Abstract
In this work, we assess both the morphological and genetic diversity of 68 important olive cultivars from three Southern Italian regions: Calabria, Campania and Sicily. Twenty-five phenotypic traits were evaluated and 12 simple sequence repeat (SSR) markers were analysed. All SSR primers were polymorphic and reliable. The total number of alleles per locus varied from 5 to 19 with an average number of 13.1 and a mean polymorphic information content (PIC) of 0.81. These results suggested high genetic diversity within these three olive germplasm collections. Morphological traits also showed significant variability amongst cultivars. Two cases of identity were found and ten statistically significant cases of putative parent/sibling were discovered by performing a SSR-based parentage simulation analysis with CERVUS. The Mantel test indicated low but significant correlations between the morphological data and SSR allelic frequency, origin and SSR allelic frequency, and origin and morphology. Structure software allowed inference of relationships between the three olive germplasm collections and allowed us to obtain the most consistent grouping and to identify putative admixed or exchanged cultivars. Cluster and multivariate analysis, based on morphological traits, revealed geographic grouping in agreement with UPGMA dendrogram and structure analysis using SSRs. Sicilian cultivars showed a more homogenous genetic makeup, probably due to geographical isolation, whilst Calabrian and Campanian cultivars seemed to have a less distinct genetic structure, with a greater degree of intermixing. A correlation between the presence of certain SSR alleles and fruit size was also found.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The olive (Olea europaea L. subsp. europaea), which originated in the Eastern Mediterranean area, has been cultivated throughout the Mediterranean basin since ancient times for its fruit and oil. It is believed to have been domesticated as far back as 3500–3700 BC (Zohary and Hopf 1994), and references to the use and trade of olive oil date back to 2000–3000 BC (Baldoni and Belaj 2010). Today, the olive is one of the most widely cultivated fruit crops in the world, and the Mediterranean region, in particular, produces 95 % of the world’s olives. Italy ranks second after Spain (FAO 2004) in terms of production.
The olive (2n = 2x = 46) is an allogamous, longevous and evergreen species, and most of the cultivars are not usually considered to be self-compatible (Mookerjee et al. 2005). The great longevity of this species, the lack of replacement of traditional and well-adapted cultivars combined with its allogamous nature gave rise to very rich germplasm, which has conserved most of its variability intact (Fiorino and Lombardo 2002; Baldoni and Belaj 2010). Olive cultivars are believed to be varieties of unknown origin presumably selected by growers over the centuries from wild genotypes, perhaps those producing the larger fruits, and are generally vegetatively propagated. Hence, the high genotypic diversity of olive varieties could be explained by human selection in response to local environmental and agronomic conditions (Besnard et al. 2001a; Angiolillo et al. 2006; Bracci et al. 2009). It is also likely that crosses between wild local olive genotypes and introduced cultivars have occurred in many areas, thereby leading to new cultivars (Besnard et al. 2001b). Bartolini et al. (1998) stated that at least 1,200 varieties are cultivated, 5,300 names are known, and in Italy alone, 538 cultivars are recognised.
In Italy, each region has its own local cultivars, and many seedling trees grow spontaneously. The larger olive-producing Italian regions are Apulia, Calabria, Campania and Sicily. The use of the species, both as table and oil cultivars, is well-documented in these regions, with many archaeological and written relics dating back to ancient times, attested also by the presence of monumental trees. This species was brought to Sicily with the Phoenicians in the sixth century BC and from this region moved throughout Southern Italy. Cultivation then began to move north with the Romans, who worked intensely on developing grafting technology (Zohary and Hopf 1994; Besnard et al. 2001b; Rugini et al. 2011). In Sicily, Bottari and Spina (1952) described 29 cultivars and landraces which mostly contributed to olive regional production. In the 1980s, work began in the “Dipartimento DEMETRA” in Palermo and since then it has collected and studied, at the morphological level, 37 Sicilian accessions (Caruso et al. 2007), 25 of which have been characterised at the molecular level by using molecular markers (La Mantia et al. 2005; Caruso et al. 2007; Marchese et al. 2008). On the Island, almost 92 % of olive production is used for olive oil extraction and numerous cultivars produce extremely high quality oil (Caruso et al. 2007). Table olive production is also significant (8 %), based on the Sicilian cultivars, ‘Nocellara Etnea’, ‘Nocellara del Belice’, ‘Ogliarola Messinese’ and ‘Moresca’, as they have large-sized fruits of high commercial value. In the Calabria region, olive cultivation was developed over many centuries and Calabrian germplasm is characterised by a remarkable variety of cultivars. The first morphological characterisation of cultivars was reported by Caruso (1883) followed by many authors (Grippo 1923; Zito 1931; Catanea 1934; Pavirani 1959; Chimenti 1963; D’Amore et al. 1977; Parlati et al. 1995, 1999; Mafrica et al. 1996; Motisi et al. 2001; Lombardo et al. 2004). These studies reported that Calabrian olive cultivars, almost exclusively used for oil production, usually have vigorous growth habits, small-sized fruits and show great variability in agronomical behaviour and adaptability to environmental conditions. Generally, in these papers, the number of cultivars described is small; morphological descriptions are very schematic and conducted using different methodologies, making any comparison between genotypes difficult, especially amongst those exhibiting a high degree of similarity. Molecular screening of Calabrian cultivars, together with those of numerous Southern Italian regions, was performed by Carriero et al. (2002) and Muzzalupo et al. (2009) with SSR markers, most of which were recently discarded on the basis of the many drawbacks associated, as reported in literature (reviewed in Baldoni et al. 2009). In Campania, thanks to its orographic and climatic heterogeneity, there is a rich olive genetic heritage. Most of the traditional cultivars are exclusively used for oil production; only few cultivars, such as ‘Ortice’, ‘Ortolana’ and ‘Caiazzana’ are suitable for use both as table olives and oil production. A number of local cultivars are thought to be resistant to cold (ex. ‘Rotondella’) or drought (ex. ‘Carpellese’ and ‘Pisciottana’) conditions, and a few cultivars are putatively considered to be resistant to black mould, peacock spot and knot diseases (ex. ‘Pisciottana’) (Pugliano 2000). Campanian olive oils vary considerably and possess typical and distinctive sensory characteristics (Sacchi et al. 1999; Di Vaio et al. 2013). Pugliano (2000) morphologically described 66 olive accessions, whereas the most representative cultivars were characterised at the molecular level by means of AFLP and/or SSR markers by Rao et al. (2009), Muzzalupo et al. (2009) and Corrado et al. (2009). Considering the historical link and the geographical proximity between these three regions, it is highly likely that the local olive germplasm for each of the regions share genetic relationships.
It is known that olive cultivar differentiation based on morphological descriptions is not particularly reliable, as it can be influenced by environmental conditions and requires skilled staff (Belaj et al. 2001). Furthermore, the presence of both native and foreign olive cultivars with ambiguous naming together with the interchange of plant material over the centuries, make it difficult to ensure cultivar identification and to fully understand the pattern of geographic distribution of olive cultivars (Sarri et al. 2006). Therefore, a wide range of molecular markers has been employed in order to study genetic diversity in olive cultivars from the whole of the Mediterranean area (reviewed in Bracci et al. 2011). Of those molecular markers available, microsatellites or SSRs (simple sequence repeats), developed in olives (Rallo et al. 2000; Sefc et al. 2000; Carriero et al. 2002; Cipriani et al. 2002; De la Rosa et al. 2002; Marrazzo et al. 2002; Sabino Gil et al. 2006) enable parentage analysis and diversity studies (reviewed in Bracci et al. 2011). In this study, the diversity of major and extensively cultivated olive cultivars from three Southern Italy regions was investigated morphologically, measuring reliable and stable traits, and, at the molecular level, by using most of the currently recommended SSR markers. Our work aimed to investigate relationships in order to ascertain, for the first time, the existence of putative parents and/or siblings by parentage simulation analysis, test the putative existence of genetic structure and, finally, to clarify how genetic diversity is partitioned at the micro-scale level for regional germplasm.
Material and methods
Plant material
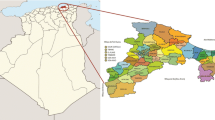
In this investigation, 68 accessions of O. europaea, representing the diversity of olive germplasm from three Southern Italian regions—Calabria, Campania and Sicily—were collected in Spring from the olive cultivar collections of the ‘Azienda Sperimentale Regionale Improsta’—Eboli (Salerno; Campania), the ‘Azienda Carboj E.S.A.’—Menfi (Agrigento; Sicily) or, as in the case of the Calabrian accessions, from local farms (Fig. 1).
Morphological characterisation and clustering analysis of the data
The description of 13 quantitative and 17 qualitative morphological traits was recorded (Table 1) for 2 years (2009–2010), from either the collection fields or farms, following indications provided in literature (Bottari and Spina 1952; Barranco et al. 2000; Bartolini et al. 2005; Caruso et al. 2007). Quantitative traits were transformed as ordinal characters using the classes reported by Caruso et al. (2007) to minimise environmental effects. Five variables (leaf dimensions, flesh to pit ratio, and fruit and pit weight) were excluded from the analysis as they were considered to be more influenced by environmental conditions.
Classical cluster analysis was carried out on the remaining 25 morphological variables (Table 1). The type of metric distance used between objects was “percent” that is the percentage of comparisons between values resulting in disagreements in two profiles. Average linkage was performed using a joining algorithm.
Canonical discriminant analysis (CDA) was also implemented in order to distinguish between groups of cultivars with the same origin. One of the major assumptions of CDA is that each predictor variable is normally distributed. Therefore, only the 15 explanatory variables that were normally distributed were selected (Table 1). A graph was built for the first two canonical functions (Can1 and Can2), illustrating the 80 % confidence ellipses of the mean vectors for each cultivar, in order to visualise multivariate trends for all treatments jointly.
All the statistical analyses based on morphological traits were performed using the Systat statistical program (SYSTAT Software Inc., Chicago, IL).
SSR analysis
Genomic DNA was extracted from fresh leaves according to the method of Doyle and Doyle (1987) with modifications. Its quantity was determined by comparing the fluorescent yield of the samples with λ DNA standards [Gibco BRL, Paisley, Scotland, UK] on 0.8 % (w/v) agarose gel in TAE (40 mM Tris–acetate, 10 mM EDTA, pH 8). The DNA was further quantified by spectrophotometer at 260 and 280 nm and diluted in order to perform SSR analysis.
A total of 12 SSR loci, chosen on the basis of their polymorphism and reproducibility, were used—OeUA-DCA: 03, 04, 07, 09, 14, 16, 17 and 18 (Sefc et al. 2000), UDO43 (Cipriani et al. 2002; Marrazzo et al. 2002), GAPU: 101 and 103 (Carriero et al. 2002), and EMO90 (De la Rosa et al. 2002) (Table 2). Nine of these microsatellites had been proposed previously as being the most suitable for olive fingerprinting studies (Baldoni et al. 2009). Polymerase chain reactions (PCRs) were performed in a final volume of 8 μL, containing 1X PCR buffer (Buffer 10x, Roche Diagnostic Indianapolis, Indianapolis, USA), 0.2 mM of each dNTP (Roche Diagnostic Indianapolis, IN, USA), 0.312 mM of each primer (Roche Diagnostic Indianapolis, IN, USA), 1.5 mM MgCl2 (2.5 nM for the primer pair UDO-043), 0.312 mM of each primer (Roche Diagnostic Indianapolis, IN, USA), 20 ng genomic DNA and 1 U/ Taq Polymerase enzyme (Roche Diagnostic Indianapolis, IN, USA), using 7300 System Thermal Cycler (Applied Biosystems, USA). Reactions were performed using the PCR cycles as described by La Mantia et al. (2005). The forward primer was labelled with one of two fluorescent dyes, namely 6-FAM or HEX (Invitrogen), and sized by capillary electrophoresis through an ABI-PRISM 3130 Genetic Analyser (Applied Biosystems). Data were collected and analysed using the GeneMapper® software v3.7 (Applied Biosystems). Peaks were considered to correspond to alleles; genotypes showing a single peak for a particular locus were considered to be homozygous.
Genetic analysis and paternity inference analysis
The number of alleles per locus (Na), the observed (Ho) and expected (He) heterozygosity, the total number of null alleles (Fnull), the polymorphic information content (PIC) and the deviation from the Hardy–Weinberg equilibrium (HWE), which was inferred by sequential Bonferroni correction, were calculated using CERVUS 3.0 software (Marshall et al. 1998; Kalinowski et al. 2007). Paternity inference analysis was performed using CERVUS 3.0, based on the “likelihood” approach of Thomson (1975; 1976) and Meagher (1986), by selecting settings for both “parents being unknown” and ”incomplete parental sampling”. Internal simulations were run (set to 10,000) to determine the significance of LOD scores (the logarithm of the likelihood ratio). Relaxed and strict confidence levels were set to 95 and to 99 % respectively, and the proportion of loci mistyped was set to 0.005.
Cluster and structure analysis based on microsatellites
Microsatellite alleles were scored as present (1) or absent (0), and the genetic distance between the cultivars was analysed with the NTSYS-pc, version 2.02k (Rohlf 1993). The similarity matrix was obtained using the simple matching coefficient (Sokal and Michener 1958) based on SIMQUAL (Similarity of Qualitative Data), available in the NTSys-Pc software, and was used to construct a UPGMA dendrogram. Starting from allele frequency data (distance method “shared allele”; tree method UPGMA), dendrogram robustness was also assessed by performing bootstrap analysis, running 1,000 iterations with the program PowerMarker (Liu and Muse 2005). Consensus tree was calculated by the “Consense” module of PHYLogeny Inference Package (Phylip, version 3.69). Mantel tests (Mantel 1967) were also performed with PowerMarker (Liu and Muse 2005) to investigate relationships and fit between genetic/morphological, morphological/geographic and genetic/geographic distances of the cultivars, using simple matching dissimilarity matrices. The p values were also calculated.
The software package Structure 2.3.1 (Pritchard et al. 2000; Falush et al. 2003, 2007; Evanno et al. 2005; Hubisz et al. 2009) was employed, using the SSR data, in order to infer relationships between the three collections of olive germplasm, obtain the most consistent grouping of the 68 cultivars studied, and identify putative admixed or exchanged cultivars. The ‘admixture’ model, specifying one to seven populations (K), a burn-in length of 10,000, followed by 90,000 runs at each K, with ten replicates for every K, were used. To select the right number of populations (K), the log likelihood for each K (L(K)) was adopted (Rosenberg et al. 2002). Cultivars with membership probabilities equal to or above 0.80 were considered to belong to the same group.
Results
Cluster and canonical discriminant analysis based on morphological traits
A good correlation among the 2 years of morphological data was observed. The dendrogram, which was constructed using 25 morphological traits (listed in Table 1) and based on the distance “percent” and the ”average linkage method”, showed two main clusters; cluster I and II (Fig. 2). Cluster I included all of the Sicilian cultivars and five Campanian cultivars: ‘Oliva Bianca’, ‘Racioppella’, ‘Cornia’, ‘Ravece’ and ‘Ortice’; cluster II grouped Calabrian and Campanian cultivars. Cluster II contained two quite well-defined sub-clusters, namely sub-cluster A, which included the Campanian cultivars, and sub-cluster B, which incorporated the Calabrian cultivars. Only two Campanian cultivars—‘Ogliarola Campana’ and ‘Pisciottana’—cultivated in areas bordering on the Calabria region, were also included in sub-cluster B (Fig. 2).
Results of canonical discriminant analysis based on the region of origin are shown in the Fig. 3. The coordinates on the two axes, which describe the complete discriminant space, correspond to the three groups and to the individual values of the 68 retained profiles. The first axis (Can 1) separated the profiles of the Sicilian genotypes sharply from those of the other two regions, which showed a complete overlap. The cultivar profiles from the Calabria region, however, were separated from Campanian cultivars by the second axis, in spite of a slight overlap. Despite this, few cultivars were misclassified. The Sicilian cultivars ‘Bottone di Gallo’ and ‘Nocellara Etnea’ were located amongst the Campanian cultivars, and the Calabrian ‘Cassanese’ and the Campanian ‘Ravece’ were classified as Sicilian cultivars. Two Campanian cultivars—‘Racioppella’ and ‘Ogliarola Campana’—were grouped with the Calabrian cultivars, whilst the classification of the Calabrian cultivars ‘Minuta Maierato’ and ‘Ottobratica Rotondella’ was uncertain.
SSR diversity
The 12 SSR primer pairs belonging to the series OeUA-DCA (Sefc et al. 2000), GAPU (Carriero et al. 2002) EMO, (De la Rosa et al. 2002) and UDO (Cipriani et al. 2002; Marrazzo et al. 2002) successfully amplified polymorphic and reproducible alleles in all 68 cultivars, allowing a unique profile to be obtained for most of them (97 %) and, in two cases, for synonymy to be revealed. A total of 157 alleles were found; the number of alleles per locus varied from five for SSR loci EMO90 to 19 for SSR locus DCA04, with an average number of 13.08 (Table 2). For 66 cultivars, excluding from the calculation the two cultivars which could represent cases of synonymy, the mean expected heterozygosity (He) was 0.84 (ranging from 0.73 for EMO-90 to 0.91 for DCA09), the mean observed heterozygosity (Ho) was 0.83 (varying from 0.53 for DCA07 to 0.985 for DCA09 and DCA18), and the mean polymorphic information content (PIC) was 0.81 (ranging from 0.678 for EMO-90 to 0.895 for DCA09) (Table 2). Only SSR locus DCA07 showed a significantly high estimated null allele frequency value +0.2284. In six cases (DCA: 03, 09, 14, 16 and 18 GAPU101) Ho was higher than He, indicating high genetic variability amongst the cultivars analysed (Table 2).
The probability of identity (PI) was estimated as being between 3.3 × 10−2 for the SSR locus DCA09 and 2.13 × 10−1 for EMO-90. The value of the total probability of identity for the 12 SSR analysed, which indicates the probability that two unrelated genotypes chosen at random from all surveyed genotypes have the same profile, was very low 7.23 × 10−14 (Table 2).
Allele frequencies varied from a minimum of 0.007 to a maximum of 0.41 for allele 186 bp at the EMO-90 locus. In many cases, the allelic frequency was very low, particularly for loci having a high number of alleles. ‘Rare alleles’ were also found: allele 216 bp of the locus DCA03 was present only in ‘Minuta Maierato’ and ‘Minuta Zungri’, allele 162 bp of DCA04 was present only in ‘Verdello’, allele 153 bp of the locus DCA07 in ‘Cassanese’, 199 bp DCA09 in ‘Carolea’, allele 150 bp of the DCA14 locus in ‘Cornia’, allele 188 bp of the locus DCA16 in ‘Salella’, 171 bp of DCA17 in ‘Ciciarello’, and allele 166 bp of the locus GAPU103 was found only in ‘Aitana’. In addition, allele 166 of the DCA07 locus was mostly restricted to the Sicilian germplasm, apart from its presence in two Calabrian cultivars (‘Cassanese’ and ‘Oliva d’ogghiu’), and allele 196 bp of the EMO90 locus was present mostly in the Calabrian germplasm, with the exception of its presence in the Sicilian cultivar ‘Santagatese’ and two Campanian cultivars ‘Cornia’ and ‘Pisciottana’ (Supplementary Table 3, available from http://dx.doi.org/10.5061/dryad.3dv00).
The non-exclusion probability between two unrelated individuals (NE-I) and two hypothetical full siblings (NE-SI), calculated with CERVUS, ranged from 0.017 (DCA09) to 0.122 (EMO90), and from 0.303 (DCA09) to 0.420 (EMO90), respectively (Table 2). These values depict the probability that the genotypes at a single locus do not differ between two randomly chosen genotypes. This probability is usually calculated in two ways by CERVUS, either by assuming that two individuals are unrelated, or by assuming that the two individuals are full sibs. Simulation of parentage analysis was used to assess the power of this set of SSR markers to assign parentage and to find putative parentage assignments. Critical LOD values were 6.22 and 3.7 for the single parent analysis, and 16.27 and 12.33 for parent pair (sexes unknown) for strict (99 %) and relaxed (95 %) confidence levels. Noteworthy, single parent simulation showed that the following ten pairs or groups of cultivars can be reciprocally parents or siblings based on LOD scores and confidence level values: ‘Minuta Zungri’/‘Minuta Maierato’ (7.24), ‘Chianota’ (identical to ‘Olivo di Mandanici’)/‘Olivo a Rappu’ (7.72), ‘Caiazzana’/‘Femminella’/‘Racioppella’ (7.88), ‘Cerasuola’/ ‘Nocellara del Belice’ (9.08), ‘Giarraffa’/‘Ravece’ (9.73), ‘Carolea’/‘Tombarella’ (10.7), ‘Minuta’/‘Vaddarica’ (11.2), ‘Cavalieri’/‘Crastu’/‘Ogliarola Messinese’ (11.2). ‘Tonda Campana’/‘Ottobratica Rotondella’/‘Rotondella’ (15) and ‘Ghiastrina’/‘Ottobratica Perciasacchi V.J.’/‘Ottobratica Perciasacchi V.T.’/‘Ottobratica std’ (17).
Cluster analysis (Fig. 4) showed the grouping of cultivars in two main clusters, one of which (cluster I) containing mainly Calabrian and Campanian cultivars, the other (cluster II) containing mostly Sicilian cultivars, confirming the results of CDA (Fig. 3). Only five Sicilian cultivars, ‘Minuta’, ‘Nasitana’, ‘Olivo di Mandanici’, ‘Santagatese’ and ‘Vaddarica’, grown in the north-eastern tip of the island, showed a closer relationship to Calabrian and Campanian cultivars, sharing a level of similarity with them of approximately 0.8. Five Calabrian cultivars, ‘Carolea’, ‘Cassanese’, ‘Oliva d’ogghiu’, ‘Imperiale’ and Tombarella’, and one Campanian cultivar, ‘Ortice’, were located inside a sub-group of cluster II, showing strict relationships with the Sicilian cultivars. Two cultivars, ‘Miseo’ and ‘Salella’, clustered together and appeared as the most isolated of cluster I.
Two cases of identity were found; ‘Olivo di Mandanici’ (Sicily)/’Chianota’ (Calabria) and the Campanian ‘Biancolilla Campana’/’Carpellese’ were undistinguishable at the molecular level with the markers used. ‘Biancolilla Campana’/’Carpellese’, widespread in Southern Campania, were grouped with the Calabrian cultivar ‘Ottobratica V.J.’ (common on the South Ionian sea side of the Calabrian region) with a coefficient of genetic similarity equal to approximately 0.99. The cultivar ‘Ottobratica V.J.’ (Calabria) differed from ‘Biancolilla Campana’/‘Carpellese’ by only one allele at the UDO43 locus.
The Calabrian ‘Ottobratica Rotondella’ and the two Campanian cultivars ‘Tonda Campana’ and ‘Rotondella’ grouped together, showing a coefficient of genetic similarity of approximately 0.982. ‘Tonda Campana’ and ‘Ottobratica Rotondella’ differed by only one allele at the DCA7 locus (149 and 147 bp, respectively) (Supplementary Table 3, available from http://dx.doi.org/10.5061/dryad.3dv00). For this locus, ‘Rotondella’ was found to be homozygous for allele 128 bp and differed from the other two cultivars regarding SSR alleles at locus UDO43. The genotypes ‘Ghiastrina’, ‘Ottobratica Perciasacchi V.J.’, ‘Ottobratica Perciasacchi V.T.’ and ‘Ottobratica std’, grown in Central and Southern Calabria, were found to group together. ‘Ghiastrina’, ‘Ottobratica Perciasacchi V.J.’ and ‘Ottobratica Perciasacchi V.T.’ differed by only one allele at the locus DCA4 (194, 186 and 190, respectively) (Supplementary Table 3, available from http://dx.doi.org/10.5061/dryad.3dv00). The cultivar ‘Ottobratica V.J.’ from Calabria showed all SSR profiles as being distinct from the remaining cultivars of the ‘Ottobratica’ group.
Interestingly, the UPGMA clustering based on SSRs reflected all ten putative parent/sibling groups as indicated by the parentage simulation performed by Cervus. Generally, these groups were confirmed by their bootstrap values (Fig. 4).
Clustering based on SSR diversity differed from clustering built on morphological trait variability, with the following exceptions: the four Calabrian accessions (‘Ghiastrina’, ‘Ottobratica Perciasacchi V.J.’, ‘Ottobratica Perciasacchi V.T.’ and ‘Ottobratica std’); ‘Biancolilla Campana’ and ‘Carpellese’ which showed a close morphological distance of about 0.37, and two pairs (‘Cavalieri’/‘Crastu’ and ‘Cerasuola’/‘Nocellara del Belice’) of the ten groups of putative parent/sibling cultivars (Fig. 2). The Sicilian cultivar ‘Olivo di Mandanici’ and the Calabrian cultivar ‘Chianota’, sharing the same SSR profiles, were shown to differ at the morphological level and clustered in two different groups (Figs. 2, 3 and 4).
A Mantel test (Mantel 1967) was conducted to determine the correlation between SSR profiles, morphological data and geographic origins. We found low, but significant correlations, between morphological data vs SSR allelic frequency (r = 0.24; p < 0.001), between origin vs SSR allelic frequency (0.22; p < 0.001), and between origin and morphological data (0.50; p < 0.001).
To assess the genetic structure of the three olive germplasm collections, we used the software Structure 2.3.1. It is well-known that when K is approaching a true value, L (K) reaches a plateau and has high variance between runs (Rosenberg et al. 2002); in our analysis the plateau was reached at K = 3. In Fig. 5, the three groups were marked as group 1 (black), group 2 (light grey) and group 3 (dark grey). Nineteen of the Sicilian cultivars were found in the black group (which appeared strongly structured); in this group, only two Sicilian cultivars, ‘Bottone di Gallo’ and ‘Santagatese’, had membership value lower than 0.8. The Sicilian cultivar ‘Olivo di Mandanici’, identical at the SSR level to ‘Chianota’, belonged to the light grey group, which comprised thirteen Calabrian and twelve Campanian cultivars. Five Sicilian cultivars (‘Brandofino’, ‘Minuta’, ‘Nasitana’, ‘Vaddarica’ and ‘Verdello’) belonged to the dark grey group, which also included ten Calabrian cultivars (‘Cassanese’, ‘Carolea’, ‘Imperiale’, ‘Oliva d’ogghiu’, ‘Tombarella’, ‘Sinopolese’, ‘Ciciarello’, ‘Minuta Maierato’, ‘Minuta Zungri’ and ‘Miseo’) and three Campanian cultivars (‘Cornia’, ‘Ortice’ and ‘Salella’). Grouping from the structure analysis reflected the UPGMA clustering exactly. The Calabrian and the Campanian germplasm shared most of their allelic makeup, contrary to the Sicilian cultivars, which tended to separate from them. It is likely that cultivars with membership values lower than 0.8 may be crossbred. Overall, the genetic diversity was consistent with the geographic area of origin.
Genetic structure of 68 olive accessions, considering K = 3. Each vertical bar represents the olive cultivar, reported under the respective bar. Colours (black, pale grey and dark grey) represent the three groups, defined by the K value. Olive cultivars showing more than one colour may have an intermixed genetic makeup, resulted from crossing. The vertical axis indicates the membership value
A further canonical discriminant analysis on molecular data and fruit size, as grouping variables, was also performed (data not shown). The analysis was able to discriminate genotypes having large (>6.1 g) or small (<2.0 g) fruit sizes. Moreover, a correlation was found between the presence of some alleles of the loci DCA03, DCA04 and Gapu101, and the fruit size. In particular, alleles 240 and 244 bp of DCA03, allele 152 bp of DCA04 and alleles 185 and 191 bp of Gapu101 occurred in cultivars which had a small fruit size, whilst alleles 250, 252 and 254 bp of DCA03, allele 166 bp of DCA04 and alleles 199, 201 and 207 bp of Gapu101 were present in cultivars which had a very large fruit size.
Discussion
The 68 olive cultivars were analysed morphologically for the 2 years and phenotypically they were considered extremely representative of the olive germplasm variability present in these three Southern Italian regions.
Fifteen leaf, fruit and endocarp traits can be considered the most important to discriminate cultivars at the phenotypic level. Cluster analysis based on morphological traits showed that the grouping of the cultivars reflected their geographical origin. In the light of this finding, it seems that phenotypic traits are moulded by the area of origin and cultivation. Of these three regions, the Sicilian cultivar group was found to be the most compact. Therefore, it is likely that olive cultivars from Sicily were subjected to limited exchange or diffusion from the areas of origin, most probable due to geographical isolation, although, historical links exist between these regions. Geographical closeness between Calabria and Campania may have favoured the exchange of plant material within the area and/or inter-crosses. Canonical discriminant analysis confirmed the cluster analysis indicating an even stronger grouping, which reflected the area of origin and cultivation. In particular, two endocarp characters, pit base shape and pit shape, were found to be the most important on the second canonical function, whereas longitudinal curvature of the blade; leaf apex angle and fruit apex shape displayed more discriminant power on the first canonical function. Most of the Sicilian genotypes, which were divided by the first canonical function, presented epinastic leaves, acute leaf apex angle and rounded fruit apex shape.
The selected 12 SSR loci, nine of which were reported to be the most highly resolving SSRs for olive (Baldoni et al. 2009), were shown to be highly polymorphic in this investigation and gave reproducible amplification patterns for all 68 olive cultivars analysed. The average number of alleles per locus (Na), reported in this study was 13, higher than that obtained by La Mantia et al. (2005), 7.8 (Na) using 12SSR and a set of 50 olive accessions by Lopes et al. (2004), 9.6 (Na) with 14 SSR screened in 130 accessions by Belaj et al. (2012), 11.35 (Na) analysing 23 SSR in 361 cultivars from 19 different countries. The average expected heterozygosity (He) 0.84 was higher in comparison to other published works, 0.76 (La Mantia et al. 2005), 0.68 (Lopes et al. 2004), 0.62 (Belaj et al. 2012), indicating remarkably large genetic variation amongst the germplasm studied from these three Southern Italian regions. For all 12 utilised SSR markers, PIC values of at least 0.67 were recorded, indicating that all loci were highly informative and suitable for individual identification. The value of the total probability of identity (PI) was very low (7.23 × 10−14), demonstrating that the 12 SSR markers used in our study were extremely powerful at genotyping olive cultivars. The presence of “rare alleles” also revealed great genetic variability within the olive germplasm of Southern Italian regions.
The groups identified by morphological analysis did not correspond to the clustering based on molecular analysis, except for the cluster of four Calabrian cultivars (‘Ghiastrina’, ‘Ottobratica Perciasacchi V.J.’, ‘Ottobratica Perciasacchi V.T.’ and ‘Ottobratica std’), which shared from 78 to 84 % of morphological traits. They also showed very few differences at the DCA04 locus and only one at the locus DCA09 occurring in ‘Ottobratica std’, and the pair of cultivars ‘Rotondella’ and ‘Tonda Campana’ which differed by only one allele at the locus DCA07. These differences seemed of somatic origin and can be considered insufficient to have originated by sexual reproduction in a species such as olive, predominantly allogamous and with a high degree of heterozygosity at the genomic level (Zohary and Spiegel-Roy 1975). However, these putative cases of synonymy deserve further investigation with additional molecular markers. A close proximity has also been observed in the morphological analysis between ‘Biancolilla Campana’/‘Carpellese’, that were undistinguishable at the SSR level, but it was not retained among ‘Olivo di Mandanici’ (Sicily) and ‘Chianota’ (Calabria) also identical at the SSR level. Twelve of the 25 morphological characteristics analysed differed for ‘Olivo di Mandanici’ and ‘Chianota’, whilst ‘Biancolilla Campana’ and ‘Carpellese’ differed in eight of them. Concerning ‘Biancolilla Campana’ and ‘Carpellese’, the differences were minutiae regarding the pit shape and pit bundles. Regarding ‘Olivo di Mandanici’ and ‘Chianota’, with the exception of the parameter fruit veraison and fruit shape (length/width), other differences concerned four fruit traits and five pit traits. However, as these cultivars were from extremely close areas of cultivation it is likely that they share a strict relationship, further analysis is needed to clarify this point.
Noteworthy is the fact that the Mantel tests showed significant, albeit weak, correlations between genetic/morphological, morphological/geographic and genetic/geographic data. As already stated by several authors and confirmed by our results, it is likely that multi-local selection and breeding of olive cultivars occurred in each area of origin (Besnard et al. 2001a; Owen et al. 2005; Dominguez-Garcia et al. 2012).
Parentage simulation performed with CERVUS allowed identification, for the first time, of ten statistically significant cases of putative parent/sibling. In most of the cases, these cultivars originated within the same region from very close areas of cultivation. Therefore, it is evident that olive diversification arose not simply through the occurrence of somatic mutations but also through sexual reproduction. Concerning the Sicilian cultivar ‘Giarraffa’ and the Campanian ‘Ravece’, which were found to be related, as well as ‘Olivo di Mandanici’ (Sicily), ‘Chianota’ and ‘Olivo a Rappu’ from Calabria, the Campanian ‘Tonda Campana’ and the Calabrian ‘Ottobratica Rotondella’, our analysis confirmed the occurrence of hybridization between cultivars or exchanges of plant material. These results were partially confirmed by the canonical discriminant analysis performed on morphological variables. Regarding ‘Minuta Zungri' and ‘Minuta Maierato’, we can assume there is a certain degree of relatedness, as they are from the same area of cultivation—the Southern Tyrrhenian coast of Calabria. Local growers have also commented on their morphological similarity, together with the cultivar ‘Ciciarello’. ‘Tombarella’ and ‘Carolea’, both having large fruit and originate in the Ionian side of Calabria, are likely to be strictly related and possibly derive from landraces or genotypes introduced by the Greeks. For similar reasons the Campanian cultivars ‘Caiazzana’/‘Femminella’/‘Racioppella’, and the three Sicilian cultivar pairs ‘Cerasuola’/‘Nocellara del Belice’, ‘Minuta’/‘Vaddarica’ and ‘Crastu’/‘Ogliarola Messinese’ may also be related. As regard, ‘Ghiastrina’/‘Ottobratica Perciasacchi V.J.’/‘Ottobratica Perciasacchi V.T.’/‘Ottobratica std’ and ‘Ottobratica Rotondella’/‘Rotondella’/‘Tonda Campana’ (also indicated as putative siblings by CERVUS), we are more inclined to presume that these are bud sport mutations, originating from somatic mutation events, and not a result of sexual reproduction. The UPGMA dendrogram showed relationships between putative parent/sibling groups, which were supported by bootstrap values. Although we are aware that variants of the same length may be homoplasic, we are quite confident that these putative parent/sibling cases may be genuine, especially because none of the SSRs used in our study presented the drawback of allelic homoplasy, as demonstrated by Baldoni et al. (2009) through SSR sequencing.
In Italy, olive cultivars with large fruits are used as table olives, whilst small-fruited cultivars are used for olive oil production. In some of previously published molecular analyses, groupings of olive cultivars based on their usage and fruit size were found (Fabbri et al. 1995; Besnard et al. 2001b; Belaj et al. 2001). In our study, we found an association between the presence of some alleles of the loci DCA03, DCA04 and GAPU101. However, so far no association between olive fruit size and SSRs or other molecular markers has been reported in any of the published linkage maps (De la Rosa et al. 2003; Wu et al. 2004; Khadari et al. 2010; Dominguez-Garcia et al. 2012).
The structure analysis, with a K value equal to 3, gave us an understanding of genetic relationships at the micro-scale level between and within the three olive germplasm collections. Sicilian cultivars had the highest membership value and a more homogenous genetic makeup, probably due to geographical isolation. The Calabrian and the Campanian cultivars seemed to have a less distinct genetic structure, more intermixed, possibly because they shared putative ancestors. Six Campanian cultivars presented genetic affinity with the Sicilian germplasm, and six Sicilian cultivars had a membership value lower than 0.1, confirming plant material exchanges between regions. This may explain the non-grouping of ‘Brandofino’, ‘Bottone di Gallo’, ‘Caiazzana’, ‘Ravece’ and ‘Ortice’ with cultivars of the alleged germplasm sources in the cluster analysis based on morphological traits. However, on the other hand, the presumable different genetic background of ‘Minuta’, ‘Nasitana’, ‘Olivo di Mandanici’, ‘Santagatese’ and ‘Verdello’ was not evident from the cluster analysis and the CDA based on morphological traits.
The lack of complete correspondence between morphological and molecular data indicates that although morphological characterisation is useful when describing cultivars, it is not sufficient to reflect olive genetic diversity. It is possible that cultivars with different genetic backgrounds tend to assume similar forms under the pressure exerted by human selection and agronomic conditions. DNA marker analysis is, therefore, fundamental in order to depict the level of genetic structure and to discriminate between olive varieties showing similar phenotypes, given that morphological characterisation provides different information, often leading to overestimate or underestimate the real level of genetic diversity.
Our results will be used to construct an olive database based on morphological, phenological and molecular data of most Southern Italian regions, which will allow comparison and identification of cultivars and the exchange of reliable genetic material among institutes for future research.
References
Angiolillo A, Reale S, Pilla F, Baldoni L (2006) Molecular analysis of olive cultivars in the Molise region of Italy. Genet Resour Crop Ev 53(2):289–295
Baldoni L, Belaj A (2010) Olive. In: Vollmann J, Rajean I (eds) Oil crops. Handbook of plant breeding, vol 4. Springer Science Business Media, New York, pp 397–421. doi:10.1007/978-0-387-77594-4_13
Baldoni L, Cultrera NG, Mariotti R, Ricciolini C, Arcioni S, Vendramin GG, Buonamici A, Porceddu A, Sarri V, Ojeda MA, Trujillo I, Rallo L, Belaj A, Perri E, Salimonti A, Muzzalupo I, Casagrande A, Lain O, Messina R, Testolin R (2009) A consensus list of microsatellite markers for olive genotyping. Mol Breed 24:213–231. doi:10.1007/s11032-009-9285-8
Barranco D, Trujillo I, Rallo P (2000) Are ‘Oblonga’ and ‘Frantoio’ olives the same cultivar? HortSci 35(7):1323–1325
Bartolini G, Prevost G, Messeri C, Carignani G, Menini U (1998) Olive germplasm: cultivars and world-wide collections. FAO, Rome
Bartolini G, Prevost G, Messeri C, Carignani C (2005) Olive germplasm: cultivars and world-wide collections. FAO, Rome, Italy. http://www.oleadb.it/.
Belaj A, Trujillo I, De la Rosa R, Rallo L, Gimenez MJ (2001) Polymorphism and discrimination capacity of randomly amplified polymorphic markers in an olive germplasm bank. J Am Soc Hortic Sci 126(1):64–71
Belaj A, Dominguez-García MC, Atienza SG, Martin Urdiroz N, De la Rosa R, Satovic Z, Martin A, Kilian A, Trujillo I, Valpuesta V, del Rio C (2012) Developing a core collection of olive (Olea europaea L.) based on molecular markers (DarTs, SSRs, SNPs) and agronomic traits. Tree Genet Genomes 8:365–378. doi:10.1007/s11295-011-0447-6
Besnard G, Baradat P, Berville A (2001a) Genetic relationships in the olive (Olea europaea L.) reflect multilocal selection of cultivars. Theor Appl Genet 102(2–3):251–258. doi:10.1007/s001220051642
Besnard G, Baradat P, Breton C, Khadari B, Berville A (2001b) Olive domestication from structure of oleasters and cultivars using nuclear RAPDs and mitochondrial RFLPs. Genet Sel Evol 33:S251–S268
Bottari V, Spina P (1952) Le varietà di olivo coltivate in Sicilia. Annuario Sperimentale Agricoltura 7:937–1004
Bracci T, Sebastiani L, Busconi M, Fogher C, Belaj A, Trujillo I (2009) SSR markers reveal the uniqueness of olive cultivars from the Italian region of Liguria. Sci Hortic-Amsterdam 122(2):209–215. doi:10.1016/j.scienta.2009.04.010
Bracci T, Busconi M, Fogher C, Sebastiani L (2011) Molecular studies in olive (Olea europaea L.): overview on DNA markers applications and recent advances in genome analysis. Plant Cell Rep 30:449–462. doi:10.1007/s00299-010-0991-9
Carriero F, Fontanazza G, Cellini F, Giorio G (2002) Identification of simple sequence repeats (SSRs) in olive (Olea europaea L.). Theor Appl Genet 104(2–3):301–307. doi:10.1007/s001220100691
Caruso G (1883) Monografia dell'olivo. Enciclopedia Agraria Italiana vol. III, parte V. UTET edizioni Torino, Italy
Caruso T, Cartabellotta D, Motisi A, Campisi G, Occorso G, Bivona G, Cappello A, Pane G, Pennino G, Ricciardo G, Patti M, La Mantia M, Lain O, Testolin R, Finoli C, Cacioppo L, Corona O, Catagnano L, Savino V, Saponari M (2007) Cultivar Di Olivo Siciliane. Identificazione validazione, caratterizzazione morfologica e molecolare e qualità degli oli. Contiene manuale per la caratterizzazione primaria di cultivar di olivo siciliane. Palermo, Italy.
Catanea A (1934) L’olivicoltura e l’industria olearia nel reggino. Catania, Italy
Chimenti V (1963) Ricerche ed identificazione delle varietà di olivo in provincia di Cosenza. A.G.A editore, Cosenza, Italy
Cipriani G, Marrazzo MT, Marconi R, Cimato A, Testolin R (2002) Microsatellite markers isolated in olive are suitable for individual fingerprinting and reveal polymorphism within ancient cultivars (Olea europaea L.). Theor Appl Genet 104:223–228. doi:10.1007/s001220100685
Corrado G, La Mura M, Ambrosino O, Pugliano G, Varricchio P, Rao R (2009) Relationships of Campanian olive cultivars: comparative analysis of molecular and phenotypic data. Genome 52:692–700. doi:10.1139/G09-044
D'Amore R, Iannotta N, Perri L (1977) Contributo allo studio delle principali cultivar di olivo presenti in Calabria. Annali Istituto Sperimentale per l'Olivicoltura. Numero speciale: vol. I, Cosenza, Italy
De la Rosa R, James CM, Tobutt KR (2002) Isolation and characterization of polymorphic microsatellites in olive (Olea europaea L.) and their transferability to other genera in the Oleaceae. Mol Ecol Notes 2:265–267. doi:10.1046/j.1471-8286.2002.00217.x
De la Rosa R, Angiolillo A, Guerrero C, Pellegrini M, Rallo L, Besnard G, Bervillé A, Martín A, Baldoni L (2003) A first linkage map of olive (Olea europaea L.) cultivars using RAPD, AFLP, RFLP and SSR markers. Theor Appl Genet 106:1273–1282. doi:10.1007/s00122-002-1189-5
Di Vaio C, Nocerino S, Paduano A, Sacchi R (2013) Characterization and bio-agronomic evaluation of olive germplasm cultivars in Southern Italy. J Sci Food Agr. doi:10.1002/jsfa.6057
Dominguez-Garcia MC, Belaj A, De la Rosa R, Satovic Z, Heller-Uszynska K, Kilian A, Martin A, Atienza SG (2012) Development of DArT markers in olive (Olea europaea L.) and usefulness in variability studies and genome mapping. Sci Horti 136:50–60
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. doi:10.1111/j.1365-294X.2005.02553.x
Fabbri A, Hormaza JI, Polito VS (1995) Random amplified polymorphic DNA analysis of olive (Olea europaea L.) cultivars. J Am Soc Hort Sci 120(3):538–542
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null allele. Mol Ecol Notes 7:574–578. doi:10.1111/j.1471-8286.2007.01758.x
Fiorino P, Lombardo N (2002) Germoplasma, materiale vivaistico e certificazione. Atti Convegno Internazionale Olivicoltura, Spoleto, Italy, pp 82–90
Grippo N (1923) Studio di alcune varietà di olivi nel catanzarese. Tipografia Silipo, Catanzaro, Italy
Kalinowski ST, Taper ML, Marshall TC (2007) Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16(5):1099–1106. doi:10.1111/j.1365-294X.2007.03089.x
Khadari B, El Aabidine AZ, Grout C, Sadok IN, Doligez A, Moutier N, Santoni S, Costes E (2010) A genetic linkage map of olive based on amplified fragment length polymorphism, intersimple sequence repeat and simple sequence repeat markers. J Am Soc Hort Sci 135(6):548–555
Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9:1322–1332. doi:10.1111/j.1755-0998.2009.02591.x
La Mantia M, Lain O, Caruso T, Testolin R (2005) SSR-based DNA fingerprints reveal the genetic diversity of Sicilian olive (Olea europaea L.) germplasm. J Hortic Sci Biotech 80:628–632
Liu K, Muse SV (2005) PowerMarker: integrated analysis environment for genetic marker data. Bioinformatics 21(9):2128–2129. doi:10.1093/bioinformatics/bti282
Lombardo N, Perri E, Muzzalupo I, Madeo A, Godino G, Pellegrino M (2004) Il germoplasma olivicolo calabrese. C.R.A.—Istituto sperimentale per l'olivicoltura, Rende, Italy
Lopes MS, Mendoca D, Sefc KM, Sabino Gil F, Da Câmara MA (2004) Genetic evidence of intra-cultivar variability within Iberian olive cultivars. HortSci 39:1562–1565
Mafrica R, Zappia R, Continella G (1996) Indagine preliminare sul germoplasma di olivo nell’area grecanica della provincia di Reggio Calabria. Atti III Giornate Scientifiche SOI, Erice (TP), Italy, March10-14.
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Marchese A, Costa F, Marra FP, Campisi G, Cutuli M, Ricciardo P, Tobutt KR, Caruso T (2008) Determination of intra-cultivar variation among Sicilian olive germplasm by using microsatellite markers. VI International Symposium on Olive Growing. Resumos, Evora, Portugal, September 9–13, pp. 67
Marrazzo T, Cipriani G, Marconi R, Cimato A, Testolin R (2002) Isolation and characterisation of microsatellite DNA in Olive (Olea europaea L.). Acta Horticulturae 586:61–64
Marshall TC, Slate J, Kruuk LEB, Pemberton JM (1998) Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol 7(5):639–655. doi:10.1046/j.1365-294x.1998.00374.x
Meagher TR (1986) Analysis of paternity within a natural population of Chamaelirium luteum. I. Identification of most-likely male parents. Am Nat 128:199–215
Mookerjee S, Guerin J, Collins G, Ford C, Sedgley M (2005) Paternity analysis using microsatellite markers to identify pollen donors in an olive grove. Theor Appl Genet 111(6):1174–1182. doi:10.1007/s00122-005-0049-5
Motisi A, Marra FP, Zappia R, Mafrica R (2001) Indagine sulle caratteristiche biometriche di 18 biotipi di olivo del germoplasma calabrese. Atti VI Convegno Nazionale "Biodiversità: opportunità di sviluppo sostenibile". Bari, Italy, September 6–7.
Muzzalupo I, Stefanizzi F, Perri E (2009) Evaluation of olives cultivated in Southern Italy by SSR markers. HortSci 44:582–588
Owen CA, Bita E, Banilas G, Hajjar SE, Sellianakis V, Aksoy U, Hepaksoy S, Chamoun R, Talhook SN, Metzidakis I, Hatzopoulos P, Kalaitzis P (2005) AFLP reveals structural details of genetic diversity within cultivated olive germplasm from eastern Mediterranean. Theor Appl Genet 110:1169–1176. doi:10.1007/s00122-004-1861-z
Parlati MV, Perri E, Rizzuti B, Palopoli A (1995) Selezione dell'olivo in Calabria clone “Carolea Cefaly”: un interessante clone caratterizzato da precocità di maturazione e pezzatura del frutto superiore alla media. Atti del Convegno “L'Olivicoltura mediterranea: stato e prospettive della coltura e della ricerca”. Rende, Italy, January 26–28.
Parlati MV, Perri E, Rizzuti B, Pellegrino M (1999) Caratterizzazione della cv. Pennulara o Nostrale di Caccuri: ecotipo interessante per le caratteristiche carpologiche e qualitative dell'olio. Atti 5° Convegno Nazionale sulla Biodiversità. Caserta, Italy, September 9–10.
Pavirani M (1959) L'Olivicoltura in provincia di Catanzaro. Ispettorato Provinciale Agrario, Catanzaro, Italy
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pugliano G (2000) La risorsa genetica dell’olivo in Campania. Se.S.I.R.C.A, Regione Campania
Rallo P, Dorado G, Martin A (2000) Development of simple sequence repeats (SSRs) in olive tree (Olea europaea L.). Theor Appl Genet 101:984–989. doi:10.1007/s001220051571
Rao R, La Mura M, Corrado G, Ambrosino O, Foroni I, Perri E, Pugliano G (2009) Molecular diversity and genetic relationships of southern Italian olive cultivars as depicted by AFLP and morphological traits. J Hortic Sci Biotech 84(3):261–266
Rohlf EJ (1993) NTSYS-pc: numerical taxonomy and multivariate analysis system, version 1.80. Applied Biostatistics Inc, Setauket, New York
Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW (2002) Genetic structure of human populations. Science 298:2381–2385
Rugini E, De Pace C, Gutierrez-Pesce P, Muleo R (2011) Olea. In: Chittaranjan K (ed) Wild crop relatives: genomic and breeding resources, 1st edn. Springer, Heidelberg, pp 79–117
Sabino Gil F, Busconi M, Da Câmara MA, Fogher C (2006) Development and characterization of microsatellite loci from Olea europaea. Mol Ecol Notes 6:1275–1277. doi:10.1111/j.1471-8286.2006.01513.x
Sacchi R, Ambrosino ML, Della Medaglia D, Paduano A, Spagna Musso S (1999) Gli oli della Campania. Se.S.I.R.C.A, Regione Campania, Italy
Sarri V, Baldoni L, Porceddu A, Cultrera NGM, Contento A, Frediani M, Belaj A, Trujillo I, Cionini PG (2006) Microsatellite markers are powerful tools for discriminating among olive cultivars and assigning them to geographically defined populations. Genome 49(12):1606–1615. doi:10.1139/g06-126
Sefc KM, Lopes MS, Mendonça D, Rodrigues Dos Santos M, Laimer Da Câmara Machado M, Da Câmara MA (2000) Identification of microsatellite loci in olive (Olea europaea) and their characterization in Italian and Iberian olive trees. Mol Ecol 9:1171–1173. doi:10.1046/j.1365-294x.2000.00954.x
Sokal R, Michener C (1958) A statistical method for evaluating systematic relationships. Univ KansSci Bull 38:1409–1438
Thompson EA (1975) The estimation of pairwise relationships. Ann Hum Genet 39:173–188
Thompson EA (1976) Inference of genealogical structure. Soc Sci Inform 15:477–526
Wu S, Collins G, Sedgley M (2004) A molecular linkage map of olive (Olea europaea L.) based on RAPD, microsatellite and SCAR markers. Genome 47:26–35
Zito F (1931) Le varietà di olivo da olio in Italia. Contributo alla conoscenza delle varietà della zona di Palmi. Roma, Italy
Zohary D, Spiegel-Roy P (1975) Beginnings of fruit growing in the Old World. Science 187:319–327
Zohary D, Hopf M (1994) Domestication of plants in the old world, 2nd edn. Clarendon Press, Oxford
Acknowledgments
We thank the anonymous reviewer for his/her most helpful comments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E. Dirlewanger
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table 3
SSR allelic profiles of the 68 olive cultivars analysed at 12 SSR loci. (Rare alleles are in bold. Restricted alleles are underlined. 1Sicilian; 2Calabrian;3Campanian) (DOCX 44 kb)
Rights and permissions
About this article
Cite this article
Marra, F.P., Caruso, T., Costa, F. et al. Genetic relationships, structure and parentage simulation among the olive tree (Olea europaea L. subsp. europaea) cultivated in Southern Italy revealed by SSR markers. Tree Genetics & Genomes 9, 961–973 (2013). https://doi.org/10.1007/s11295-013-0609-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-013-0609-9