appropriate conservation strategies for T. contorta in Pakistan.
Materials and methods
Population sampling
We used a total of 129 individuals from seven populations, which covered almost the entire distribution range of T. contorta in Pakistan (Fig. 1 and Table 1). Silica-gel dried leaf materials of 13–20 trees per population were sampled (Shah et al. 2008a) and stored in the Germplasm Bank of Wild Species, Kunming Institute of Botany. The corresponding vouchers deposited in the herbarium of Kunming Institute of Botany, Chinese Academy of Sciences (KUN) were also consulted for taxonomic validation of each sample used for genetic analysis here.
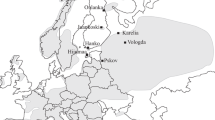
Sampling locations and the distribution frequencies of cpDNA haplotypes (top) and cluster assignment inferred by STRUCTURE (bottom) for Taxus contorta. The top left inset highlights the study area in a map of South Asia. The bottom left inset depicts a median joining phylogenetic network among the cpDNA haplotypes with mutational steps shown by dashes across the branches. For population codes, refer to Table 1
DNA isolation, PCR, sequencing and genotyping
Total DNA was extracted from silica-gel dried leaf material using a modified CTAB method (Liu and Gao 2011). PCR amplification and sequencing of the cpDNA trnL-F region followed Shah et al. (2008b). Sequences were edited in Sequencher v5.0 (Gene Codes Corp., Ann Arbor, MI, USA) and aligned with Clustal W in Bioedit v7.0.9.0 (Hall 1999) with manual adjustments. Ten nuclear microsatellite loci (TG111, TA116, Tax86, TC04, TG06, TW01, TG34, TG47, TS03 and TG141) described in Liu et al. (2011) were used to genotype the samples following Liu et al. (2013a). Allele size was scored and checked for possible genotyping errors like stuttering, large allele dropouts and null alleles in MICRO-CHECKER (Van Oosterhout et al. 2004).
Chloroplast data analysis
The cpDNA haplotypes were defined in DnaSP v5 (Librado and Rozas 2009). The haplotypes identified in each population were plotted on the map of Pakistan using ArcGis v9.3 (Environmental Systems Research Institute, Redlands, CA, USA). Phylogenetic relationships between the haplotypes were inferred through a reduced median network applied to the dataset using Network v4.6 (Bandelt et al. 1999). Indices of haplotype diversity (H d) (Nei et al. 1983) and nucleotide diversity (π) (Nei 1987) were calculated in ARLEQUIN v3.5 (Excoffier and Lischer 2010). The total haplotype diversity (H T), within population diversity (H S) and population differentiation coefficients (G ST and N ST) were estimated using PERMUT (available at: http://www.pierrotoninrafr/genetics/labo/Software/Permut) with 1,000 permutations. A significantly larger N ST than G ST implies the presence of significant phylogenetic structure (Pons and Petit 1996). Geographic structure among the haplotypes was assessed through spatial analysis of molecular variance (SAMOVA) with SAMOVA v10 (Dupanloup et al. 2002). Genetic differentiation within and between populations was investigated by an analysis of molecular variance (AMOVA) using 10,000 permutations in ARLEQUIN.
Microsatellite data analysis
Genetic diversity statistics, i.e. the total number of private alleles (Pa), number of different alleles (Na), observed (H O) and expected (H E) heterozygosity, was estimated using GenAlEx v6 (Peakall and Smouse 2012). The allelic richness (AR), based on a minimum population size of 12 diploid individuals and Wright’s inbreeding coefficient (F IS), was calculated for all populations at each locus. Within each population, Hardy-Weinberg equilibrium and linkage disequilibrium were assessed by applying an adjusted sequential Bonferroni correction. These analyses were performed in FSTAT v2.9.3.2 (Goudet 2002). Null alleles were estimated to re-compute the expected heterozygosity (H E Null) and the population differentiation index (F ST Null) using FreeNA (Chapuis and Estoup 2007). To test the association between pairwise genetic differentiation and geographic distances, a Mantel test was implemented in GenAlEx. Both F ST values, corrected (Chapuis and Estoup 2007) and uncorrected (F ST) (Weir and Cockerham 1984) for null alleles, were used in the Mantel test. Genetic differentiation among the populations was calculated using pairwise F ST between populations and overall F ST across all populations in FSTAT. The F ST is highly likely to underestimate the genetic differentiation between populations for markers with high levels of allelic variations. Thus, standardized measures of genetic differentiation (G' ST) were calculated to account for the influence of high within population heterozygosity (Hedrick and Goodnight 2005; Jost 2008) using SMOGD (Crawford 2010).
To understand the genetic variation among and between populations, an AMOVA was carried out using ARLEQUIN with 10,000 permutations. Nei’s (Nei et al. 1983) inter-population genetic distance (D A) was calculated in Microsatellite Analyser v4.05 (MSA) (Dieringer and Schlötterer 2003), and the resultant matrix used to construct an unrooted neighbour-joining (NJ) tree using 1,000 bootstrap replicates across each population. The tree was visualized using TreeView 1.6.6 (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). A Bayesian clustering program, STRUCTURE v2.3.3 (Pritchard et al. 2000), was used to investigate genetic differentiation and assignment of individuals. With an admixture model and model for correlated allele frequencies between populations, the length of the burn-in and Monte Carlo Markov chain was set to 100,000 each. For the whole microsatellite dataset (ten loci over 129 individuals), initially 15 runs were performed, but later, based on the initial results, the simulation was run ten times for each value of K from 2–10. The best K was selected and used according to Evanno et al. (2005).
Population demographic history
To unravel the demographic history of T. contorta populations in Pakistan, Tajima’s D (Tajima 1989) and Fu’s F S (Fu 1997) values were calculated for the trnL-F sequence data using neutrality tests in ARLEQUIN. The significance of both tests was examined using 1,000 permutations. The purpose of the neutrality tests was to determine whether or not the populations are in mutation-genetic drift equilibrium or not. Tajima’s D and Fu’s F S tend to have significant negative values when populations are not in equilibrium. Further, a pairwise mismatch distribution analysis (MDA) was conducted under a model of demographic and spatial expansion using a total of 1,000 parametric bootstrap replicates. In both models, expected distributions were generated in ARLEQUIN. In the case of populations having undergone expansions in the past, the mismatch distribution is expected to be unimodal, whereas subdivided populations or populations at demographic equilibrium are expected to exhibit multimodal distributions (Slatkin and Hudson 1991; Rogers and Harpending 1992). The sum of squared deviation (SSD) and the Harpending′s raggedness index (HRI) between observed and expected mismatch distributions were used as test statistics. Where the null hypothesis was not rejected, the formula t = τ/2u was used to estimate the timing of expansion (t), where u = μkg (with μ the mutation rate of the sequence, k the number of nucleotides in the matrix and g the generation time in years). The mutation rate for the trnL-F region for Taxus species was taken from Liu et al. (2013a) and was 8.08 × 10−10 s/s/y. The generation time for Taxus followed Wang et al. (2006) and was set to 25 years.
To corroborate our findings of the population expansion of T. contorta in Pakistan as suggested by cpDNA, we tested four plausible scenarios of demographic changes using approximate Bayesian computation (ABC) framework in DIYABC v2 (Cornuet et al. 2014) using the nuclear microsatellite data: (1) old expansion (approximately 13 thousand years ago, kya), (2) recent expansion (approx. 6 kya), (3) old expansion followed by bottleneck to current effective size and (4) old expansion that later bottlenecked and recently recovered back to a large current population size through sudden expansion (Fig. 2). DIYABC allows a selection of the demographic scenario that best fits the data and parameters of interest. We set uniform priors for all demographic parameters that were bound by specified minimum and maximum values and conditions (Table S1, online resource). DIYABC draws a value from prior distributions and performs coalescent-based simulations to generate simulated samples with the same number of gene copies and loci per populations. Time was number of generations with a generation length of 25 years. The mutation model for microsatellite loci, mutation rate of individual locus, mean coefficient, individual locus coefficient, SNI rate, individual locus SNI rate and possible range of allelic state for each locus were set to default values of the software (Table S1, online resource). We pooled all populations into a single sample and for each of the four scenario, we simulated 1 × 106 datasets. After each simulation, Euclidean distances between observed and simulated datasets were drawn from uniform prior distributions. We used the ‘pre-evaluation scenario prior combination’ option in DIYABC to check in which of the scenarios the combinations of priors have produced simulated data sets similar to observed data. Logistic regression analyses were performed to estimate the posterior probability of the best explanatory scenario and subsequent parameters of interest. The scenario with the highest posterior probability was selected, and the associated parameters were determined. Confidence in choice of scenario was assessed by evaluating type I and type II errors (Cornuet et al. 2010). Type I errors were estimated as proportions of instances where the chosen scenario did not exhibit the highest posterior probability compared to the competing scenarios. While type II errors were estimated by counting the proportion of data sets that resulted in the highest posterior probability of the chosen scenario, although simulated with other scenarios. Finally, we used the model checking option of DIYABC for our best fitting scenario. Model checking was carried out by simulating 10,000 pseudo observed datasets under each studied model to check the ability of a given scenario to produce data sets similar to the real data set.
Schematic representation of the four demographic scenarios including model parameters of Taxus contorta in Pakistan tested by the approximate Bayesian computation (ABC) approach. For the demographic parameters, see Table S1. Times and effective population sizes are not to scale
Results
Chloroplast haplotype diversity and differentiation
When the cpDNA variation was examined over all populations, we identified a total of six haplotypes (H1 to H6), with single nucleotide differences for each from the most common haplotype H1 (GenBank accession JX680636), which occurred in all populations and was present in 117 individual yew trees. The haplotype frequencies for each population and their geographical distribution are given in Table 1 and Fig. 1. Two populations, SWAT valley (SW) and Hazara valley (HZ), showed the highest number of haplotypes (four in each population). The populations Tirah (TH), Palas valley (PV) and Chitral (CH) each possessed two haplotypes, one of them being the dominant haplotype H1, while the peripheral populations Neelam valley (KK) and Kurram valley (KV) were monomorphic and fixed for haplotype H1. Haplotypes H3 (KF765507), H4 (KF765508) and H5 (JX680642) were all shared haplotypes. Two private haplotypes (H2– JX680641 and H6– KF765509) were present in population SW and HZ respectively. The haplotype (H d) and nucleotide (π) diversities ranged from 0.000 to 0.602 and 0.000 to 0.099 respectively. The haplotype diversity was highest in population SW, followed by HZ, with no diversity found in the two marginal populations KK and KV. The total intraspecific diversity (H T = 0.201) across the populations was slightly higher than the average within-population diversity (H S = 0.182).
The phylogenetic relationships among the haplotypes using a median-joining method formed a ‘star-like’ network (Fig. 1). The most common and widespread haplotype H1 was located in the centre of the network, which indicates that it likely represents the ancestral haplotype for this species in this geographic region. The permutation test revealed a higher value for G ST (0.096) than N ST (0.043). However, the difference was not significant in the test, indicating an absence of phylogenetic structure among the populations. The SAMOVA gave one or more single population groups without reflecting geographic pattern of distribution (data not shown). A significant genetic differentiation was observed among all populations (F ST = 0. 04, P < 0.01), with most chloroplast diversity occurring within populations (95.28 % of the total variance) compared to only 4.72 % partitioned among populations.
Microsatellite diversity and differentiation
We did not find scoring errors, stuttering or large allele dropouts in the microsatellite dataset. No linkage disequilibrium was detected between any pair of loci in any of the populations. All populations deviated significantly from Hardy-Weinberg (HW) equilibrium. A total of 74 alleles, of which 14 were private, were found for the ten loci among the seven populations (Table S2, online resource). An estimate of null allele frequency for each locus in all seven populations is given in Table S2 (online resource). Allelic richness, based on 12 minimum samples, ranged from 3.48 (SW) to 4.83 (CH). A strong variability among loci was observed across the populations with a mean observed heterozygosity (H O) ranging from 0.016 in locus TG34 to 0.791 in locus TA116. The observed and expected heterozygosity per population varied from 0.304 to 0.435 and 0.486 to 0.584, respectively. The expected heterozygosity was slightly higher when adjusted for null alleles (H E = 0.541 ± 0.034, H E null = 0.586 ± 0.041, P = 0.041) (Table S2, online resource). Across all loci, the mean value of total genetic diversity (H T = 0.601) was higher than that of the mean genetic diversity within populations (H S = 0.546). The distribution of genetic diversity and allelic richness among populations did not reveal any geographic pattern (Table 1 and Fig. 1). The inbreeding coefficient F IS ranged from 0.219 to 0.418 and was significant (P < 0.05) for all populations.
All pairwise comparisons of genetic differentiations (F ST) between populations were significant and ranged from 0.038 to 0.226 (Table 2). A higher population differentiation was observed particularly between populations SW and HZ, KV and KK. Although there were some differences between the unadjusted pairwise F ST and those adjusted using the ENM algorithm, which corrects for null alleles, the overall data indicated no consistent bias among the data sets (Table 2). The estimated F ST values for T. contorta at each locus, both with and without adjustment for null alleles, were similar (F ST = 0.0985, F ST(Null) = 0.0954). However, the standardized genetic differentiation, G' ST, was higher than the F ST across all loci (Table S2, online resource).
The result obtained from the AMOVA indicated that 89.33 % of the genetic variation could be attributed to differences within populations, the remaining 10.67 % explained variation among populations (F ST = 0.107, P < 0.01). The NJ tree based on Nei’s D A illustrated clear differences among populations (Fig. 3). However, none of the proximal populations, except PV and CH, showed genetic similarities. The Bayesian analysis with STRUCTURE did not suggest clear genetic structures for the seven populations (Figs. 1 and 4). Although the optimum number of clusters was suggested to be K = 2, the admixture proportions of each cluster differed among the populations showing no geographic pattern. Population TH and SW showed high proportions of one (green) cluster group and grouped together in the NJ tree with maximum bootstrap support (100 %). The two populations PV and CH had a similar proportion of members of the two cluster classes. They were geographically relatively close and clustered together in the NJ tree (66 %). Similarly, the two geographically most distant populations KV and KK with more than 60 % red cluster members, grouped together in the NJ tree, albeit with relatively low bootstrap support (56 %). Population from Hazara valley (HZ) that had highest proportion of red cluster showed clear difference with all the other populations both in STRUCTURE and NJ tree (Figs. 1, 3 and 4). For both, null alleles adjusted and unadjusted genetic differentiation (F ST), no significant correlation was observed between the genetic distance and geographic distance (r 2 = 0.028, P = 0.289 and r 2 = 0.021, P = 0.325, respectively).
Bayesian inferences of the number of clusters (K) of 129 individuals of Taxus contorta, inferred by STRUCTURE analyses of ten nuclear microsatellites. The graph to the right shows the value of ΔK that suggests the appropriate number of clusters for the STRUCTURE analysis of Taxus contorta in Pakistan. The graph to the left shows the cluster assignments of the 129 individuals at the population level using K = 2
Population demographic history
In the demographic model of mismatch distribution analysis based on the cpDNA trnL-F sequence data, the null model of rapid population expansion was not rejected for T. contorta since the graph showed a distinct unimodal distribution (Fig. 5). Likewise, the mismatch distribution analysis was not significant for SSD (demographic expansion 0.00014, P = 0.457 and spatial expansion 0.00003, P = 0.544) and HRI (demographic expansion 0.464, P = 0.669 and spatial expansion 0.464, P = 0.608), indicating that a sudden expansion event had likely happened in the past. The neutrality test revealed significant negative values in Fu’s F S test (−5.812, P = 0.001) and Tajima’s D test (−1.771, P = 0.006).
The DIYABC analysis on microsatellite data also supported a scenario of a past population expansion (scenario 1, Fig. 2). The posterior probability of this scenario was with 53 % (52.61–53.13 %) significantly higher than those of scenario 2 with 16.51 % (15.94–17.27 %), scenario 3 with 30.36 % (29.43–30.79 %) and scenario 4 with 0.027 % (0.014–0.056 %). The results point to a scenario that assumed a demographic expansion of Taxus populations in Pakistan from small ancestral populations (Na) to a presently large effective population size (N1) (scenario 1, Fig. 2). Despite the markedly higher posterior probability obtained for scenario 1 compared to the other three scenarios, it was difficult to distinguish between the four scenarios based on an analysis of confidence performed to evaluate the choice of the best scenario. However, the average type I or alpha error rate (i.e. probability that data sets simulated under the true scenario were assigned to other scenarios) of scenario 1 was only 11.4 % that gave the highest, 88.6 % statistical power for our model choice procedure (Table S3, online resource). Model check values and marginal posterior probability densities for the demographic parameters of the best-supported demographic scenario are provided in supplementary material (Table S4 and S5, online resources). The parameter estimates provided reasonable values, indicating that a historic population with an average effective size of N e = 291 individuals increased extensively to a present average population size of 2,830 individuals. This expansion is estimated to have occurred during, on average, 1,500 generations ago (i.e. about 37.5 kya) (Table S5, online resource).
Most interestingly, the estimates of population expansion time obtained from the DIYABC analysis on microsatellite data fits within the range suggested by the tau (τ) values and corresponding population expansion time estimates as 140.06–6.71 kya for the studied populations of T. contorta in Pakistan based on cpDNA sequence data.
Discussion
Chloroplast and nuclear microsatellite diversity in T. contorta
This study reports the first detailed analysis of comprehensive chloroplast DNA and nuclear microsatellite data for T. contorta from Pakistan. Shah et al. (2008a), using RAPDs on the same populations (as T. fuana Nan Li & R.R. Mill) came to somewhat different conclusions (low genetic diversity within populations, low within and high among population variation and no information regarding the demographic history of the species), which will likely have been due to the differences in marker technique applied. Particularly, the sample size can have a significant effect on the estimation of expected heterozygosity for RAPD and nSSR markers (Nybom 2004). Usually, two to ten times more individuals per RAPD loci are needed to be sampled to get comparable results based on the minimum number of loci required for co-dominant markers (Lynch and Milligan 1994).
In the present study, the total haplotype diversity in T. contorta (H T = 0.201) was found to be lower than the average value for other gymnosperms (Petit et al. 2005) and even compared to its congeners T. wallichiana (H T = 0.824) (Liu et al. 2013a) and T. mairei (H T = 0.550) (Zhang 2010) distributed in the eastern Himalaya and South China respectively. However, T. contorta populations from Pakistan harboured a slightly higher total haplotype diversity compared with the populations that are distributed in the eastern extent of its distribution range, namely in Nepal and southwest Xizang of China (H T = 0.106) (Poudel et al. 2014). Interestingly, the nuclear microsatellite diversity in T. contorta (H E = 0.541) was, although lower than the mean value for outcrossing gymnosperms (H E = 0.650) (Nybom 2004) and a European species T axus baccata (H E = 0.655) (González-Martínez et al. 2010), still higher than in T. wallichiana (H E = 0.478) (Liu et al. 2013a) and T. mairei (H E = 0.538) (Zhang and Zhou 2013), the two yew species distributed along the Himalayas. Within T. contorta, populations from Nepal and southwest Xizang of China revealed slightly lower diversity estimates (H E = 0.539) compared to those in Pakistan (Poudel et al. 2014). There could be several factors for the observed discrepancies between species and the chloroplast and nuclear microsatellites diversity estimates within species, such as different evolutionary rates and mode of inheritance between the markers (Sunnucks 2000; Hamilton et al. 2003), different evolutionary histories of the species (Schaal et al. 1998), and/or species-specific impacts of human induced habitat degradation and fragmentation.
Genetic structure and population differentiation in T. contorta
In this study, though most of the total genetic variation was partitioned within populations (95.28 and 89.33 % for cpDNA and nSSRs, respectively), a significant genetic differentiation among the populations of T. contorta in Pakistan was estimated for both cpDNA and nuclear microsatellite data. The genetic variation was not structured geographically, and even nearest populations especially from the SWAT valley (SW) and Hazara valley (HZ) were highly differentiated (F ST = 0.226) (Table 2). However, the overall population differentiation observed in the analyzed T. contorta populations was still lower than the average F ST observed in gymnosperms (Nybom 2004; Petit et al. 2005) and congeneric species, e.g. T. baccata (F ST = 0.155, González-Martínez et al. 2010), T. wallichiana (F ST = 0.110, Liu et al. 2013a) and T. mairei (F ST = 0.159, Zhang and Zhou 2013). Nevertheless, the value for differentiation obtained for the seven populations in Pakistan were much higher when compared with the nine populations of T. contorta from its eastern range (overall F ST = 0.073 and pairwise F ST = 0.037–0.132) in Nepal and SW Xizang of China (Poudel et al. 2014). In general, endemic species and species with scattered isolated populations often show low within and high among population variation (Nybom 2004), due to a limited gene flow between the isolated populations and the higher impact of genetic drift. The significant population differentiation, observed heterozygote deficiency, and high level of inbreeding in T. contorta indicated a restricted gene flow among the populations with stronger genetic drift in the small isolated populations (Shah et al. 2008a; Dubreuil et al. 2010). High levels of inbreeding, as estimated in T. contorta here, were also observed in its congeners T. baccata (nSSRs: Dubreuil et al. 2010), T. brevifolia (allozymes: El-Kassaby and Yanchuk 1994), T. cuspidata (allozymes: Chung et al. 1999), T. mairei (nSSR: Zhang and Zhou 2013) and T. wallichiana (nSSR: Poudel 2012). The restricted gene flow may be attributable to the complex mountainous areas and human-induced habitat fragmentation (Poudel et al. 2012), limited wind-mediated pollen flow (Wheeler et al. 1995) and limited seed dispersal range in Taxus (Gao et al. 2007).
Population demographic history of T. contorta in Pakistan
The demographic history of the T. contorta populations in Pakistan was addressed for the first time in this study and indicated that a rapid population expansion in this species has likely occurred in the past. A demographic expansion has possibly taken place in the late Pleistocene, after the penultimate ice age (marine oxygen isotope stage (MIS) 6), and before the local glaciation MIS 3, both of which were the most extensive among the four Pleistocene glaciations in northwest Pakistan (Su 2006; Kamp and Owen 2011). Our inference from the ABC analysis, the star-like network, the mismatch distribution analysis with unimodal frequency distribution and the highly significant negative values of Fu’s F S and Tajima’s D, all indicated a sudden demographic expansion of populations of T. contorta happened in past in Pakistan. The absence of significant relationships between genetic distance and geographic distance is consistent with such a scenario. A similar demographic history was found in the HM lineage of T. wallichiana that is confined to the Hengduan Mountain region (Liu et al. 2013a). Studies on glacial chronologies have shown that glaciation in north Pakistan was more extensive during MIS 3, and that there were multiple glacial advances during the last glacial maxima (LGM) and in the late Holocene (Kamp and Owen 2011). For example, the Chitral valley was covered by an over 270-km-long ice sheet that advanced as low as 1,300 m a.s.l. 30–60 kya at the Dros glacial stage (MIS 3). The ice sheet during the following glaciation in the early Holocene (MIS 2), about 24 kya, was less extensive, with around 250 km length, terminating at 1,670 m a.s.l. in the south of the Chitral valley (Kamp and Owen 2011). The glacial geology of the Nanga Parbat, the Deosai plateau and the Indus valley, that lies at the northeast border of Pakistan was extensively glaciated 56 kya and 35 kya (during MIS 3) and during multiple glacial advances in the late Holocene and the Neoglacial (Phillips et al. 2000; Richards et al. 2000; Owen et al. 2008; Kamp and Owen 2011). Furthermore, studies based on different proxy data (marine cores, lake sediments and pollen and peat deposits) also revealed that the climate during the Pleistocene and Holocene in the western Himalaya was likely highly variable, with several consecutive cold and dry events (Ansari and Vink 2007; Gupta 2008). The available palynological evidence from northwest India suggests the presence of Taxus there as early as 30 kya (Gupta 2008; Kotlia et al. 2000). In the context of these events, we can infer that populations of T. contorta in Pakistan most possibly underwent a rapid demographic expansion during the Pleistocene, from ancient small populations, which survived in valleys, such as Swat and Hazara.
Paleoecological data studied by Miehe et al. (2009) demonstrated that an earlier human impact in north Pakistan (Shukan) has caused the decline of conifer forest before 5.7 kya. Such an extensive landscape disturbance coupled with climate fluctuations during the recent evolutionary history of T. contorta might have caused the observed low diversity and high population differentiation in the extant populations of yews in Pakistan.
Conservation of T. contorta
Based on satellite data, Pandit et al. (2007) projected the loss of >50 % of the remaining forest in the western Himalaya by the year 2100. Forest encroachment to expand farmland and overharvesting of forest resources for commercial or domestic purposes have severe negative impacts on the demography of yew populations (Shah et al. 2008a; Poudel et al. 2013). Habitat fragmentation and forest destruction remove natural stands, causing demographic fluctuations that are ultimately manifested genetically in low diversity levels, high population differentiation and high levels of inbreeding. Consanguineous mating in small, fragmented populations may cause loss of genetic variation, which strongly influences the fitness of threatened species to cope with changing environments (Bellard et al. 2012; Kremer et al. 2012). Therefore, maintaining an effective population size and restoring genetic variation among the remnant populations of T. contorta should be a priority in conservation plans (Frankham 2005). Our data on chloroplast and microsatellite variation has increased our understanding of the possible major factors that have influenced the genetic make-up of the remnant populations of T. contorta in Pakistan. The findings have supplemented a previous genetic study (Shah et al. 2008a) and provided information beyond this initial study to design conservation priorities for populations that need specific attention.
Because the western Himalayan climate fluctuated frequently in the past (Owen et al. 2008), it is possible that the ancestral population was genetically flexible to adapt accordingly, and that current populations may contain genetic remnants of this adaptability (Sork et al. 2013). In situ conservation would thus be a priority to safeguard the genetic variation present in T. contorta. Protection and a proper management of the natural forests through controlled animal grazing and minimising the disturbance by local people is further necessary to allow the regeneration of wild yew populations. The community forest approach is widely practised in the neighbouring Himalayan country Nepal and encompasses a set of policies that empower local communities to manage forests nearby their villages following both customary and statutory laws. Given the highly isolated populations of yew, a community forest approach is equally applicable in Pakistan, where, if managed properly, the remaining patches of forest isolated by farming terraces may work as small forest islands acting as stepping stones for gene flow from one community forest to another. In in situ conservation of threatened species, a detailed inventory of different demographic parameters (age, size class distribution, male/female ratio) and quantification of anthropogenic and environmental threats for each population are the most basic requirements (Poudel et al. 2014). Populations that contain a high number of private haplotypes and high microsatellite diversity, such as CH, HZ, PV and SW in Pakistan, should be considered as distinct evolutionary significant units (ESUs). These should be given a higher in situ conservation priority because there is a greater likelihood that the genomes of these plants have a wider genetic base and may contain traits that may become significant in future adaptive scenarios, e.g. involving climate shifts. They can also be used as source material for ex situ seed banks, botanic gardens, tissue culture units, plant nurseries and to conduct research for yield improvements (WHO 1993).
Local people in Pakistan use Taxus for fodder, fuel-wood and furniture making (Shah et al. 2008a; Poudel et al. 2013), but surprisingly not for the production of paclitaxel (‘Taxol’), a highly effective anticancer compound. Commercial collection of yews to extract paclitaxel, which is one of the major reasons behind the extirpation of several natural populations of yews along the Himalayas, has not been reported yet from Pakistan. Pakistan could learn lessons on the negative consequences of unscientific collections from neighbouring countries and design schemes to sustainably use this species with its tremendous economic potential. Planting Taxus for commercial use can provide a sustainable income to rural subsistence farmers (Nüsser 2001) of rural villages in north Pakistan. The establishment of micro-enterprises/-nurseries in villages for the provision of supplying propagation materials and technical support for the cultivation, value addition and a marketing consultation service could encourage rural people to grow Taxus on private farms or marginal lands (Hamilton 2004). Taxus can relatively easily be propagated en masse from cuttings (Nandi et al. 1996; Kaul 2008; Muñoz-Gutiérrez et al. 2009). Plant materials used for cultivation are generally selected based on the inherent superiority of an individual for the desirable characters and propagated clonally. Suitable cultivars can give high yield and exhibit pest resistance and adaptability to specific localities. However, cultivars are genetically homogeneous, and if the aim is to restock wild populations of yews, emphasis should be given to use a sufficient number of different propagation material (seed, seedlings and/or stem cuttings), collected from different populations following both mix and match approaches (Lesica and Allendorf 1999). To avoid inbreeding and further genetic depauperation in the future, the selection of planting material must proceed with caution.
In conclusion, our analyses of chloroplast DNA and nuclear microsatellite marker datasets have provided detailed information on the population genetics and demographic history of T. contorta in Pakistan, beyond the previous study of Shah et al. (2008a). We observed low chloroplast and moderate nuclear microsatellite diversity and a high level of inbreeding in T. contorta. This pattern can mainly be attributed to climate changes in the late Quaternary and, more recently, due to an unprecedented level of human-induced habitat destruction and fragmentation in the western Himalayas. Declining population sizes and an increased level of consanguineous mating in T. contorta is likely to have elevated the extinction risks for the remnant populations, especially in the current rapid climate change scenario. From this perspective, conservation relevant insights into the genetic composition of the remnant populations need to be appropriately interpreted and converted to clearly defined in situ and ex situ conservation approaches.
References
Ali J, Benjaminsen TA, Hammad AA, Dick ØB (2005) The road to deforestation: an assessment of forest loss and its causes in Basho valley, northern Pakistan. Glob Environ Chang 15:370–380
Ansari MH, Vink A (2007) Vegetation history and palaeoclimate of the past 30 kyr in Pakistan as inferred from the palynology of continental margin sediments off the Indus Delta. Rev Palaeobot Palyno 145:201–216
Bai WN, Liao WJ, Zhang DY (2010) Nuclear and chloroplast DNA phylogeography reveal two refuge areas with asymmetrical gene flow in a temperate walnut tree from East Asia. New Phytol 188:892–901
Bandelt H, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Barbier S, Gosselin F, Balandier P (2008) Influence of tree species on understory vegetation diversity and mechanisms involved—a critical review for temperate and boreal forests. Forest Ecol Manag 254:1–15
Bellard C, Bertelsmeier C, Leadley P, Thuiller W, Courchamp F (2012) Impacts of climate change on the future of biodiversity. Ecol Lett 15:365–377
Benítez-Malvido J, Martínez-Ramos M (2003) Impact of forest fragmentation on understory plant species richness in Amazonia. Conserv Biol 17:389–400
Chapuis MP, Estoup A (2007) Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol 24:621–631
Chung MG, Oh GS, Chung JM (1999) Allozyme variation in Korean populations of Taxus cuspidata (Taxaceae). Scand J Forest Res 14:103–110
Collins D, Mill RR, Möller M (2003) Species separation of Taxus baccata, T. canadensis, and T. cuspidata (Taxaceae) and origins of their reputed hybrids inferred from RAPD and cpDNA data. Am J Bot 90:175–182
Cornuet JM, Ravigne V, Estoup A (2010) Inference on population history and model checking using DNA sequence and microsatellite data with the software DIYABC (v1.0). BMC Bioinforma 11:401
Cornuet JM, Pudlo P, Veyssier J, Dehne-Garcia A, Gautier M, Leblois R, Marin JM, Estoup A (2014) DIYABC v2.0: a software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics, doi:10.1093/bioinformatics/btt763.
Crawford NG (2010) SMOGD: software for the measurement of genetic diversity. Mol Ecol Resour 10:556–557
Dieringer D, Schlötterer C (2003) MICROSATELLITE ANALYSER (MSA): a platform independent analysis tool for large microsatellite data sets. Mol Ecol Notes 3:167–169
Dubreuil M, Riba M, González-Martínez SC, Vendramin GG, Sebastiani F, Mayol M (2010) Genetic effects of chronic habitat fragmentation revisited: strong genetic structure in a temperate tree, Taxus baccata (Taxaceae), with great dispersal capability. Am J Bot 97:303–310
Dupanloup I, Schneider S, Excoffier L (2002) A simulated annealing approach to define the genetic structure of populations. Mol Ecol 11:2571–2581
El-Kassaby YA, Yanchuk AD (1994) Genetic diversity, differentiation, and inbreeding in pacific yew from British Columbia. Heredity 85:112–117
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Frankham R (2005) Genetics and extinction. Biol Conserv 126:131–140
Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925
Gao LM, Möller M, Zhang XM, Hollingsworth ML, Liu J, Mill RR, Gibby M, Li DZ (2007) High variation and strong phylogeographic pattern among cpDNA haplotypes in Taxus wallichiana (Taxaceae) in China and North Vietnam. Mol Ecol 16:4684–4698
González-Martínez SC, Dubreuil M, Riba M, Vendramin GG, Sebastiani F, Mayol M (2010) Spatial genetic structure of Taxus baccata L. in the western Mediterranean basin: past and present limits to gene movement over a broad geographic scale. Mol Phylogenet and Evol 55:805–815
Goudet J (2002) FSTAT: a program to estimate and test gene diversities and fixation indices, Version 2.9.3.2. Available from http://www2.unil.ch/popgen/softwares/fstat.htm.
Gupta A (2008) Late Quaternary vegetation and climate from temperate zone of the Kumaun Himalaya, India (with remarks on neotectonic disturbance). Acta Palaeontol Sinica 48:325–333
Hall T (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hamilton AC (2004) Medicinal plants, conservation and livelihoods. Biodivers Conserv 13:1477–1517
Hamilton MB, Braverman JM, Soria-Hernanz DF (2003) Patterns and relative rates of nucleotide and insertion/deletion evolution at six chloroplast intergenic regions in new world species of the Lecythidaceae. Mol Biol Evol 20:1710–1721
Haq F (2012) The critically endangered flora and fauna of district Battagram Pakistan. Adv Life Sci 2:118–123
Hedrick PW, Goodnight C (2005) A standardized genetic differentiation measure. Evolution 59:1633–1638
Hoffmann AA, Sgro CM (2011) Climate change and evolutionary adaptation. Nature 470:479–485
Hughes AR, Inouye BD, Johnson MTJ, Underwood N, Vellend M (2008) Ecological consequences of genetic diversity. Ecol Lett 11:609–623
Jadwiszczak K, Banaszek A, Jabłońska E, Sozinov O (2012) Chloroplast DNA variation of Betula humilis Schrk. in Poland and Belarus. Tree Genet Genomes 8:1017–1030
Jost L (2008) GST and its relatives do not measure differentiation. Mol Ecol 17:4015–4026
Jump AS, Marchant R, Peñuelas J (2009) Environmental change and the option value of genetic diversity. Trends Plant Sci 14:51–58
Jump AS, Peñuelas J (2006) Genetic effects of chronic habitat fragmentation in a wind-pollinated tree. Proc Natl Acad Sci U S A 103:8096–8100
Kamp U, Owen LA (2011) Chapter 66:Late Quaternary glaciation of northern Pakistan. In: Ehlers J, Gibbard PL, Hughes PD (eds) Quaternary glaciations-extent and chronology:a closer look. Elsevier, Amsterdam, pp 909–927
Kaul K (2008) Variation in rooting behaviour of stem cuttings in relation to their origin in Taxus wallichiana Zucc. New For 36:217–224
Kotlia BS, Sharma C, Bhalla MS, Rajagopalan G, Subrahmanyam K, Bhattacharyya A, Valdiya KS (2000) Palaeoclimatic conditions in the late Pleistocene Wadda Lake, eastern Kumaun Himalaya (India). Palaeogeogr Palaeocl 162:105–118
Kremer A, Ronce O, Robledo-Arnuncio JJ, Guillaume F, Bohrer G, Nathan R, Bridle JR, Gomulkiewicz R, Klein EK, Ritland K, Kuparinen A, Gerber S, Schueler S (2012) Long-distance gene flow and adaptation of forest trees to rapid climate change. Ecol Lett 15:378–392
Lesica P, Allendorf FW (1999) Ecological genetics and the restoration of plant communities: mix or match? Restor Ecol 7:42–50
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Liu J, Gao LM (2011) Comparative analysis of three different methods of total DNA extraction used in Taxus. Guihaia 31:244–249
Liu J, Gao LM, Li DZ, Zhang DQ, Möller M (2011) Cross-species amplification and development of new microsatellite loci for Taxus wallichiana (Taxaceae). Am J Bot 98:e70–e73
Liu J, Möller M, Provan J, Gao LM, Poudel RC, Li DZ (2013a) Geological and ecological factors drive cryptic speciation of yews in a biodiversity hotspot. New Phytol 199:1093–1108
Liu W, Kang M, Tian H, Huang H (2013b) A range wide geographic pattern of genetic diversity and population structure of Castanea mollissima populations inferred from nuclear and chloroplast microsatellites. Tree Genet Genomes 9:975–987
Lynch M, Milligan BG (1994) Analysis of population genetic structure with RAPD markers. Mol Ecol 3:91–99
Meng HH, Zhang ML (2013) Diversification of plant species in arid Northwest China: species-level phylogeographical history of Lagochilus Bunge ex Bentham (Lamiaceae). Mol Phylogenet Evol 68:398–409
Miehe G, Miehe S, Schlütz F (2009) Early human impact in the forest ecotone of southern High Asia (Hindu Kush, Himalaya). Quaternary Res 71:255–265
Mohapatra K, Sehgal R, Sharma R, Mohapatra T (2009) Genetic analysis and conservation of endangered medicinal tree species Taxus wallichiana in the Himalayan region. New For 37:109–121
Möller M, Gao LM, Mill RR, Li DZ, Hollingsworth ML, Gibby M (2007) Morphometric analysis of the Taxus wallichiana complex (Taxaceae) based on herbarium material. Bot J Linn Soc 155:307–335
Muñoz-Gutiérrez L, Vargas-Hernández JJ, López-Upton J, Soto- Hernández M (2009) Effect of cutting age and substrate temperature on rooting of Taxus globosa. New For 38:187–196
Nandi SK, Palni LMS, Rikhari HC (1996) Chemical induction of adventitious root formation in Taxus baccata cuttings. Plant Growth Regul 19:117–122
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Nei M, Tajima F, Tateno Y (1983) Accuracy of estimated phylogenetic trees from molecular data. Mol Evol 19:153–170
Nüsser M (2001) Understanding cultural landscape transformation: a re-photographic survey in Chitral, eastern Hindukush, Pakistan. Landsc Urban Plan 57:241–255
Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol 13:1143–1155
Nybom H, Bartish IV (2000) Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Perspect Plant Ecol 3:93–114
Owen LA, Caffee MW, Finkel RC, Seong YB (2008) Quaternary glaciation of the Himalayan–Tibetan orogen. J Quaternary Sci 23:513–531
Pandit M, Sodhi N, Koh L, Bhaskar A, Brook B (2007) Unreported yet massive deforestation driving loss of endemic biodiversity in Indian Himalaya. Biodivers Conserv 16:153–163
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Petit RJ, Duminil J, Fineschi S, Hampe A, Salvini D, Vendramin GG (2005) Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol Ecol 14:689–701
Phillips WM, Sloan VF, Shroder JF, Sharma P, Clarke ML, Rendell HM (2000) Asynchronous glaciation at Nanga Parbat, northwestern Himalaya Mountains, Pakistan. Geology 28:431–434
Pons O, Petit RJ (1996) Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144:1237–1245
Poudel RC (2012) Species delimitation and conservation genetics of Taxus (Taxaceae) along the Hindu Kush-Himalaya region. PhD dissertation. Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
Poudel RC, Möller M, Gao LM, Ahrends A, Baral SR, Liu J, Thomas P, Li DZ (2012) Using morphological, molecular and climatic data to delimitate yews along the Hindu Kush-Himalaya and adjacent regions. PLoS ONE 7:e46873
Poudel RC, Gao LM, Möller M, Baral SR, Uprety Y, Liu J, Li DZ (2013) Yews (Taxus) along the Hindu Kush-Himalayan region: exploring the ethnopharmacological relevance among communities of Mongol and Caucasian origins. J Ethnopharmacol 147:190–203
Poudel RC, Möller M, Liu J, Gao LM, Baral SR, Li DZ (2014) Low genetic diversity and high inbreeding of the endangered yews in Central Himalaya: implications for conservation of their highly fragmented populations. Divers Distrib (in press)
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Qi XS, Chen C, Comes HP, Sakaguchi S, Liu YH, Tanaka N, Sakio H, Qiu YX (2012) Molecular data and ecological niche modelling reveal a highly dynamic evolutionary history of the East Asian Tertiary relict Cercidiphyllum (Cercidiphyllaceae). New Phytol 196:617–630
Richards BW, Owen LA, Rhodes EJ (2000) Timing of late Quaternary glaciations in the Himalayas of northern Pakistan. J Quaternary Sci 15:283–297
Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9:552–569
Ruan Y, Huang BH, Lai SJ, Wan YT, Li JQ, Huang S, Liao PC (2013) Population genetic structure, local adaptation, and conservation genetics of Kandelia obovata. Tree Genet Genomes 9:913–925
Schaal BA, Hayworth DA, Olsen KM, Rauscher JT, Smith WA (1998) Phylogeographic studies in plants: problems and prospects. Mol Ecol 7:465–474
Shah A, Li DZ, Gao LM, Li HT, Möller M (2008a) Genetic diversity within and among populations of the endangered species Taxus fuana (Taxaceae) from Pakistan and implications for its conservation. Biochem Syst Ecol 36:183–193
Shah A, Li DZ, Möller M, Gao LM, Hollingsworth M, Gibby M (2008b) Delimitation of Taxus fuana Nan Li RR Mill (Taxaceae) based on morphological and molecular data. Taxon 57:211–222
Slatkin M, Hudson R (1991) Pairwise comparisons of mitochondrial-DNA sequences in stable and exponentially growing populations. Genetics 129:555–562
Sork VL, Aitken SN, Dyer RJ, Eckert AJ, Legendre P, Neale DB (2013) Putting the landscape into the genomics of trees: approaches for understanding local adaptation and population responses to changing climate. Tree Genet Genomes 9:901–911
Spielman D, Brook BW, Frankham R (2004) Most species are not driven to extinction before genetic factors impact them. Proc Natl Acad Sci U S A 101:15261–15264
Su Z (2006) Quaternary glaciations in the Karakorum mountains. In: Shi Y, Cui Z, Su Z (eds) The Quaternary glaciations and environmental variations in China. Hebei Science and Technology Publishing House, China
Sunnucks P (2000) Efficient genetic markers for population biology. Trends Ecol Evol 15:199–203
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Wang W, Jiang Y, Wang D, Jing Y (2006) Biological and ecological characteristics of Taxus yunnanensis. J West China Forest Sci 35:33–39
Weir B, Cockerham C (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Wheeler NC, Jech KS, Masters SA, O'Brien CJ, Stonecypher RW, Timmons DW, Lupkes A (1995) Genetic variation and parameter estimates in Taxus brevifolia (Pacific yew). Can J Forest Res 25:1913–1927
WHO (1993) Guidelines on the conservation of medicinal plants. The world health organization (WHO), The world conservation union (IUCN) and World wide fund for nature (WWF)
Yin A, Harrison TM (2000) Geologic evolution of the Himalayan-Tibetan orogen. Annu Rev Earth Pl Sc 28:211–280
Zhang DQ (2010) Phylogeography of the Taxus wallichina complex around the Sichuan Basin. PhD dissertation, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
Zhang DQ, Zhou N (2013) Genetic diversity and population structure of the endangered conifer Taxus wallichiana var. mairei (Taxaceae) revealed by simple sequence repeat (SSR) markers. Biochem Syst Ecol 49:107–114
Acknowledgments
We are grateful to Jie Liu, Jun-BoYang, Hong-Tao Li, Wei Jiang and Santiago C. González-Martínez for their help. We thank two anonymous reviewers and the Associate Editor for their very constructive comments. This study was supported by the National Key Basic Research Program of China (2014CB954100), Key Research Program of the Chinese Academy of Sciences (KJZD-EW-L07) and the National Natural Science Foundation of China (31370252, 30700042). The Royal Botanic Garden Edinburgh is supported by the Rural and Environment Science and Analytical Services division (RESAS) in the Scottish Government.
Data archiving statement
Chloroplast DNA trnL-F sequences have been submitted to GenBank. Microsatellite data is available from the Dryad Digital Repository: http://doi.org/10.5061/dryad.3gq01.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by S. C. González-Martínez
Supplementary information (online resources)
Below is the link to the electronic supplementary material.
Table S1
(Online resource). Priors for the ABC modelling. (DOCX 21 kb)
Table S2
(Online resource). Locus-wise genetic statistics estimated for the seven populations of Taxus contorta from Pakistan analyzed. (DOC 65 kb)
Table S3
(Online resource). Type I and type II error probabilities for each of the four scenarios estimated under the ABC approach (DOCX 17 kb)
Table S4
(Online resource). Model checking performed under the ABC approach for the four demographic scenarios. (DOCX 17 kb)
Table S5
(Online resource). Marginal posterior distributions for the demographic parameters and estimates of bias and precision of the best supported demographic scenario 1 under the ABC approach. (DOCX 21 kb)
Rights and permissions
About this article
Cite this article
Poudel, R.C., Möller, M., Li, DZ. et al. Genetic diversity, demographical history and conservation aspects of the endangered yew tree Taxus contorta (syn. Taxus fuana) in Pakistan. Tree Genetics & Genomes 10, 653–665 (2014). https://doi.org/10.1007/s11295-014-0711-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-014-0711-7