Abstract
Acacia senegal (L.) Willd. is the main species producing the internationally traded gum arabic. Genetic studies of this species are rare and until now the chromosome number was thought to be diploid (2n = 2x = 26). Here, using chromosome number counting, we demonstrate for the first time that tetraploids (2n = 4x = 52) also occur in A. senegal. Nuclear and chloroplast microsatellite markers were used to estimate and compare genetic variation within this newly described polyploidy complex in the Sudano-Sahelian region in Africa. Genetic diversity was higher in diploids, suggesting that the formation of tetraploids is recent and that mutation–drift equilibrium has not yet been reached. The two cytotypes do not have the same genetic structure and are genetically differentiated. Among tetraploids, populations are greatly differentiated and do not share the same chlorotypes. Based on these results, we discuss recurrent formation of tetraploids from different diploid progenitors across the distribution range of A. senegal in the Sudano-Sahelian zone.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Acacia senegal is a multipurpose tree that belongs to the Leguminosae family and to the Acacia genus (Bentham 1875; Vassal 1972). A. senegal covers extensive areas especially in drier areas south of the Sahara, throughout the Sahelian zone from Senegal to Somalia, and in regions stretching across Arabia to India and Pakistan. In its natural range, annual rainfall ranges from 100 to 1,000 mm (Nicholson et al. 1988) and soil types from poor sandy soils to sandy clays (Fagg and Allison 2004). A. senegal is mainly used for the production and sale of gum arabic (gum hashab) but is also extensively used for soil fertilization and as animal fodder. Gum arabic is a natural exudate obtained from the stems and branches of A. senegal and is recognized to be nontoxic and safe for food use (FAO 1998). It is used in the food processing industry, where it is known as food additive E414, and has adhesive, anti-crystallization, thickening, and protective properties. It is widely used as an emulsifier and suspensoid in beverages. The same properties are also exploited in pharmaceutical products and in many technical applications. The quality of gum arabic depends on its chemical characteristics and molecular structure and varies with the provenance (Motlagh et al. 2006). Indeed, provenance characteristics have significant effects on the gum arabic trade. Sudan is the world’s biggest producer of gum arabic and the main source of gum in international trade. The region of Kordofan has traditionally been the region that produced the most gum hashab (Fagg and Allison 2004). Nigeria has long been the second biggest producer, followed by Chad.
A. senegal is a very variable species and its delimitation is not yet clear (Ross 1975, 1979; Maslin et al. 2003; Miller and Bayer 2003; Robinson and Harris 2000). Remaining uncertainties concern not only the status of the species, especially with respect to closely related species, but also the definition, classification, and phylogeny of the Acacia genus. A diploid chromosome number (2n = 2x = 26) has been reported by several authors (Ross 1979; Obunga 1995; Bukhari 1997), but to our knowledge, no level of polyploidy has been recorded for this species.
Despite its wide distribution and its economic and ecological importance, studies of the genetic diversity and structure of A. senegal are rare (Chevallier et al. 1994; Maslin et al. 2003). The highly contrasted ecological conditions observed in the natural range of A. senegal lead us to hypothesize that these variations have had an impact on the genetic diversity of the species. Analysis of the genetic variability at the scale of the whole natural area could provide useful information both for the preservation and management of biodiversity and for selection and genetic improvement. Exploring the genetic bases of such variation could provide powerful tools to improve both the quantity and quality of the product.
Here, using chromosome counting, we demonstrate for the first time that polyploidy (2n = 4x = 52) occurs in A. senegal. To understand the relationship between diploids and tetraploids, we used nuclear and chloroplast microsatellite markers for analysis and we discuss genetic variation within the polyploid complex across the Sudano-Sahelian range of the species.
Materials and methods
Sampling
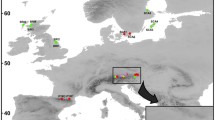
Samples of provenances were chosen in the natural distribution area of A. senegal var senegal. Leaves were collected from 11 populations in six African countries distributed throughout the Sudano-Sahelian distribution range (Table 1, Fig. 1). Two comparative trials of provenances and four natural stands were used to represent the maximum genetic diversity of A. senegal. For the first trial, leaves were collected in Bambey (Senegal) and for the second in Sorey (Niger), the natural stands were located in Niger. The genetic resources of A. senegal used for these trials were mainly acquired by the Oxford Forestry Institute and were exchanged in consultation and collaboration with international agencies (CIRAD-Forêt, DFSC, FAO, ICRAF) and national research institutes throughout Africa (Barnes et al. 1999). For the provenances tested, seeds were harvested on 20–30 trees respecting a minimum distance of 100 m between trees. Leaf samples from five populations, Sodera (Ethiopia), El Obeid (Sudan), Tourba (Chad), Somo (Mali), and Kidira (Senegal), were collected at Bambey, Senegal. Leaf samples from populations originating from Kordofan (Sudan) and Tendjé (Mali) were collected from the 10-year-old provenance trial at Sorey. The leaves for simple sequence repeat (SSR) analysis of these two provenances were harvested randomly, without selection, on 33 trees in Kordofan and 36 trees in Tendjé. Leaf samples of the four remaining populations (Tabé, Bader, Téra, and Kiki) were collected from natural populations located on a rainfall gradient from the east to west of Niger (Table 1). Here, the objective was to complete the observation of genetic variability for the whole natural distribution range by observations along an environmental cline. The aim of the sampling procedure in natural stands was to obtain as broad a representation of the full range of genetic distribution as possible at each site. Trees were sampled randomly without selection based on growth form but respecting a minimum distance of 20 m between sampled trees. Young leaves were removed from trees, dried in silica gel, and kept in sealed plastic bags until DNA extraction and molecular analysis. A total of 364 trees were sampled, but sampling was not balanced as the number of individuals per population varied from 15 (Sodera) to 86 (Téra). The method used to harvest seeds for the provenances trials is strictly identical to the collection of leaves in the natural stands, i.e., distant trees, contrasted areas in which selection has taken place, multiple generations, and spatial genetic structure.
Among the 11 populations studied, three (Kordofan, Tendjé, and Sodera) had more than two alleles at almost all nuclear SSR loci. Chromosomes were then counted to check if this was due to partial duplication of the genome or to a different ploidy level in the three populations. The plant material used came from two populations (Kordofan and Tendjé) that had three or four alleles per microsatellite locus and, as control, one population (Téra) that had two alleles. A total of 15 seeds were collected on five mother trees genotyped with SSR markers and presumed to be polyploidy in each provenance studied.
SSR genotyping and revelation of polymorphism
DNA extraction, PCR conditions, and electrophoresis conditions are described in another paper (Assoumane et al. 2009). Nuclear genetic analysis was performed using 11 nuclear microsatellites: mAsCIRB09, mAsCIRB10, mAsCIRC07, mAsCIRE06, mAsCIRE07, mAsCIRE08, mAsCIRE10, mAsCIRF02, mAsCIRF03, mAsCIRH01, and mAsCIRH09, designed specifically for A. senegal (L.) Willd. var. senegal. All samples collected were analyzed using these markers. We used universal chloroplast microsatellites to complete the information provided by the nuclear microsatellites; 7 CCMP markers described by Weising and Gardner (1999) and 20 NTCP markers designed by Bryan et al. (1999) were tested. Genotyping with chloroplast microsatellites was performed on a subset of 96 individuals: 12 from one population in Niger (Téra) and 12 individuals from each population in the Sudano-Sahelian range. PCR with chloroplast microsatellites was performed in duplex, in a 10-μl reaction mix with 1 μl of DNA (5 ng/μl), 5 μl of Qiagen buffer, 0.08 μl (10 μM) of each of the two reverse primers, 0.1 μl (10 μM) of each of the two forward primers, and 0.1 μl (10 μM) of oligos labeled with M13; the volume was completed with sterile water. All reactions were performed with an Eppendorf Mastercycler gradient. Optimal amplification conditions were: one 15-min cycle at 95 °C (initial denaturation), 1 min 30 s at 67 °C (annealing), and 1 min at 72 °C (extension); followed by eight 30-s cycles at 94 °C (denaturation), 1 min 30 s at 65 °C (annealing), and 1 min at 72 °C (extension); then another 24 cycles as follows: 30 s at 94 °C (denaturation), 1 min 30 s (annealing) at 51 °C, and 1 min at 72 °C. A final 30-min step at 72 °C ensured full extension of all amplified products. The PCR products (for nuclear and chloroplast microsatellites) were analyzed by electrophoresis on acrylamide gel (LI-COR 4200 sequencer and IR-DNA analyzer) at the Montpellier Languedoc-Roussillon Genopole genotyping platform. Genotyping was performed with SAGAGT software (LI-COR, Inc).
Ploidy determination
The number of chromosomes was determined in two stages: chromosome preparation and in situ hybridization. To be completely visible and accurately counted, the chromosomes must be in metaphase, and for this reason, dividing tissues have to be used. We chose the root meristem. Seed dormancy was counteracted by treatment in sulfuric acid for 15 min at 95 °C. The pretreated seeds were then placed on blotting paper in a Petri dish and incubated for 48 h at 37 °C. The meristem of young roots was harvested with fine forceps and placed directly in a solution of 0.04 % 8-hydroxyquinoline for 4 h in the dark. Root meristems were then fixed in a solution of ethanol/acetic acid (3:1) for 48 h and stored in 70 % ethanol at room temperature (RT). The fixed roots were hydrolyzed for 10 min in 0.25 N HCl, rinsed for 10 min in dH2O, and immersed in digestion buffer (10 mM trisodium citrate dihydrate, 500 mM citric acid monohydrate, pH 4.5) for 10 min. The distal (1–1.5 mm) portion of the root tip was then removed with a fine scalpel and placed in a digestion enzyme solution (1 % of cellulose Onozuka R-10, 1 % of Pectolyase Y-23, and 1 % cytohelicase in digestion citrate buffer at pH 4.5) at 37 °C. To optimize incubation, a range of tests was performed, and in A. senegal, the highest rate of digestion was obtained after 45 min.
Roots were rinsed twice with pure H2O, and then, using a Pasteur pipette, they were carefully removed and placed on a watch glass containing pure H2O and left for 60 min. They were then transferred on a slide previously cleaned with acid (one root per slide); one drop of 3:1 fixative was added and the tip was immediately spread out with a fine forceps. After air drying, the slides were stored at RT.
The principle of in situ hybridization is to allow hybridization of labeled probes with their target chromosome, to select only the strong homologies by conducting stringent washes, and to detect and amplify the signal (Gerlach and Bedbrook 1979; Salvo-Garrido et al. 2001). Slides were treated with RNase (1 μg/ml) for 45 min at 37 °C, denatured for 3 min in a solution of 70 % formamide in 2× SSC at 80 °C, then dehydrated through an ethanol series (70, 90, and 100 %) at −20 °C. Probe consisted of pTA 71 (rDNA genes 18 S-5.8 S-25 S), labeled by random priming (Fisher Bioblock Scientific) with digoxigenin-11-dTTP. The 18 S-5.8 S-25 S rDNA sequence was located at the terminal position on the chromosome and sometimes appeared as satellites. In most species, it is present on one site per chromosome set, which makes it a good indicator of ploidy level. The hybridization mixture (50 μl per slide) consisted of 50 % formamide, 10 % dextran sulfate, 2× SSC, 1 % SDS, 2 μg/ml labeled probe, and 0.5 μg/μl salmon sperm DNA as a distant competitor to saturate the sites and avoid nonspecific hybridization. The hybridization mixture was denatured for 10 min in a boiling water bath and deposited on each slide. Hybridization was performed overnight in a moist chamber at 37 °C. After hybridization, slides were washed for 10 min in 2× SSC, 0.5× SSC, and 0.1× SSC at 42 °C. Labeled probe has to be amplified to enable detection: 50 μl of detection mix (Anti Dig FITC 10 ng/μl, BSA 4× SSC Tween was added on each slide and slides were incubated for 45 min at 37 °C). The signal was amplified with 50 μl of a mix composed of rabbit-anti-sheep-FITC (1.5 μg/μl goat serum 4× SSC Tween). Slides were incubated for 45 min at 37 °C and washed with a solution of 4× SSC/Tween at 42 °C. Slides were then dehydrated in an ethanol series (70, 90, and 100 %) and dried. Finally, slides were mounted in Vectashield antifade solution (Vector Laboratories) containing 2.5 μg/ml 4′,6-diamidino-2-phenylindole as counterstaining. The slides were examined under a Leica DMRAX2 fluorescence microscope and the images of blue and green fluorescence were acquired separately with a cooled high-resolution black and white CCD camera, ORCA-Hamamatsu 1394. The camera was connected to a PC running the Volocity software (Perkin Elmer).
Genetic data analysis
As in our samples, the molecular markers and ploidy determination revealed a mixture of three tetraploids and eight diploid provenances; we used six additional methods to analyze genetic variability and structure among populations of A. senegal. Analyses were performed using two types of data coding previously used to describe diversity in the case of polyploids (Sampson and Byrne 2012; Jørgensen et al. 2008; Kloda et al. 2008; Robertson et al. 2010).
Estimating genetic similarity between mixed polyploid populations with codominant markers remains difficult. Obbard et al. (2006) suggested combining all alleles detected at each locus to form multilocus allele phenotypes for each individual. Allelic diversity was measured as the total number of alleles over all loci (A), as the number of different alleles in a given population, averaged over loci (A′), the percentage of polymorphic loci (P), the information index (I) often used in studies of polyploid complexes, expected heterozygosity (H e) corrected for sample size (Nei 1978), the number of alleles in an individual, averaged over loci (H′), and the number of different single-locus allele phenotypes observed, averaged over loci (N′p). Computations of these statistics were performed using Fstat 2.9.3 (Goudet 2001), SpageDi 1.2b (Hardy and Vekemans 2002), and GenAlex 6.3 (Peakall and Smouse 2006). Student’s t tests were used to test for significant differences between diploid and tetraploid populations for these data. The number of different allele phenotypes was counted in each population and the average calculated across loci. Thus, the specific number of alleles found in eight diploid populations (N S Di), the specific number of allele in three tetraploid populations (N S Tetra), and the number of common alleles were calculated between diploid and tetraploid populations. For 11 SSR loci, the variation in the number of alleles observed is given for the tetraploid populations from Kordofan, Tendjé, and Sodera. A phylogeographic pattern occurs when gene copies sampled at nearby locations carry alleles that are, on average, more closely related than in gene copies sampled at greater distances from one another. Phylogeographic structure can be tested with R ST (Slatkin 1995) estimated as Michalakis and Excoffier (1996) and the software SpageDi.
SSR markers were treated as dominant and were scored as present (1) or absent (0) for each allele in each individual. Variations in the data set were visualized using principal coordinate analysis (PCO) based on the similarity measure of Dice (1945). The first 72 of the resulting 372 eigenvectors explained 100 % of the variation in the data set, and these were then analyzed with canonical variate analysis (CVA) and multivariate analysis of variance in PAST version 2.13 (Hammer et al. 2001) following the procedure of Anderson and Willis (2003), of a priori dividing the specimens into groups according to the 11 provenances of A. senegal. For the purpose of comparison, Bruvo’s distance (2004) was also calculated and analyzed by PCA with the software Polysat (Clark and Jasieniuk 2011). This measure, which was developed for polyploids, takes the mutational distance between alleles into account and is advocated if it can be assumed that no allele from one ancestral allopolyploid genome is within three or four mutation steps of any allele from the other ancestral genome (Clark and Jasieniuk 2011; Sampson and Byrne 2012).
Pairwise genetic distances (F ST/1 − F ST values) between pairs of provenances were computed using SpageDi software with 1,000 replications. The distance tree was constructed using the neighbor-joining method (Saitou and Nei 1987) and the robustness of each node was evaluated using the Seqboot program in the Phylip 3.67 package (Felsenstein 2005). The consensus tree was visualized with Darwin (Perrier and Jacquemoud-Collet 2006).
The Bayesian method was used to describe the genetic structure using Structure version 2.3. (Pritchard et al. 2010, 2000). Each cytotype was analyzed separately to avoid artifacts stemming from analyzing both diploids and tetraploids simultaneously. We tested the admixture and no-admixture model with a set of K values ranging from 1 to 13 for diploids and one to seven for tetraploids. Five independent runs of each K were performed with 200,000 MCMC repetitions and 40,000 burn-in periods. The number of clusters was determined by combining the recommendations of Pritchard et al. (2000) and the approach developed by Evanno et al. (2005).
Analyses of molecular variance (AMOVA) were performed among populations of each cytotype using GenAlex version 6.3 (Peakall and Smouse 2006). For diploid populations, the genetic distance matrix was calculated for codominant data using the following rules: for a single locus, with ith, jth, kth, and lth different alleles, a set of squared distances was defined as d 2(ii, ii) = 0, d 2(ij, ij) = 0, d 2(ii, ij) = 1, d 2(ij, ik) = 1, d 2(ij, kl) = 2, d 2(ii, jk) = 3, and d 2(ii, jj) = 4. For tetraploid populations, the codominant SSR data were converted into binary data by denoting the presence of the allele as “1” and its absence as “0”. Population genetic differentiation was calculated via ϕ PT , a measure that allows intraindividual variation to be suppressed and is therefore ideal for comparing codominant and binary data when no intraindividual variation (heterozygosity) is present. Statistical tests were performed with 10,000 permutations.
V AP and V WP are the estimate of variance among and within populations, respectively.
Differences in genetic diversity between cytotypes were estimated by calculating and comparing total gene diversity between diploids (H Tdip) and tetraploids (H Ttet). ANOVA analyses were performed on I values for the same purpose, with the cytotype as the main effect, using SPAGeDi version 1.3 (Hardy and Vekemans 2002) and Xlstat-Pro (Addinsoft© 1995-2007). Of the 27 chloroplast microsatellites tested, 11 were shown to be amplified after the PCR. Among these, only one (CCMP5, which in fact amplifies the same microsatellite as NTCP 12) was polymorphic. Chlorotypes (i.e., a combination of the different alleles at each locus) were analyzed at the only polymorphic locus, but it was not possible to calculate chlorotype diversity and genetic structure parameters due to the small number of polymorphic loci.
Results
Ploidy variation in A. senegal
In the Tendjé population, 18 metaphases were analyzed, 10 metaphases clearly displayed 52 chromosomes. In eight metaphases, it was difficult to distinguish all the chromosomes because some overlapped; however, these eight metaphases showed quite a large number of chromosomes, i.e., more than 50. In the Kordofan population, 11 metaphases were examined of which six unambiguously displayed 52 chromosomes (Fig. 2) and five metaphases displayed between 49 and 51. In the control population from Téra, six metaphases were counted and unambiguously displayed 26 chromosomes. In populations with 52 chromosomes, four rDNA sites were identified (Fig. 2a). This was interpreted as being due to the presence of four copies of one locus (i.e., four chromosomes), confirming that these populations are tetraploids 2n = 4x = 52. In the control population, only two rDNA sites were indentified (Fig. 2b).
In situ hybridization of the rDNA probe (pTA 71) to chromosome preparations of A. senegal counterstained with DAPI. a Samples from the Tendjé population are tetraploid with 2n = 4x = 52 chromosomes; b samples from Téra population are diploid with 2n = 2x = 26 chromosomes. Green fluorescence (FITC), indicated by arrows, shows hybridization of pTA 71 probe to rDNA site
Within-population and within-ploidy genetic diversity
A total of 117 alleles were amplified from the 11 SSR loci. All 11 loci were found to be highly polymorphic and very simple to score in eight populations (Bader, Kidira, Kiki, El Obeid, Somo, Tabé, Téra, and Tourba) which presented only one allele (homozygote) or two alleles (heterozygote) in all individuals (general mean = 1.49 ± 0.50). In the populations from Kordofan, Sodera, and Tendjé, the 11 loci were also found to be highly polymorphic and the number of alleles varied between one and four (general mean = 2.51 ± 0.85). This is more problematic because in the case of two or three alleles, the genotype is not fully resolved (Table 2). Diploid and tetraploid populations presented 37 and 23 specific alleles, respectively. In the tetraploid provenances of A. senegal, the frequencies of the specific alleles ranged from 0.5 to 53.4 %. Five loci (E08, F02, E06, C07, and H01) displayed a total number of specific alleles ranging from 6 to 13, the maximum frequency was observed at locus E08 in tetraploid populations, and at locus E06 in diploids, the latter locus displayed a high frequency of specific alleles both in diploids and tetraploids. Among the tetraploids, one locus (E07) in the Sodera population and three loci (E07, B09, and E10) in the Kordofan population were monomorphic (Table 2). Fixed heterozygosity was observed in this cytotype (Fig. 3) at all the other remaining loci.
The mean number of alleles per locus ranged from 3.46 to 5.18 and from 2.55 to 4.09 in 2× and 4×, respectively (Table 3). The genetic diversity was highly significant between diploid and tetraploid populations for H′ and P and significant for I and N′p. Diploid populations presented a higher percentage of polymorphic loci and a higher information index than tetraploid populations, but a smaller number of alleles in an individual and of different single-locus allele phenotypes than in tetraploid populations. Comparison of H e among populations revealed no significant differences within each cytotype.
The phylogeographic signals within and among populations were not significant; the p value was 0.08 and 0.09 for the global R ST and the slope of pairwise R ST, respectively. Distinct alleles were not more closely related to nearby populations than to distant populations.
Among 11 universal chloroplast microsatellites, only one was polymorphic with two different alleles, and this variation was checked by sequencing. Two tetraploid populations (Sodera, Ethiopia and Tendjé, Mali) had two alleles whereas all the diploid populations and one tetraploid population (Kordofan, Sudan) had one and the same allele (Fig. 4). This allele was rare in the Sodera and Tendjé populations.
Population differentiation and genetic structure
PCO analysis of all samples separated tetraploid A. senegal from diploid A. senegal along axis 1 (spanning 15.7 % of the total variation; Fig. 5). The eight diploid provenances formed only one group. Axis 2 (14.8 %) separated the tetraploid populations from Kordofan and Sodera. Axis 3 (8.2 %) separated the tetraploid population from Tendjé. Diploids were distinguished from tetraploids, except one individual from Tendjé, which clustered with a few individuals from Téra, El Obeid, and Kiki populations. CVA analysis (data not shown) separated the four previously defined groups completely and significantly (Wilk’s lambda = 9.42 E-5; Pillai trace = 4.2 were highly significant). PCA based on Bruvo’s genetic distance (2004) separated the diploid and tetraploid populations more clearly (Fig. 6).
Principal coordinate analysis (PCA) of Bruvo’s genetic distance (Bruvo et al. 2004) between 11 provenances of A. senegal
The phylogram of 11 provenances of A. senegal based on pairwise F ST/1 − F ST clearly separated provenances from Kordofan, Tendjé, and Sodera with a p value = 1. The provenances from Niger and Mali grouped together and were located further away from provenances from Senegal, Chad, and Sudan (Fig. 7). Moreover Wright’s F statistics F ST (Wright 1951) showed that differentiation was greater between tetraploid populations than between diploids, indicating that more gene flow occurred in the latter. F IS and F ST were higher in tetraploids than in diploids (mean F IS equal to −0.566 ± 0.044 and −0.005 ± 0.027, mean F ST equal to 0.353 ± 0.034 and 0.095 ± 0.014, respectively, for tetraploids and diploids). According to the results of AMOVA (Table 4), in which cytotypes were analyzed separately, up to 83 % (p < 0.001) of total variation was found within diploid populations, whereas in tetraploid populations, only 7 % (p < 0.001) of variation was found within populations. Most of the variation was divided among populations. When ploidy level was analyzed, up to 21 % (p < 0.001) of the variation was distributed among diploids and tetraploids, 29 % among populations, and 50 % within populations.
When populations were analyzed with Structure software, the two models, admixture and no-admixture, gave the same results. In diploid populations, the ln[Pr(X|K)] of the software and the magnitude of ΔK calculated with the method of Evanno et al. (2005) gave the same K value for four populations (Fig. 8a). Two neighboring populations from Niger (Bader and Tabé) were grouped in the same cluster. Three populations from Senegal (Kidira), Chad (Tourba), and Sudan (El Obeid) formed the second cluster. The third cluster mainly comprised Téra from Niger, and the two remaining populations, Kiki from Niger and Somo from Mali, were shared between the third and the fourth cluster (Fig. 8c). Although the four clusters were relatively easy to distinguish, more individuals were shared between them. This is in agreement with the results obtained with F ST (0.09) estimates indicating gene flow among diploid populations. Concerning tetraploids, optimal partitioning of individuals was obtained with K = 3 (Fig. 8b). The three populations clearly differed from one another in their allelic composition (Fig. 8c). Only the Tendjé population from Mali included some individuals from Sodera in Ethiopia. This confirmed F ST (0.353) results for tetraploids.
Bayesian inferences of the K clusters estimated with Structure using the no-admixture model. a A. senegal 2x and b A. senegal 4x. ln[Pr(X|K)] and ΔK values are plotted against K to identify the most probable K in each cytotype. In diploid populations (a), four clusters of genotypes were identified, whereas in tetraploid populations (b), three clusters of genotypes were identified. c Inference of individuals in clusters. Clusters are represented by colors, and individuals are represented by columns. In each column (individual), the extent of the component colors indicates the magnitude of the membership of each cluster
Discussion
Genetic diversity of diploids and tetraploids
The H e values (Table 3) of diploid populations showed that the genetic diversity of A. senegal is comparable with that of other forest species, as already revealed using the same types of markers (England et al. 2002; Novick et al. 2003; Omondi et al. 2010). Expected heterozygosity (H e) and total gene diversity (Wright) were higher in tetraploids than in diploids, whereas the mean number of alleles per locus (A), the percentage of polymorphic loci (P) and the Shannon index (I) were higher in diploids. Under a neutral model of evolution (i.e., assuming population sizes and histories are equal and equilibrium between genetic drift and mutation has been reached), one would expect genetic diversity to be higher in tetraploids than in diploids (Moody et al. 1993). Some authors (Soltis and Soltis 1989; Brown and Young 2000; Hardy and Vekemans 2001; Luttikhuizen et al. 2007) reported higher genetic diversity in outcrossing autotetraploids than in their diploid counterparts. H Ttet could be higher due to effective population size, which is larger in tetraploids, as each individual harbors twice the number of gene copies. However, H Ttet could also be higher due to the larger number of alleles collected in tetraploid populations than in diploid ones. But in A. senegal, H Ttet (0.73) was higher than H Tdip (0.53), while A dip (4.6) was higher than A tet (3.6). So the greater diversity found in A. senegal tetraploids is not linked to the number of alleles but to fixed heterozygosity, which was observed at almost all loci in each tetraploid population (Fig. 3). Results shown by chloroplast microsatellites suggest that populations diverged recently as 1 locus out of 11 was seen to be polymorphic and all diploid populations had a single allele. Two alleles were found in tetraploids and two populations (Sodera and Tendjé) had these two alleles, whereas one population (Kordofan) had only one allele that was the same as the one in the diploid populations. This result shows that the three tetraploid populations do not share the same cytoplasm and consequently are not of the same origin. This result implies that at least one tetraploid population is allotetraploid, which is consistent with the fixed heterozygosity observed using nuclear microsatellites. Some loci displayed a bimodal distribution of alleles (data not shown). One mode was alleles shared by diploids and tetraploids and the other mode was private alleles in tetraploids, knowing that each tetraploid population has its own private alleles. This result suggests tetraploids have multiple origins, probably with A. senegal as one parent and the other parent coming from another species or variety, probably within the A. senegal complex. Given that similar diploid relatives of A. senegal species are known, allopolyploidy is possible.
If tetraploid A. senegal are allotetraploids, expected heterozygosity H e is no longer a suitable parameter for comparing levels of genetic diversity between diploids and tetraploids because of disomic inheritance and fixed heterozygosity. In this case, we can only use the mean allele number per locus (A), the percentage of polymorphic loci (P), and the Shannon index (I) to compare the two cytotypes, and these parameters are higher in diploids. In the case of allotetraploids, Bruvo’s genetic distance (Bruvo et al. 2004) clearly separated all the tetraploid provenances, also supporting the hypothesis that no allele from one ancestral allopolyploid genome is within three or four mutation steps of any allele from the other ancestral genome. But this hypothesis still needs to be confirmed. Hence, tetraploid A. senegal showed lower genetic diversity than diploid A. senegal. Lower genetic diversity might be harbored by recently formed polyploid populations if polyploid formation is a rare and/or recent event (Wagner 1970). When polyploids formed only recently, they started out with limited genetic diversity, and it would take a considerable amount of time to reach equilibrium between mutation and drift, and hence higher levels of genetic diversity. Population bottlenecks associated with the recent establishment of a new population would also have led to loss of genetic diversity in tetraploids. Each of these events would result in lower genetic diversity in A. senegal tetraploids than in diploids. But this hypothesis should be considered with caution as tetraploid samples were collected in two provenance comparison trials. For SSR genotyping, our sample comprised 15 % from Sodera and around 4 % from Tendjé and Kordofan. In our samples of these three provenances, tetraploidy observed by SSR genotyping was confirmed by chromosome counting in 15 descendants of each provenance. Even though some provenances displayed high rates of tetraploidy, it is difficult to affirm that all the populations are tetraploids. In natural populations, both diploids and tetraploids may coexist and a provenance seed lot may show a different rate of polyploidy than that found at natural sites. Furthermore, the distribution of tetraploids observed suggests that they are likely to have multiple independent origins, potentially through hybridization with different local parent species. The fact that different chloroplast haplotypes were found in tetraploid samples suggests that this is the case and that earlier reporting of Acacia laeta as a triploid hybrid is the first observed case of polyploidy in this complex. More intensive sampling in the entire natural Sudano-Sahelian range is required to improve these results.
Gene dispersal efficiency of diploid and tetraploid A. senegal
F IS did not significantly differ from zero in diploids whereas it was highly significant and negative in tetraploids (F IS = −0.56) reflecting an excess of heterozygotes that could be explained by self-incompatibility or asexual reproduction. Accordingly, either there was no breakdown of the self-incompatibility system in A. senegal in response to low hybrid fitness (Petit et al. 1999) or these populations are still in the process of asexual reproduction (e.g., apomixis), which may enable polyploids to survive until conditions become favorable. Such a situation allows ecological speciation (Hegarty and Hiscock 2008).
Population differentiation and genetic structure were not the same in diploid and tetraploid A. senegal in the Sudano-Sahelian belt. The high F ST value (F ST = 0.35) among tetraploids was confirmed by the dendrogram (Fig. 7), while differentiation among diploids was shown to be weak (F ST = 0.09). This result suggests that diploid and tetraploid A. senegal do not have the same gene dispersal capabilities. But to draw such a conclusion, it has to be assumed that migration–drift equilibrium has been reached by both cytotypes, whereas the above analyses of genetic diversity suggest that equilibrium has not yet been reached in tetraploid populations. According to the results of AMOVA, the proportion of gene diversity distributed within diploid populations is larger than that distributed between them, whereas in tetraploid populations the reverse is true, only 7 % of diversity was shared by tetraploids and only 21 % of variation was observed among cytotypes. Taken together, these results suggest that diploids and tetraploids are reproductively isolated. As is generally true in polyploid complexes, partial barriers to intercytotype gene flow may explain these patterns of variation (Gauthier et al. 1998; Husband and Schemske 1998, 2000). Gene flow may occur through intercytotypes via the fertilization of a tetraploid by unreduced gametes from a diploid and/or via backcrosses between a hybrid triploid and one parental cytotype (Ramsey and Schemske 1998) or through the recurrent formation of tetraploids from existing diploid species (Soltis et al. 1993). The latter hypothesis appears to correspond to A. senegal tetraploids as they are highly differentiated and do not have the same cytoplasm. Some studies of polyploid plants emphasized that the formation of polyploids is a continually repeated process, at rates close to those of genic mutation (Ramsey and Schemske 1998), and Soltis and Soltis (2000) stressed that multiple origins of polyploids is the rule rather than the exception. The recurrent formation of polyploids within a species can create a mosaic of existing diploid populations and provide independently established polyploid populations throughout the range of the species concerned (Soltis et al. 1993). The establishment of newly formed polyploids will depend on their ability to either colonize new habitats or to persist in sympatry with the parental populations. In the latter situation, they can suffer from the “minority cytotype disadvantage” (Husband 2000), whereby mating opportunities with other polyploids or backcrosses with one or both parental taxa will be rare. Allopolyploids may have more success in establishing themselves than autopolyploids thanks to their hybrid origin, which allows them to easily overcome barriers such as competition with progenitor species (Baack and Stanton 2005; Rausch and Morgan 2005). Polyploids may also become established if they are able to occupy new habitats where the progenitor taxa are not present. To identify the strategy used by tetraploids A. senegal, further investigations with an appropriate sampling plan will be required to address the many questions raised by the present study.
Occurrence of tetraploids in A. senegal
Polyploidy, i.e., the possession of more than two basic chromosome sets in nuclei, is regarded as a major force driving plant evolution and speciation (Grant 1981; Levin 2002; Rieseberg and Willis 2007). It is estimated that about 50 % of angiosperms are polyploids (Grant 1981; Masterson 1994), and cytogenetic studies indicate that many diploids were in fact previously polyploids (Leitch and Bennett 1997). This study is the first report of tetraploidy in A. senegal and this finding confirms the difficulty involved in delimiting the species. In addition to wide individual variability, A. senegal is able to hybridize naturally with other species and produce new viable tetraploid offspring or new viable and fertile diploid species. A. laeta, which has 2n = 39 chromosomes, is a triploid hybrid between A. senegal var. senegal x Acacia mellifera subsp. mellifera (Khan 1951; El Amin 1976; Ross 1979), even though both presumed parents are diploid. The occurrence of tetraploids in A. senegal could fuel the ongoing debate about the new systematic of the genus Acacia (Maslin et al. 2003; Newmaster and Ragupathy 2009).
Several questions arise including the origin of A. senegal tetraploids. The results of our molecular analyses suggest the occurrence of allopolyploidy. Allopolyploids represent permanent hybrids and possess the genes and gene products of both diploid progenitors. They thus have the capacity to produce novel heterodimeric proteins that are not produced by either diploid parent (Soltis et al. 2004). If A. senegal tetraploids are allotetraploids, gum arabic (i.e., water soluble proteinaceous polysaccharides) from different tetraploid populations may differ in composition and hence in quality. A number of molecular studies at the genetic, transcriptional and epigenetic levels have provided insights into possible mechanisms behind the phenotypic variability of polyploid species (Hegarty and Hiscock 2008). Importantly, recent evidence from proteomic studies suggests that the protein level is impacted by transcriptional changes observed in microarray experiments. While studying synthetic Brassica allopolyploids, Albertin et al. (2007) identified several proteins (305 in the stem, 200 in roots) that displayed nonadditive changes to translation compared with the parental taxa.
The impact of polyploidy on the population phenotype is potentially vast and polyploidy is often accompanied by adaptive advantages (Soltis et al. 2004; Prentis et al. 2008). Field-based model experiments should allow the adaptive significance of A. senegal tetraploids to be demonstrated, and future studies of association genetics are thus required to determine whether the genetic variability observed in tetraploids is indeed adaptive.
Conclusion
The present study revealed the presence of tetraploids in A. senegal. We compared genetic diversity and structure among populations and cytotypes within the polyploidy complex. Based on the results of our analyses, a significant fraction of diversity in this species is found in diploid populations. When these populations are maintained across the natural distribution range, the generation of new tetraploid lineages is possible. Once several polyploidy populations have been established, mating among polyploids of independent origin can create novel genetic combinations that may subsequently increase genetic diversity of A. senegal. These results are only based on our sample but nevertheless add a new level of complexity to gum trees. They also raise questions whose answers could influence sustainable management of genetic diversity, genetic improvement, and the productivity of the gum belt. Exact knowledge of the original genetic pool and its variation is crucial for effective management as well as for genetic improvement of A. senegal. Larger-scale studies are now needed to better understand the origin and extent of tetraploid populations in this complex of this species.
References
Addinsoft© (1995-2007) XLSTAT Version 2007.8.04. Available at http://wwwxlstatcom
Albertin W, Alix K, Balliau T, Brabant P, Davanture M, Malosse C, Valot B, Thiellement H (2007) Differential regulation of gene products in newly synthesized Brassica napus allotetraploids is not related to protein function nor subcellular localization. BMC Genomics 8(56):15p
Anderson MJ, Willis TJ (2003) Canonical analysis of principal coordinates: a useful method of constrained ordination for ecology. Ecol 84(2):511–525
Assoumane AA, Vaillant A, Mayaki AZ, Verhaegen D (2009) Isolation and characterization of microsatellite markers for Acacia senegal (L.) Willd., a multipurpose arid and semi-arid tree. Mol Ecol Resour 9(5):1380–1383. doi:10.1111/j.1755-0998.2009.02669.x
Baack EJ, Stanton ML (2005) Ecological factors influencing tetraploid speciation in snow buttercups (Ranunculus Adoneus): niche differentiation and tetraploid establishment. Evol 59(9):1936–1944
Barnes RD, Marunda CT, Maruzane D, Zirobwa M (1999) African Acacias: genetic evaluation. Oxford Forestry Institute Department of Plant Sciences United Kingdom and Forest Research Centre Zimbabwe Forestry Commission Zimbabwe
Bentham G (1875) Revision of the suborder Mimoseae, vol 30. Transactions of the Linnean Society of London vol 30
Brown AHD, Young AG (2000) Genetic diversity in tetraploid populations of the endangered daisy Rutidosis leptorrhynchoides and implications for its conservation. Hered 85(2):122–129
Bruvo R, Michiels NK, D’Souza TG, Schulenburg H (2004) A simple method for the calculation of microsatellite genotype distances irrespective of ploidy level. Mol Ecol 13(7):2101–2106. doi:10.1111/j.1365-294X.2004.02209.x
Bryan GJ, McNicoll J, Ramsay G, Meyer RC, De Jong WS (1999) Polymorphic simple sequence repeat markers in chloroplast genomes of Solanaceous plants. Theor Appl Genet 99(5):859–867
Bukhari YM (1997) Cytoevolution of taxa in Acacia and Prosopis (Mimosaceae). Aust J Bot 45(5):879–891
Chevallier MH, Brizard JP, Diallo I, Leblanc JM (1994) La diversité génétique dans le complexe Acacia senegal. Bois For Trop 240(2):5–12
Clark LV, Jasieniuk M (2011) Polysat: an R package for polyploid microsatellite analysis. Mol Ecol Resour 11(3):562–566. doi:10.1111/j.1755-0998.2011.02985.x
Dice LR (1945) Measures of the amount of ecologic association between species. Ecol 26(3):297–302. doi:dx.doi.org/10.2307/1932409
El Amin HM (1976) Acacia laeta (R. Br.) ex. Benth., considered as a species of hybrid origin. Sudan Silva 3 (21):14-23
England PR, Usher AV, Whelan RJ, Ayre DJ (2002) Microsatellite diversity and genetic structure of fragmented populations of the rare, fire-dependent shrub Grevillea macleayana. Mol Ecol 11(6):967–977. doi:10.1046/j.1365-294X.2002.01500.x
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software Structure: a simulation study. Mol Ecol 14(8):2611–2620
Fagg CW, Allison GE (eds) (2004) Acacia senegal and the gum arabic trade. Tropical Forestry Papers 42. Oxford Forestry Institute
FAO (ed) (1998) Gum arabic. Compendium of food additive specifications. Food and Nutrition Paper 52, addendum 6, FAO, Rome
Felsenstein J (2005) PHYLIP (Phylogeny Inference Package) version 3.6. Department of Genome Sciences, University of Washington, Seattle, distributed by the authors at (http://evolution.gs.washington.edu/phylip.html)
Gauthier P, Lumaret R, Bédécarrats A (1998) Genetic variation and gene flow in Alpine diploid and tetraploid populations of Lotus (L. alpinus (D.C.) Schleicher/L. corniculatus L.). I. Insights from morphological and allozyme markers. Hered 80(6):683–693
Gerlach WL, Bedbrook JR (1979) Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res 7(7):1869–1885
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices. Version 2.9.3. Department of Ecology & Evolution, Biology Building, UNIL, CH-1015 Lausanne, Switzerland, distributed by the authors at (http://www2.unil.ch/popgen/softwares/fstat.htm)
Grant V (ed) (1981) Plant speciation 2nd edn. Columbia University Press, New York
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: Paleontological statistics software package for education and data analysis. Palaeontologia Electronica 4 (1):9 pp. Available at http://folk.uio.no/ohammer/past/
Hardy OJ, Vekemans X (2001) Patterns of allozyme variation in diploid and tetraploid Centaurea jacea at different spatial scales. Evol 55(5):943–954
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2(4):618–620. doi:10.1046/j.1471-8286.2002.00305.x
Hegarty MJ, Hiscock SJ (2008) Genomic clues to the evolutionary success of polyploid plants. Curr Biol 18(10):435–444. doi:10.1016/j.cub.2008.03.043
Husband BC (2000) Constraints on polyploid evolution: a test of the minority cytotype exclusion principle. Proc R Soc Biol Sci 267(1440):217–223. doi:10.1098/rspb.2000.0990
Husband BC, Schemske DW (1998) Cytotype distribution at a diploid-tetraploid contact zone in Chamerion (Epilobium) angustifolium (Onagraceae). Am J Bot 85(12):1688–1694
Husband BC, Schemske DW (2000) Ecological mechanisms of reproductive isolation between diploid and tetraploid Chamerion angustifolium. J Ecol 88(4):689–701. doi:10.1046/j.1365-2745.2000.00481.x
Jørgensen MH, Carlsen T, Skrede I, Elven R (2008) Microsatellites resolve the taxonomy of the polyploid Cardamine digitata aggregate (Brassicaceae). Taxon 57(3):882–892
Khan IR (1951) Study of somatic chromosomes in some Acacia species and hydrids. Pak J For 1(4):326–341
Kloda JM, Dean PDG, Maddren C, MacDonald DW, Mayes S (2008) Using principle component analysis to compare genetic diversity across polyploidy levels within plant complexes: an example from British Restharrows (Ononis spinosa and Ononis repens). Hered 100(3):253–260. doi:10.1038/sj.hdy.6801044
Leitch IJ, Bennett MD (1997) Polyploidy in angiosperms. Trends Plant Sci 2(12):470–476
Levin DA (ed) (2002) The role of chromosomal change in plant evolution. Oxford Series in Ecology and Evolution. Oxford University Press, New York
Luttikhuizen PC, Stift M, Kuperus P, Van Tienderen PH (2007) Genetic diversity in diploid vs. tetraploid Rorippa amphibia (Brassicaceae). Mol Ecol 16(17):3544–3553. doi:10.1111/j.1365-294X.2007.03411.x
Maslin BR, Miller JT, Seigler DS (2003) Overview of the generic status of Acacia (Leguminosae: Mimosoideae). Aust Syst Bot 16(1):1–18. doi:10.1071/SB02008
Masterson J (1994) Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Sci 264(5157):421–424. doi:10.1126/science.264.5157.421
Michalakis Y, Excoffier L (1996) A genetic estimation of population subdivision using distances between alleles with special references for microsatellite loci. Genet 142(3):1061–1064
Miller JT, Bayer RJ (2003) Molecular phylogenetics of Acacia subgenera Acacia and Aculeiferum (Fabaceae: Mimosoideae), based on the chloroplast matK coding sequence and flanking trnK intron spacer regions. Aust Syst Bot 16(1):27–33
Moody ME, Mueller LD, Soltis DE (1993) Genetic variation and random drift in autotetraploid populations. Genet 134(2):649–657
Motlagh S, Ravines P, Karamallah KA, Ma Q (2006) The analysis of Acacia gums using electrophoresis. Food Hydrocoll 20(6):848–854
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genet 89(3):583–590
Newmaster SG, Ragupathy S (2009) Testing plant barcoding in a sister species complex of pantropical Acacia (Mimosoideae, Fabaceae). Mol Ecol Resour 9(supplement s1):172–180. doi:10.1111/j.1755-0998.2009.02642.x
Nicholson SE, Kim J, Hoopingarner J (eds) (1988) Atlas of African rainfall and its interannual variability. Florida State University, Dept. of Meteorology
Novick RR, Dick CW, Lemes MR, Navarro C, Caccone A, Bermingham E (2003) Genetic structure of Mesoamerican populations of Big-leaf mahogany (Swietenia macrophylla) inferred from microsatellite analysis. Mol Ecol 12(11):2885–2893. doi:10.1046/j.1365-294X.2003.01951.x
Obbard DJ, Harris SA, Pannell JR (2006) Simple allelic–phenotype diversity and differentiation statistics for allopolyploids. Hered 97(4):296–303. doi:10.1038/sj.hdy.6800862
Obunga EO (1995) A study of genetic systems of four African species of Acacia. DPhil, University of Sussex
Omondi SF, Kireger E, Dangasuk OG, Chikamai B, Odee DW, Cavers S, Khasa DP (2010) Genetic diversity and population structure of Acacia senegal (L) Willd. in Kenya. Trop. Plant Biol 3(1):59–70
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6(1):288–295
Perrier X, Jacquemoud-Collet JP (2006) DARwin software. Cirad-Bios UPR Genetic Improvement of Vegetatively Propagated Crops, distributed by the authors at (http://darwin.cirad.fr/darwin)
Petit C, Bretagnolle F, Felber F (1999) Evolutionary consequences of diploid–polyploid hybrid zones in wild species. Trends Ecol Evol 14(8):306–311
Prentis PJ, Wilson JRU, Dormontt EE, Richardson DM, Lowe AJ (2008) Adaptive evolution in invasive species. Trends Plant Sci 13(6):288–294
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genet 155(2):945–959
Pritchard JK, Wen X, Falush D (2010) Documentation for the structure software: Version 2.3.Available at: http://pritch.bsd.uchicago.edu/structure_software/release_versions/v2.3.3/structure_doc.pdf
Ramsey J, Schemske DW (1998) Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu Rev Ecol Syst 29(1):467–501
Rausch JH, Morgan MT (2005) The effect of self-fertilization, inbreeding depression, and population size on autopolyploid establishment. Evol 59(9):1867–1875
Rieseberg LH, Willis JH (2007) Plant speciation. Sci 317(5840):910–914. doi:10.1126/science.1137729
Robertson A, Rich TCG, Allen AM, Houston L, Roberts C, Bridle JR, Harris SA, Hiscock SJ (2010) Hybridization and polyploidy as drivers of continuing evolution and speciation in Sorbus. Mol Ecol 19(8):1675–1690
Robinson J, Harris SA (2000) A plastid DNA phylogeny of the genus Acacia Miller (Acacieae, Leguminoseae). Bot J Linn Soc 132(3):195–222. doi:10.1111/j.1095-8339.2000.tb01527.x
Ross JH (1975) The Acacia complex. Bothalia 11(4):453–462
Ross JH (ed) (1979) A conspectus of the African Acacia species. Memoirs of the Botanical Survey of South Africa volume 44 Botanical Research Institute, Dept. of Agricultural Technical Services
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Salvo-Garrido H, Travella S, Schwarzacher T, Harwood WA, Snape JW (2001) An efficient method for the physical mapping of transgenes in barley using in situ hybridization. Genome 44(1):104–110
Sampson JF, Byrne M (2012) Genetic diversity and multiple origins of polyploid Atriplex nummularia Lindl. (Chenopodiaceae). Biol J Linn Soc 105(1):218–230
Slatkin M (1995) A measure of population subdivision based on microsatellite allele frequencies. Genet 139(1):457–462
Soltis DE, Soltis PS (1989) Genetic consequences of autopolyploidy in Tolmiea (Saxifragaceae). Evol 43(3):586–594
Soltis DE, Soltis PS, Rieseberg LH (1993) Molecular data and the dynamic nature of polyploidy. Crit Rev Plant Sci 12(3):243–273
Soltis DE, Soltis PS, Tate JA (2004) Advances in the study of polyploidy since Plant speciation. N Phytol 161(1):173–191. doi:10.1046/j.1469-8137.2003.00948.x
Soltis PS, Soltis DE (2000) The role of genetic and genomic attributes in the success of polyploids. Proc Natl Acad Sci USA 97(13):7051–7057
Vassal J (1972) Apport des recherches ontogéniques et sémiologiques à l'étude morrphologique, taxonomique et phylogénique du genre Acacia. Bull Soc Hist Nat Toulouse 108:125–247
Wagner WH Jr (1970) Biosystematics and evolutionary noise. Taxon 19(2):146–151
Weising K, Gardner RC (1999) A set of conserved PCR primers for the analysis of simple sequence repeat polymorphisms in chloroplast genomes of dicotyledonous angiosperms. Genome 42(1):9–19
Wright S (1951) The genetical structure of populations. Ann Eugenics 15:323–354
Acknowledgments
We are grateful to the Faculty of Sciences of Abdou Moumouni University (Niger) for help in collecting A. senegal samples in its natural range in Niger. We are also grateful to CIRAD-Bios (Centre de Coopération Internationale en Recherche Agronomique pour le Développement, Department Systèmes Biologiques, France), Research Unit 39, where molecular analyses were carried out. This work was supported by Acaciagum project (FP6-INCO-32233) and by IRD (Institut de Recherche pour le Développement, France).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Byrne
Rights and permissions
About this article
Cite this article
Assoumane, A., Zoubeirou, A.M., Rodier-Goud, M. et al. Highlighting the occurrence of tetraploidy in Acacia senegal (L.) Willd. and genetic variation patterns in its natural range revealed by DNA microsatellite markers. Tree Genetics & Genomes 9, 93–106 (2013). https://doi.org/10.1007/s11295-012-0537-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-012-0537-0