Abstract
Genetic parameters for stem diameter and wood density were compared at selection (4–5 years) and harvest (16–17 years) age in an open-pollinated progeny trial of Eucalyptus globulus in Tasmania (Australia). The study examined 514 families collected from 17 subraces of E. globulus. Wood density was assessed on a subsample of trees indirectly using pilodyn penetration at both ages and directly by core basic density at harvest age. Significant additive genetic variance and narrow-sense heritabilities (\( h_{\text{op}}^2 \)) were detected for all traits. Univariate and multivariate estimates of heritabilities were similar for each trait except harvest-age diameter. Comparable univariate estimates of selection- and harvest-age heritabilities for diameter masked changes in genetic architecture that occurred with stand development, whereby the loss of additive genetic variance through size-dependent mortality was countered by the accentuation of additive genetic differences among survivors with age. Regardless, the additive genetic (r a) and subrace (r s) correlations across ages were generally high for diameter (0.95 and 0.61, respectively) and pilodyn penetration (0.77 and 0.96), as were the correlations of harvest-age core basic density with selection- and harvest-age pilodyn (r a −0.83, −0.88; r s −0.96, −0.83). While r s between diameter and pilodyn were close to zero at both ages, there was a significant change in r a from adverse at selection age (0.25) to close to zero (−0.07) at harvest age. We argue that this change in the genetic correlation reflects a decoupling of the genetic association of growth and wood density with age. This result highlights the need to validate the use of selection-age genetic parameters for predicting harvest-age breeding values.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Reliable genetic parameters are important for estimating breeding values and gains from selection in forest tree breeding programs. Nearly all traits that are used to select trees (selection traits) are indirect estimates of the actual traits for which breeders seek to obtain breeding values. For example, diameter is a surrogate for whole-tree volume, core density for whole-tree density, and core predicted pulp yield for whole-tree pulp yield (Greaves et al. 1997a). In addition, the relatively long rotation time of many forest plantation crops means that selection for harvest-age traits is usually done much earlier than harvest age, in order to reduce the generational interval for breeding and speed the capture of genetic gains (Cotterill and Dean 1990; White et al. 2007). Effective selection of harvest-age traits in this manner requires significant heritability of the selection and harvest-age traits and a genetic correlation between them (Falconer and Mackay 1996; White et al. 2007). A further complication is introduced by the general requirement of breeding to improve two or more traits simultaneously. This introduces the requirement to know the strength, sign, and stability of the genetic correlation between traits at both selection and harvest age. In practice, only early-age genetic parameters are usually known and genetic parameters for harvest age are assumed to be the same by necessity (White et al. 2007).
Obtaining accurate, unbiased estimates of harvest-age parameters in forest tree populations is difficult as they may be affected by the increased competitive environment in older field trials (Franklin 1979; Bouvet et al. 2005), and the fact that mortality through artificial culling or natural attrition means that at harvest age, the population is a subset of that at earlier age. Mortality may cause a loss of additive genetic variance when it is selective, for example, size dependent (Chambers and Borralho 1997). Thus, latter age parameters may underestimate the additive genetic variance which resided in the original parental population. Multivariate analysis using both early- and late-age measurements to account for selective mortality offers the potential to better estimate genetic parameters for harvest-age traits and to better understand the manner in which these change with age (Wei and Borralho 1998; Dieters et al. 1999).
Eucalyptus globulus Labill. is the major eucalypt grown in industrial pulpwood plantations in temperate regions of the world (Potts et al. 2004). Breeding objectives for kraft pulp production in this species have been defined based on the key harvest-age traits of stem volume per hectare, wood basic density, and pulp yield (Borralho et al. 1993; Greaves et al. 1997a). The rotation length of E. globulus pulpwood plantations range from 8 to 20 years, depending on growth rates (Borralho et al. 1992a). Trees are frequently reproductively mature by 4 years of age (Chambers et al. 1997; Tibbits et al. 1997), and early selection for breeding and deployment usually occurs between 4 and 6 years of age (Borralho et al. 1992a; McRae et al. 2003). The selection-age traits assessed as predictors of harvest breeding objective traits are stem diameter at breast height (DBH; Borralho et al. 1992a; Lopez et al. 2002); wood specific gravity (density), assessed from a breast height wood core (Muneri and Raymond 2000) or using pilodyn penetration (MacDonald et al. 1997; Sanhueza et al. 2002); and pulp yield, estimated from powdered basal core wood using near-infrared chemometrics (Raymond et al. 2001; Apiolaza et al. 2005; Downes et al. 2007). Heritability estimates for growth of E. globulus, particularly early age estimates from open-pollinated trials, are commonly reported (Potts et al. 2004). However, there are fewer heritability estimates for wood quality traits and fewer still genetic correlations that include wood properties other than basic density (Raymond 2002). The published genetic correlations mainly involve selection (early)-age traits, leaving a paucity of reported correlations either between selection- and harvest-age traits, or between different traits at harvest age (Borralho et al. 1992a; Lopez et al. 2002; Greaves et al. 2003).
We used a harvest-age E. globulus base population trial to determine (a) the correlations between selection- and harvest-age diameter and wood density, (b) whether there is a genetic basis to mortality which occurred over this time interval, (c) whether this mortality is genetically correlated with any selection traits and has thus biased harvest-age genetic parameter estimates, and (d) whether the genetic parameters estimated from the selection-age population are the same as those in the surviving harvest-age population.
Materials and methods
Trial and measurements
The study is based on a field trial established with open-pollinated seedlots collected from individual trees from throughout the geographic range of E. globulus (Potts et al. 2004). These seedlots have been classified into a geographic hierarchy of 13 races and 20 subraces (Dutkowski and Potts 1999) with several additional races or subraces subsequently recognized (http://members.forestry.crc.org.au/globulus/). Trials established from these seed collections have been the subject of numerous studies (e.g., Potts et al. 2004; Costa e Silva et al. 2005). The present study focuses on one of the largest trials established from this collection. The trial is located near Latrobe in northern Tasmania, Australia (41°13′ S, 146°23′ E) and was planted in 1989. It includes 561 open-pollinated families, established in a randomized incomplete block design comprising five replicates of 24 incomplete blocks each. Each incomplete block contained 30 plots each containing two trees from a family. Each family was thus represented by ten trees at establishment. The trial includes races and subraces from the Gippsland and Otway districts of Victoria, the Bass Strait Islands, and Tasmania (Dutkowski and Potts 1999). However, the atypical races, West Tasmania, Dromedary, and Recherche Bay (all from Tasmania) as well as Wilson’s Promontory Lighthouse (Victoria) were excluded from the present study (Table 1).
Diameter at breast height (1.3 m above ground level) was measured on all trees alive at ages 4 (DBH4), 8 (DBH8), and 16 (DBH16) years. Diameter increment during the periods 4–8 years (ΔDBH4-8) and 8–16 years (ΔDBH8-16) was calculated. Survival of each tree was recorded as 0 (dead) and 1 (alive) at each age. A subset of trees was sampled for assessments of wood density and a subsample of these taken for assessment of harvest-age pilodyn. Pilodyn penetration was measured with a pilodyn model 6J with a 2.5-mm-diameter flat-faced striker pin. The tool operates by driving the pin into the specimen using a fixed force. Pilodyn penetration was measured in the spring of 1995 at the age of 6 years (PIL6) on one tree randomly chosen per two-tree plot in replicates 1 and 4, as well as the trees with the largest diameter (top 10%) throughout the trial (MacDonald et al. 1997). Pilodyn penetration was also measured in the summer (January) of 2006 at age 17 years (PIL17) from the same trees that were cored for basic density at age 16 years, mainly in replicates 1 (440 trees) and 2 (378 trees) and 42 trees from replicate 3 of any family not represented by two individuals from replicates 1 and 2. Bark was removed from a 5-cm-wide × 10-cm-high window on the west aspect where possible and pilodyn penetration measured at two positions in the same window, one above the other directly against the cambium and averaged.
Two months after the age 16-year-diameter measurement, basal wood cores were collected from one tree per plot in at least four of the five replicates. Trees were effectively chosen at random, with the first tree in each two-tree plot sampled, except where the diameter of that tree was less than 10 cm, or the tree was absent or dead, which triggered the selection of the second tree if suitable by the same criteria. Cambium-to-cambium, 12-mm-diameter cores that aimed to pass through the pith of each tree were taken at 1.1 m above ground level using a motorized increment corer (Raymond and Muneri 2001). Cores were immediately wrapped in plastic, kept cool in the field, and subsequently frozen until laboratory processing. Each core was randomly allocated to one of 73 30-core batches and batches processed in numerical order. Each core was sectioned lengthwise in the horizontal plane with one half randomly selected and set aside for chemical analysis. Basic density (DEN16) of the remaining half of each core was determined gravimetrically, whereby the oven-dried weight (105°C, 24 h) of the half-core was divided by its fresh volume as determined by the weight of water it displaces (TAPPI 1989).
In order to validate the use of cores for assessing basic density, the correlation with whole-tree basic density was measured at age 17 years. Twenty trees were randomly selected for sampling, five trees of which were selected from each of the Otways, Strzelecki, Furneaux, and Tasmanian regions. In each region, one tree was picked at random from each 20 percentile for diameter. Trees were felled, and disks of equal thickness were recovered from the 10%, 30%, 50%, 70%, and 90% of total tree height. For each tree, discs were debarked, chipped, and bulked, and two samples were taken for determination of basic density on the chips (APPITA 2002). Regression of whole-tree density to that measured from cores was reasonably high with an R 2 of 0.61, which validated the core density as being a good indicator of whole-tree density (Fig. 1).
Statistical model and estimation of variance components and heritabilities
The following mixed model was fitted to the data:
where Y is the vector of observations, MEAN is the mean, REP is the effect of replicate fitted as a fixed effect, SUBRACE is the subrace effect fitted as a fixed effect (subrace as defined by Dutkowski and Potts (1999) with subsequent modifications), IBLOCK(REP) is the random variance between incomplete blocks within replicates, FAMILY(SUBRACE) is the random variance between open-pollinated families nested within subrace, PLOT is the random variation between the two tree plots and is fitted only for diameter traits, and RESIDUAL is the vector of residuals. The fixed subrace differences were tested with the type III sums of squares using the FAMILY(SUBRACE) term as the error using PROC MIXED in SAS 9.1 (SAS Institute Inc. 2002).
Variance components for random effects were estimated with both univariate and multivariate versions of model 1 using ASReml (Gilmour et al. 2001). Univariate estimates of variance components for DBH were based on (a) all plants alive at each age or (b) a restricted data set of only those individuals alive at age 16 years. The multivariate estimates of variance components for DBH were based on a three-trait analysis involving DBH4, DBH8, and DBH16 (data presented), whereas multivariate estimates for the wood property traits were based on a five-trait analysis: DBH4, DBH16, PIL6, PIL17, and DEN16. Variance components estimated in the univariate analyses were used as starting values for multivariate analyses. In these analyses, dead or unmeasured trees (in the case of pilodyn and DEN) were treated as missing values. The variance components for DBH16 were thus estimated from only the surviving population (univariate) as well as by using the information from related individuals and genetic correlations to the age 4- and 8-year-diameter measurements in the full multivariate model to account for the effect of mortality over the 4- to 16-year interval. Testing of the significance of the variance components from zero was undertaken using one-tailed likelihood ratio test (LRT; Self and Liang 1987), comparing the difference in log likelihood of the full model vs one constrained by removing the term from the analysis (univariate case) or constraining the variance component to zero (multivariate case). Variance components for survival at age 16 years were calculated using a binary model with probit link function following Chambers and Borralho (1997) and testing the significance of the variance components with the Z test (Zar 1974).
The narrow-sense heritability (\( h_{\text{op}}^2 \)) was calculated following MacDonald et al. (1997) using variance components estimated from the univariate (full and restricted data sets) and multivariate models as:
where \( \sigma_{{{\text{family}}\left( {\text{subrace}} \right)}}^2 \) is the between open-pollinated families within subrace variance component, \( \sigma_{\text{plot}}^2 \) is the between plot within family variance component, and \( \sigma_{\text{e}}^2 \) is the error variance component. Following Griffin and Cotterill (1988), the coefficient of relatedness (r) assumed for these open-pollinated progenies was 0.4. This value of r accounts for the mixed mating system of E. globulus and assumes that 30% of the seed in each open-pollinated family is the result of self fertilization. The \( \sigma_{\text{plot}}^2 \) was only included for DBH traits, as only a single tree per plot was assessed for the other traits. The additive genetic variance (\( \sigma_{\text{a}}^2 \)) was estimated as \( \sigma_{{{\text{family}}\left( {\text{subrace}} \right)}}^{\text{2}} \) divided by r. Differences in heritabilities were tested with Z tests, using the pooled standard error as the denominator.
To compare the levels of additive genetic variance in each trait independent of their means, the additive genetic coefficient of variation (CVa) was calculated following Cornelius (1994) as:
where \( \overline{x} \) is the trait phenotypic mean in the trial and σ a was defined previously.
Estimation of genetic correlations
The genetic correlations among traits were calculated at both the family within subrace, hereafter referred to as the additive genetic correlation (r a) and subrace levels (r s) as:
where r 1,2 is the correlation between traits 1 and 2 at a defined genetic level (either the additive genetic or subrace level), σ 1,2 is the covariance between the traits, and \( \sigma_1^2 \) and \( \sigma_2^2 \) are the variance components for each trait (Jordan et al. 1999). The additive genetic correlations (r a) were estimated directly in ASReml using the CORR directive to structure the G matrix in five-trait multivariate analyses fitting model 1. The subrace correlations used variance components estimated from a model similar to (1) but treating subrace as a random effect. However, in this form, the five-trait multivariate model did not converge, and it was necessary to use bivariate analyses to estimate pair-wise r s values. The multivariate analyses were undertaken for DBH measurements as well as using incremental DBH data (ΔDBH4–8 and ΔDBH8–16) to better match the growth response period to the pilodyn assessment age (Zamudio 1995). Following Chambers et al. (1996), bivariate analyses were also undertaken to estimate r a of DBH4 and PIL6 with the binary survival data at age 16 years (SURV16). Phenotypic correlations were estimated using PROC CORR in SAS.
The significance of additive genetic and subrace correlations was tested using the LRT. The probability of differences from test correlations were obtained by fitting the statistic to the chi-square distribution with one degree of freedom where the test is two-tailed against zero and 0.5 degrees of freedom when the test is one-tailed against |1| (Stram and Lee 1994; Costa e Silva et al. 2006). One-tailed tests are appropriate when high correlations are anticipated (e.g., age–age correlations for similar traits DBH4 vs DBH16; PIL6 vs PIL17; PIL6 vs DEN16), whereas tests from zero were appropriate for intertrait correlations (e.g., DBH4 vs PIL6) where there was no a priori reason to assume they should be correlated. The stability of trait–trait genetic correlations over time (e.g., PIL6 and DBH4 vs PIL17 and DBH16) was tested using the two-tailed LRT, where the difference in log likelihood between (a) the optimal five-trait multivariate model and (b) a model restricted by the directive to seek a solution whereby the above correlations would be equal. These multivariate analyses and tests were undertaken using the full and restricted data sets (as with the heritabilities) and also replacing DBH measures with the DBH incremental data. The standard errors of these correlations and \( h_{\text{op}}^2 \) were also estimated using a truncated Taylor series (Gilmour et al. 2001).
Results
Over the period studied, the annual diameter increment of stems declined from a mean of 17 mm/year for the first 4 years to 12 mm/year between 5 and 8 years to only 8 mm/year over the 8–16-year period. The mean pilodyn penetration decreased from 11.8 mm at 5.5 years to 9.9 mm at 17 years (Table 2). The considerably reduced diameter increment over the latter period indicates that unlike the age 4 pilodyn reading, more than one annual growth ring was penetrated in the later pilodyn measurement. Significant subrace and family within subrace effects were detected for all traits at all ages (P < 0.003; Table 3), indicating significant genetic variation at both levels. At the subrace level, survival at age 16 was not significantly correlated with early diameter (r s = 0.25; Table 4). However, at the family level, these two variables were significantly and positively correlated (r a = 0.92; Table 4), indicating that within subraces, trees with larger diameter at early age had a higher probability of surviving to age 16 years.
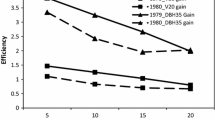
The univariate heritability estimates of diameter were not significantly different across the three measurement ages (Fig. 2a), giving the impression that the heritability of diameter is stable with increasing age. However, this apparent stability masked a dynamic genetic architecture in these open-pollinated progenies, where size-dependent mortality resulted in a loss of additive genetic variation by harvest age. This reduction was evident from the change in heritabilities in the different cohorts and analyses shown in Fig. 2 and Table 3. Univariate heritability estimated for DBH4 with all trees was 0.18, and this declined significantly (Z = 2.22, P = 0.028) to 0.07 when estimated using only the cohort of trees that survived to age 16 years. The coefficient of additive genetic variation (CVa) for diameter traits also decreased with increasing age (7.6% to 4.3%; Table 3). This reduction in additive genetic variation for growth that was evident in successive univariate analyses was countered by the increasing expression of genetic variation of DBH with age. The evidence for this was the increase in heritability of diameter between DBH4 and DBH16 (0.07 to 0.15) when the analysis included only the cohort of trees that survived to age 16 years (Fig. 2b). This trend occurred regardless of whether the analyses were univariate (Z = 2.55, P = 0.011; Fig. 2) or multivariate including DBH4 and DBH16 (Z = 2.19, P < 0.02; data not shown). Indeed, the same heritability estimate was obtained for the age 16 data regardless of whether the univariate or multivariate analysis was used. An increase was similarly observed for CVa when analyses were restricted to survivors at age 16 (univariate—DBH4 = 4.3% and DBH16 = 7.1%). Using the multivariate analysis to account for the size-dependent mortality that occurred between ages 4 and 16 years allowed a true estimate of the level of genetic variation that occurred in the original parental population for DBH16 as it projects the genetic variation lost through mortality to later ages based on genetic correlations and performance of surviving relatives. In this case, the multivariate heritability for DBH16 was 0.29 compared to 0.16 for DBH4, a significant increase (Z = 2.66 P = 0.005). The univariate heritability estimate of 0.15 for DBH16 underestimated the proportion of additive genetic variation in DBH16 in the overall parental population, but indicated the amount of variation that remained at age 16 years.
Age trend in the heritability (\( h_{\text{op}}^2 \)) of diameter (DBH) from open-pollinated families of E. globulus at the ages of 4, 8, and 16 years. a Univariate heritability (open squares) shows no significant change with advancing age, while b indicates the trend for increasing univariate heritability with age (filled triangles), for only the cohort of trees which survived. c The trajectory of heritability for DBH from the multivariate analysis that includes all data (filled squares) and projects the \( h_{\text{op}}^2 \) that may be realized for DBH at age 16 years, in the absence of mortality
A detailed analysis of temporal dynamics for wood density was not possible as only a subset of the trees was sampled for pilodyn or cored in each sampling period. However, aging and mortality appeared to have had little impact on heritability or CVa estimated for the wood density traits. The heritability of pilodyn did not change significantly with age, although the range was greater in the univariate (\( h_{\text{op}}^{\text{2}} = 0.35 - 0.55 \)) than in the multivariate analysis (\( {\text{h}}_{\text{op}}^{\text{2}} = 0.44 - 0.49 \)). The heritability of density (DEN16) was 0.52 in both uni- and multivariate analysis (Table 3).
Within subraces, the age–age additive genetic correlations (r a) for diameter were high, as were those for wood density, although almost all were significantly less than 1 (Table 4). The highest age–age correlation was for DBH4–DBH16 (r a = 0.95). The additive genetic correlation between PIL6 and PIL16 was also high (r a = 0.77), as was the correlation between early pilodyn and harvest-age core density (r a = −0.83). At the subrace level, the age–age correlations for wood density were higher than that for DBH. The age–age correlation for pilodyn penetration was not significantly different from 1 (r s = 0.96) but was for DBH (r s = 0.61; Table 4). The lower age–age subrace correlation for DBH was mainly due to the rank of the subraces from the Otway region increasing with age and that of the Furneaux and northeast Tasmanian subraces declining with age (Fig. 3). The phenotypic age–age correlations were all significantly different from zero (DBH4–DBH16 r p = 0.76, PIL6–PIL17 r p = 0.54; Table 4). The genetic correlation between DBH and pilodyn declined with age (Fig. 4). There was a significant (P < 0.05) adverse additive genetic correlation between DBH4 and PIL6 (r a = 0.26), but the correlation between DBH16 and PIL17 was effectively zero (r a = −0.07), as was the correlation between DBH16 and DEN16 (r a = 0.08; Table 4). This loss of correlation was robust and occurred whether full or restricted data sets were used, whether multivariate models that incorporated all five traits DBH4, PIL6, DBH16, PIL17, and DEN16 (Table 4) or separate bivariate analyses were undertaken (DBH4–PIL6 compared with DBH16–PIL17; data not shown), or whether different growth intervals or increments were analyzed (Fig. 4). These differences in selection and harvest-age genetic correlations between traits were statistically significant for the full data set (r a DBH4–PIL6 vs r a DBH16–PIL17; \( \chi_1^2 = 4.7 \), P = 0.03). The differences had higher significance levels when using 8-year DBH rather than 4-year DBH (e.g., r a DBH8–PIL6 vs r a DBH16–PIL17; \( \chi_1^2 = 9.0 \), P = 0.003), as well as when growth increments of 4–8 and 8–16 years were correlated with PIL6 and PIL17, respectively (full data \( \chi_1^2 = 9.5 \), P = 0.002; restricted dataset \( \chi_1^2 = 5.3 \), P = 0.021; Fig. 4). The only significant phenotypic correlation between growth and density was the weak negative correlation between DBH16 and PIL17 (r p = −0.08; Table 4).
Relationship between best linear unbiased predictors (BLUP) of E. globulus subrace effects for diameter (DBH) at age 4 and at age 16 years. The diagonal line indicates the 1:1 relationship, with races above the diagonal being those that performed better at 16 relative to age 4, and races below the diagonal being those that performed better at age 4 relative to age 16. The circled subraces are discussed in the text
Trait–trait additive genetic correlations (r a ± s.e.) between stem diameter (DBH4, DBH16) or diameter increments (ΔDBHage1–age2) and wood density traits in a E. globulus base population trial. The wood density traits are pilodyn penetration at age 6 years (PIL6) and 17 years (PIL17) and core basic density at age 16 years (DEN16). Gray bars are the correlations estimated with a multivariate model in which DBH4 and DBH8 and increments thereof were restricted to only those that survived to age 16
Discussion
Our selection (4-6 years) and harvest-age (16-17 years) univariate estimates of narrow-sense heritability for diameter (\( h_{\text{op}}^2 = 0.15 - 0.18 \)) were similar to the average of published estimates for other open-pollinated progeny trials of E. globulus (Potts et al. 2004). These estimates were also comparable to those from other open-pollinated tree species as reviewed by Cornelius (1994). Our study period of 11 years is only matched by that of Borralho et al. (1992b), who similarly found stable univariate heritability for stem basal area traits in E. globulus over a 14-year interval (\( h_{\text{op}}^2 \) age 4 = 0.17 and age 18 = 0.14). Results from shorter periods vary, with Lopez et al. (2002) reporting an increase in narrow-sense heritability of stem diameter over a 2-year period as did Volker (2002) over 4 years, both studies commencing earlier with little mortality during the interval. Lopez et al. (2003) reported an increase in the expression of additive genetic variation for height growth of E. globulus from the nursery to the last measurement in the field at 16 months of age, by which time significant additive genetic effects were detected, whereas maternal and nursery effects had subsided. In our case, the apparent stability in the univariate heritability and coefficient of additive genetic variation appeared to mask a dynamic genetic architecture in these open-pollinated progenies arising from the interplay between the effects of mortality and competition. When exactly the same cohort of trees (the restricted data set) was studied, the expression of genetic variation for growth and hence heritability tended to increase with age. However, this increase was countered by size-dependent mortality that removed genetic variation and thus acted to decrease heritability estimates.
Genetic-based, size-dependent mortality within trials of E. globulus has been previously reported (Chambers et al. 1996) and has the potential to reduce levels of genetic variance for growth traits in harvest-age populations compared with that in selection-age populations. The mortality of genetically small trees from genetic trials is comparable to their artificial removal by thinning, and the effect of either process is to reduce the available genetic variability (Matheson and Raymond 1984; Wei and Borralho 1998). Therefore, with ongoing mortality occurring, the genetic parameters estimated for a trait (e.g., DBH) at different ages are not directly comparable, as they are estimated from slightly different populations, with the later age population being a subset of the former (Hadfield 2008). E. globulus is known to exhibit significant genetic variation for adaptive traits which could impact on growth and survival. These include abiotic factors such as frost (Tibbits et al. 2006) and drought resistance (Dutkowski 1995) and biotic factors such as mammalian (O’Reilly-Wapstra et al. 2002) and insect browsing (Rapley et al. 2004) and susceptibility to leaf disease (Milgate et al. 2005). The species also exhibits severe inbreeding depression for growth and survival (Hardner and Potts 1995; Hardner et al. 1998). E. globulus has a mixed mating system (Potts et al. 2004) that results in a proportion of seed produced being inbred and having lower fitness which is typically manifested as slower growth rate, which is known as inbreeding depression. This inbreeding depression coupled with variation in outcrossing rates in the wild (Hardner et al. 1998) is believed to be a major factor driving differences in the performance of open-pollinated families in common-garden trials of E. globulus (Borralho 1994; Borralho and Potts 1996; Hodge et al. 1996; Volker 2002) and other eucalypt species such as Eucalyptus grandis (Burgess et al. 1996). Such variation is believed to be the cause of inflated estimates of narrow-sense heritabilities derived from open-pollinated families (as used in the present study) compared to control-pollinated progeny trials of E. globulus (Hodge et al. 1996; Lopez et al. 2002; Volker 2002; Li et al. 2007).
Strong selection against inbred offspring has been shown to result in their elimination from field trials with time in both Eucalyptus regnans (Hardner and Potts 1997) and E. globulus (Hardner and Potts 1995), and in our case, this is likely to result in our 16-year-old population mainly comprising outcrossed individuals. Restriction of the 4-year-diameter analysis to only those trees alive at 16 years would thus be expected to focus the analysis on a population where most inbred genotypes have been removed, rendering estimates of heritability more in line with equivalent estimates from known outcrossed populations. Indeed, our age 4 diameter heritability when restricted this way (\( h_{\text{op}}^2 = 0.07 \)) was similar to estimates based on control-pollinated progenies of E. globulus (\( h_{\text{cp}}^2 \)) of 0.08 (Li et al. 2007) and 0.11-0.12 (Volker 2002). Nevertheless, inbreeding depression for survival is unlikely to be the sole explanation for the variation in heritabilities observed with age, as Volker et al. (2008) report a decline in univariate estimates of heritability for diameter in a fully outcrossed population of E. globulus from 0.19 at age 2 years to 0.10 at age 10 years.
Restricting the analysis to use only trees that survived to age 16 revealed that the heritability and additive coefficient of variation of diameter increased with age, similar to that found in other forest tree species where the survivors only are used (Balocchi et al. 1993; Wei and Borralho 1998), or where mortality was minimal (Kube et al. 2001; Osorio et al. 2001). This increase is likely due to stand development effects which can increase the expression of genetic variation and thus heritability of growth with age. Such an increase is believed to be due to differences in growth between genotypes being exacerbated in response to competition, with faster growing genotypes tending to suppress slower growing genotypes (Franklin 1979; Magnussen 1989). For example, there is some evidence from hybrid eucalypt clones to suggest that competition among genotypes may inflate the genetic variance of growth when it is estimated from single tree or line plots compared with block plantings where most competition is between trees of the same genotype (Retief et al. 2001). Indeed, Hannrup et al. (1998) consider genetic parameters for diameter to be especially susceptible to distortion due to competition and recommend early measurement of diameter for calculation of valid genetic parameters, whereas genetic parameters of wood properties are relatively unaffected over longer periods. However, in E. globulus and many eucalypts, it is difficult to separate competitive from ontogenetic effects on growth. The ontogenetic transition between juvenile and mature leaf morphology normally occurs between 2 and 5 years of age in E. globulus (Jordan et al. 1999). This ontogenetic change may impact on (Beadle et al. 1989), or be affected by growth (Jordan et al. 2000).
The use of multivariate models to account for difference in population structure through mortality (i.e., the “invisible fraction” of the population; Hadfield 2008) better reflects the true genetic variance in the parental population for the later age growth trait than using univariate models (Wei and Borralho 1998). Indeed, in our case when later age diameter heritabilities are estimated with the multivariate model, the true age trend in expression of genetic variance is revealed, with the estimated heritability increasing from 0.16 for age 4 to 0.29 for age 16 year diameter. This heritability increase is due to accounting for mortality rather than an effect of the multivariate analysis per se as when the restricted cohort is analyzed, univariate and multivariate heritability estimates are the same at age 16 years. The failure to account for mortality, coupled with differences in the genetic behavior of open-pollinated and controlled pollinated populations, is likely to explain much of the diversity of age trends in heritability estimates for growth traits reported in forest trees.
Despite the instability in genetic parameters for diameter growth, the age–age additive genetic correlations within subraces over a 12-year-period were very high (r a = 0. 95) and comparable with values reported over shorter periods for E. globulus (Lopez et al. 2002; Volker 2002), Eucalyptus nitens (Greaves et al. 1997b) and E. grandis (van Wyk et al. 1991; Osorio et al. 2003). This high correlation over a 12-year period is somewhat surprising as age–age correlations of growth traits generally decline as the intervals between measurements increase (Borralho et al. 1992b; Gwaze et al. 2000), at least within the first 20 years, but reinforces the use of early age growth measurements as predictors of harvest-age growth (Borralho et al. 1992a; Greaves et al. 2003). However, genetic correlations only account for the stability of genetic effects within genetic groups and do not account for the possibility that the performance of genetic groups such as subraces may be less stable with age, as we observed (r s = 0.61). In the present case, the lower genetic correlation at the subrace level appears to be driven by a change in the rank of several subraces between age 4 and 16, with consistent responses observed for subraces from the same geographic region or genetic lineage as defined using molecular markers (Steane et al. 2006). The relative performance of the three subraces from the Otway Range region (Western Otway, Eastern Otway, and Cape Patton) increased from 4 to 16 years, consistent with their high ranking for 4-year-diameter growth when averaged over 15 trials across Australia (Costa e Silva et al. 2006). Concomitantly, subraces from the Furneaux Group (Flinders Island and South Furneaux) and northeastern Tasmania (St. Helens and Inland northeastern Tasmania) declined in their relative performance. In the case of the Furneaux Group subraces, this decline could be caused by a diversion of resources to sexual reproduction, as these subraces tend to flower precociously (e.g., Chambers et al. 1997; McGowen 2007) and more heavily (McGowen 2007; Suitor et al. 2008) than other subraces. However, there is no quantitative genetic evidence to support this hypothesis. While slightly adverse genetic correlations between flowering precocity and early growth were detected in the present trial (Chambers et al. 1997), these declined with age and were insignificant at both the additive genetic and subrace levels by age 8 years (Stackpole, unpublished data). The decline in growth of the northeastern Tasmanian subraces could be due to other factors, such as the susceptibility of the St. Helens subrace to browsing by arboreal marsupials (O’Reilly-Wapstra et al. 2002). Some subraces, however, were stable in their ranking, notably, the Gippsland Coastal Plain and Strzelecki Foothills subraces and the southern Tasmanian subrace.
Our estimates of heritability for pilodyn (0.35–0.55) are within the range of those reported for open-pollinated E. globulus (MacDonald et al. 1997; Lopez et al. 2002), whereas our estimate of the heritability of harvest-age basic density (0.52) is at the lower end of the range of values previously reported for E. globulus (range 0.44–1.00), although these were for younger tree ages of 8 to 11 years, and some were derived from basal discs (Borralho et al. 1992b; Muneri and Raymond 2000; Apiolaza et al. 2005). However, it is consistent with the median value reported for forest trees in general 0.48 (Cornelius 1994). No study in E. globulus has looked at whether there is an age trend in the heritability of pilodyn or basic density at a single site, but when sites of different ages are compared, no age trends are evident (Dean et al. 1990; MacDonald et al. 1997; Muneri and Raymond 2000; Lopez et al. 2002; Volker 2002). Unlike growth, our heritabilities and coefficients of additive variation of pilodyn are stable with age. This is noteworthy for the multivariate estimates, where the multivariate approach is expected to have a moderating effect on the estimate (Dieters et al. 1999). This stability is particularly significant as the two pilodyn assessments are based on separate samples of the same population, with limited commonality of individuals. The stability of wood density heritability is also evident when harvest-age core density heritability is compared with that of selection-age pilodyn. Similar stability has been reported for Pinus sylvestris (Hannrup and Ekberg 1998). This stability occurs despite the small plot size and illustrates the resilience of genetic parameters for wood properties to the effect of competition. The high correlation of open-pollinated and control-pollinated pilodyn estimates in E. globulus (Volker 2002, p. 83) indicates that wood properties are little affected by inbreeding depression and mortality.
The stability of heritability estimates for successive wood density measurements is also reflected in their high age–age genetic correlations. Indeed, the stability of subrace estimates across ages where r s = |0.96| (see also Miranda et al. 2001) contrasts with that observed for growth. There are few reports of age–age genetic correlations for wood properties of E. globulus with which to compare our results (Miranda et al. 2001). Such correlations are less readily obtained than those for growth as early age sampling may impact on subsequent measures of the same individual, by for example, coring increasing the susceptibility to decay (Wiseman et al. 2006; Hamilton et al. 2007). This source of potential bias is avoided by calculating correlations from different, but related trees. Such bias is unlikely to be an issue in the present study as our age–age additive genetic (e.g., for pilodyn r a = 0.77) and subrace correlations (r s = 0.96) are high, there is little overlap between selection and harvest-age trees sampled (20%), and in this trial, decay was not evident in trees previously sampled by pilodyn. Most reported age–age genetic correlations in wood density are based on retrospective sampling of different age classes or rings in a single core or disk, and high genetic correlations have been reported in E. nitens (Greaves et al. 1997b). Reported age–age correlations of wood properties based on basal core assay are few and of short period (2-3 years). Nevertheless, the correlations reported are relatively high at the clonal level (r > 0.7) between age 3 and 6 years for E. grandis (Osorio et al. 2003) and provenance level in E. globulus (r = 0.57) between 7 and 9 years (Miranda et al. 2001).
The pilodyn only assesses the density of the outer few rings, and while the actual density of the wood may increase with age (Ballarin et al. 2007), our high age–age genetic correlations argue for stability in genotype effects on density across ages, despite a general increase in density with age (i.e., all genotypes display a similar increase in density with age). This is consistent with the high correlation between the outer ring, whole core and whole disk densities reported by Greaves et al. (1997b) in E. nitens, and the high genetic correlations between pilodyn and core estimates of density in the present study (r a > |0.8|). Indeed, the high genetic correlations between selection-age pilodyn penetration and harvest-age wood core density observed, coupled with the high phenotypic correlations reported between core density and whole-tree density (Downes et al. 1997; Raymond and Muneri 2001), vindicate the early use of pilodyn for assessment of harvest-age whole-tree density (Callister and England 2009), although it has been argued that early age coring may have other advantages (Muneri and Raymond 2000).
A key finding in our study is the subtle but significant change in the genetic relationship between growth (diameter) and wood density (pilodyn) with increasing age. Initially, the additive genetic correlation between growth and pilodyn was slightly adverse (r a = 0.26). The reported genetic correlations between growth and density in eucalypts are generally from early age assessments and include both additive (E. globulus MacDonald et al. 1997; Muneri and Raymond 2000; E. grandis Harrand et al. 2009; Eucalyptus urophylla Wei and Borralho 1997; Kien et al. 2008) and total (E. urophylla × E. grandis Bouvet and Bailleres 1995; E. urophylla Ignacio-Sánchez et al. 2005) genetic correlations. These reported genetic correlations are varied, but when significant tend to be adverse, as are those previously reported for E. globulus (e.g., for pilodyn, MacDonald et al. 1997, r a = 0.25 ± 0.06; Costa e Silva et al. 2009, r a = 0.64 ± 0.08). However, in our study, by harvest age, this weak adverse correlation was no longer different from zero and was significantly different to the early age correlation. This change occurred despite the relatively high age–age additive genetic correlations observed in growth (>0.95) and density (>0.77) traits over the same period. The change in correlation between growth and density occurred regardless of whether (a) harvest-age pilodyn or core density were used, (b) the analysis was based on the full early-age data set or that restricted to the more vigorous subset which survived to harvest age, (c) full multivariate or bivariate (bivariate data not shown) analyses were undertaken, or (d) whether diameter or diameter increments were analyzed. Indeed, using diameter increments that bracketed the relevant pilodyn measurements increased the significance of the change in this additive genetic correlation. The decline in genetic correlation observed in the full data was robust and was also evident in individual analyses of the two larger subraces (data not shown). While this change in the growth-density correlation with age is subtle, it is robust and maybe due to the uncoupling of the early physiological relationship between these two traits with age, rather than mortality of slower growing denser individuals. In addition, the decline cannot be explained by a reduction in genetic variation in the harvest-age population for one or either traits, as the coefficients of additive genetic variation and heritabilities of all traits are significantly different from zero and comparable or often greater in the harvest-age population.
While age trends in genetic parameters such as heritabilities are commonly reported, there are few reports of changing genetic correlations with age. This is no doubt due in part to the greater sample size required to detect significance in changes in genetic correlations (Klein et al. 1973; Xie and Mosjidis 1999), as well as the paucity of harvest-age studies of genetic parameters for forest trees. Nevertheless, conceptually such changes are possible and it is well recognized that genetic correlations between traits can change with, for example, differing or changing environments (Sgro and Hoffman 2004), or through ontogeny (Watkins 2001). There are even examples of changing directionality of genetic correlations (Sgro and Hoffman 2004). Such changes in genetic correlation have been previously found across sites in E. globulus and involved growth and vegetative phase change (Jordan et al. 2000) as well as growth and pilodyn penetration (MacDonald et al. 1997). Age trends have been reported in the correlation between growth and disease damage (Milgate et al. 2005), whereby the disease which was initially more prevalent on genetically faster-growing trees, adversely affected the subsequent growth of those trees relative to other genotypes, resulting in a negative genetic correlation. In the present case, the changing genetic correlations indicate that at the genetic level, trees which initially are faster growing tend to produce less dense wood, but this was not the case at later ages. A similar trend has been reported in a retrospective study of the growth rate and wood density of hybrid larch (Larix spp.) measured using X-ray densitometry (Fujimoto et al. 2006). In contrast, Osorio et al. (2003) reported the opposite trend in E. grandis with an initial positive genetic correlation between height growth and wood density which declined with age toward zero, particularly on one site. A study of Pinus taeda basal wood cores that were partitioned into juvenile and mature segments (Atwood et al. 2002) showed no genetic correlation between juvenile tree volume (10 years of age) and juvenile wood density, but a significant negative correlation between subsequent mature volume (11–17 years) and mature wood density. Clearly, among tree species, the patterns of change in these genetic correlations with age are diverse, and further study is required to elucidate the generality of our finding even at the species level.
It is difficult to differentiate or separate environmental from ontogenetic effects on changing genetic correlations in plants, particularly when grown in common garden experiments. Metamorphosis in animals is perhaps the most marked ontogenetic change that occurs and signals the possibility of substantial changes in genetic correlations following programmed, genetic-based uncoupling of correlations (Blouin 1992). The presence of ontogenetically controlled heteroblasty of foliage in E. globulus (Jordan et al. 1999) presents the possibility of ontogenetic effects on wood properties (Loo et al. 1984; Greaves et al. 1997b; Ignacio-Sanchez et al. 2005; Zamudio et al. 2005). However, progress in this area must be prefaced by satisfactory definition of ontogenetic change in wood properties, whether based on stabilization of radial density with age (Ballarin et al. 2007) or on cellular heterochrony (Olsen 2007) and the ability to nondestructively measure wood properties on a large scale. Further, the effect of competition acting together with ontogeny cannot be ruled out for wood properties, as competition has been shown to affect the temporal expression of diameter–height correlations in other eucalypts (Bouvet et al. 2005). However, the possibility of the genetic coupling and decoupling of wood properties with growth is highlighted in a quantitative trait loci (QTL) study that found colocation of one QTL for growth rate with one for wood density, whereas another QTL for wood density was not colocated with growth (Bundock et al. 2007).
The findings of this study have several important implications for tree breeding. If genetic correlations between desired traits can change between selection and harvest age, then decisions that are made at selection may prove to be unfounded in the harvest-age population. While breeders bypass this scenario through parallel but independent modeling of desired traits (Schneeberger et al. 1992), further information on the likelihood and economic significance of changing correlations needs to be gathered. Secondly, as correlated response to selection is founded on the heritabilities of the two traits and their correlation, differing conclusions about the value of a selection trait may be drawn depending on whether full or restricted data, or uni- or multivariate analyses are used. In both cases, further information is required, ideally through further trial measurements at harvest age.
Conclusion
This study emphasizes the dynamic nature of the genetic architecture of growth and wood density in open-pollinated progeny trials of eucalypts. This dynamism may be due to mortality, competition, and ontogenetic effects and affects the level of additive genetic variation of traits, as well as the stability of genetic correlations among traits, with increasing age. These changes may occur even where high age–age correlations suggest that genetic effects across ages will be relatively stable. This finding has practical importance in estimation of breeding values, as they are based on selection-age traits that aim to predict harvest-age traits. The generality of declining genetic association between growth and wood density needs to be confirmed at other sites. Further, temporal studies of growth and wood density based on outcrossed fully pedigreed parents may provide insights into the contribution of inbreeding to the finding we reported here.
References
Apiolaza LA, Raymond CA, Yeo BJ (2005) Genetic variation of physical and chemical wood properties of Eucalyptus globulus. Silvae Genet 54:160–166
APPITA (2002) Methods of test for pulp and paper—basic density of pulpwood. Australian Pulp and Paper Industry Technical Association, Australian/New Zealand Standard 1301.001s:2002, Carlton, Victoria, Australia
Atwood RA, White T, Huber D (2002) Genetic parameters and gains for growth and wood properties in Florida source loblolly pine in the southeastern United States. Can J For Res 32:1025–1039
Ballarin A, Benjamin C, Tomazello Filho MT, Lara Palma H (2007) Wood properties characterization of Eucalyptus grandis trees from 29 years-old managed plantation. In: Eucalypts and diversity: balancing productivity and sustainability, IUFRO conference, Durban, South Africa, 22–26 October 2007
Balocchi C, Bridgwater F, Zobel B, Jahromi S (1993) Age trends in genetic parameters for tree height in a non-selected population of Loblolly pine. For Sci 39:231–251
Beadle C, Mcleod D, Turnbull CRA, Ratkowsky A, McLeod R (1989) Juvenile/total foliage ratios in Eucalyptus nitens and the growth of stands and individual trees. Trees 3:117–124
Blouin MS (1992) Genetic correlations among morphometric traits and rates of growth and differentiation in the green tree frog Hyla cinerea. Evolution 46:735–744
Borralho NMG (1994) Heterogeneous selfing rates and dominance effects in estimating heritabilities from open pollinated progeny. Can J For Res 24:1079–1082
Borralho NMG, Potts BM (1996) Accounting for native stand characteristics in genetic evaluations of open pollinated progeny from Eucalyptus globulus base population. New For 11:53–64
Borralho NMG, Cotterill P, Kanowski P (1992a) Genetic control of growth of Eucalyptus globulus in Portugal. II. Efficiencies of early selection. Silvae Genet 41:70–77
Borralho NMG, Kanowski P, Cotterill P (1992b) Genetic control of growth in Eucalyptus globulus in Portugal. I. Genetic and phenotypic parameters. Silvae Genet 41:39–45
Borralho NMG, Cotterill PP, Kanowski P (1993) Breeding objectives for pulp production under different industrial cost structures. Can J For Res 23:648–656
Bouvet JM, Bailleres H (1995) Expression of some growth and wood property traits among Eucalyptus urophylla x grandis clones in Congo. In: Potts BM, Borralho NMG, Reid JB, Cromer RN, Tibbits WN, Raymond CA (eds) Eucalypt plantations: improving fibre yield and quality. Hobart, Tasmania. CRC for Temperate Hardwood Forestry, Hobart, pp 89–92
Bouvet JM, Vigneron P, Saya A (2005) Phenotypic plasticity of growth trajectory and ontogenic allometry in response to density for Eucalyptus hybrid clones and families. Ann Bot 96:811–821
Bundock P, Potts BM, Vaillancourt RE (2007) Detection and stability of quantitative trait loci (QTL) in Eucalyptus globulus. Tree Genet Genom 4:85–95
Burgess I, Williams ER, Bell JC, Harwood C, Owen J (1996) The effect of outcrossing rate on the growth of selected families of Eucalyptus grandis. Silvae Genet 45:97–100
Callister A, England N (2009) How dense is my blue gum? Methods to predict whole-tree wood density for genetic gain estimation in Eucalyptus globulus. In: Australian Forest Genetics Conference, Forest Products Commission, Perth, Western Australia, Fremantle WA Australia, 20–22 April 2009
Chambers PGS, Borralho NMG (1997) Importance of survival in short-rotation tree breeding programs. Can J For Res 27:911–917
Chambers PGS, Borralho NMG, Potts BM (1996) Genetic analysis of survival in Eucalyptus globulus ssp. globulus. Silvae Genet 45:107–112
Chambers PGS, Potts BM, Tilyard P (1997) The genetic control of flowering precocity in Eucalyptus globulus ssp. globulus. Silvae Genet 46:207–214
Cornelius J (1994) Heritabilities and additive genetic coefficients of variation in forest trees. Can J For Res 24:372–379
Costa e Silva J, Dutkowski GW, Borralho NMG (2005) Across-site heterogeneity of genetic and environmental variances in the genetic evaluation of Eucalyptus globulus trials for height growth. Ann For Sci 62:183–191
Costa e Silva J, Potts BM, Dutkowski GW (2006) Genotype by environment interaction for growth of Eucalyptus globulus in Australia. Tree Genet Genom 2:61–75
Costa e Silva J, Borralho NMG, Araujo JA, Vaillancourt RE, Potts BM (2009) Genetic parameters for growth, wood density and pulp yield in Eucalyptus globulus. Tree Genet Genom 5:291–305
Cotterill P, Dean CA (1990) Successful tree breeding with index selection. CSIRO Division of Forestry and Forest Products, Melbourne
Dean GH, French J, Tibbits W (1990) Variation in pulp and papermaking characteristics on a field trial of E. globulus. In: 44th Appita General Conference, Rotorua New Zealand. (APPITA)
Dieters MJ, Jarvis S, Gilmour AR (1999) Multivariate approach to estimation of genetic parameters. In: 25th Biennial Southern forest tree improvement conference, Louisiana State University, New Orleans, USA, 11–14 July 1999
Downes GM, Hudson I, Raymond CA, Dean GH, Michell AJ, Schimleck LR, Evans R, Muneri A (1997) Sampling plantation eucalypts for wood and fibre properties. CSIRO Australia, Melbourne
Downes G, Catela F, Meder R (2007) Developing and evaluating a global near-infrared calibration for the prediction of kraft pulp yield in eucalypts. In: Eucalypts and diversity: balancing productivity and sustainability, IUFRO Conference, Durban, South Africa, 22–26 October 2007
Dutkowski GW (1995) Genetic variation in drought susceptibility of Eucalyptus globulus ssp globulus in plantations in Western Australia. In: Eucalypt plantations: improving fibre yield and quality, CRC for Temperate Hardwood Forestry, Hobart, Tasmania, 19–24 February 1995
Dutkowski GW, Potts BM (1999) Geographic patterns of genetic variation in Eucalyptus globulus ssp globulus and a revised racial classification. Aust J Bot 47:237–263
Falconer DS, Mackay TFC (1996) Introduction to quantitative genetics. Longman, Harlow
Franklin EC (1979) Model relating levels of genetic variance to stand development of four North American conifers. Silvae Genet 28:207–212
Fujimoto T, Kita K, Uchiyama K, Kuromaru M, Akutsu H, Oda K (2006) Age trends in the genetic parameters of wood density and the relationship with growth rates in hybrid larch (Larix bmelini var. japonica x L. kaempferi) F1. J For Res 11:157–163
Gilmour AR, Thompson R, Cullis BR, Welham SJ (2001) ASREML reference manual. NSW Agriculture, Orange
Greaves BL, Borralho NMG, Raymond CA (1997a) Breeding objective for plantation eucalypts grown for production of kraft pulp. For Sci 43:465–472
Greaves BL, Borralho NMG, Raymond CA, Evans R, Whiteman P (1997b) Age–age correlations in, and relationships between basic density and growth in Eucalyptus nitens. Silvae Genet 46:264–270
Greaves BL, Borralho NMG, Raymond CA (2003) Early selection in eucalypt breeding in Australia—optimum selection age to minimise the total cost of kraft pulp production. New For 25:201–210
Griffin AR, Cotterill P (1988) Genetic variation in growth of outcrossed, selfed and open-pollinated progenies of Eucalyptus regnans and some implications for breeding strategy. Silvae Genet 37:124–131
Gwaze DP, Bridgwater F, Byram TD, Woolliams J, Williams C (2000) Predicting age–age genetic correlations in tree-breeding programs: a case study of Pinus taeda L. Theor Appl Genet 100:199–206
Hadfield J (2008) Estimating evolutionary parameters when viability selection is operating. Proc R Soc 275:723–734
Hamilton M, Greaves BL, Potts BM, Dutkowski GW (2007) Patterns of longitudinal within-tree variation in pulpwood and solidwood traits differ among Eucalyptus globulus genotypes. Ann For Sci 64:831-837
Hannrup B, Ekberg I (1998) Age-age correlations for tracheid length and wood density in Pinus sylvestris. Can J For Res 28:1373–1379
Hannrup B, Wilhelmsson L, Danell O (1998) Time trends for genetic parameters of wood density and growth traits in Pinus sylvestris L. Silvae Genet 47:214–219
Hardner CM, Potts BM (1995) Inbreeding depression and changes in variation after selfing in Eucalyptus globulus ssp. globulus. Silvae Genet 44:46–54
Hardner CM, Potts BM (1997) Post-dispersal selection under mixed-mating in Eucalyptus regnans. Evolution 51:103–111
Hardner CM, Potts BM, Gore PL (1998) The relationship between cross success and spatial proximity of Eucalyptus globulus ssp. globulus parents. Evolution 52:614–618
Harrand L, Hernández JJV, Upton JL, Valverde GR (2009) Genetic parameters of growth traits and wood density in Eucalyptus grandis progenies planted in Argentina. Silvae Genet 58:11–19
Hodge GR, Volker PW, Potts BM, Owen JV (1996) A comparison of genetic information from open-pollinated and control-pollinated progeny tests in two eucalypt species. Theor Appl Genet 92:53–63
Ignacio-Sanchez E, Vargas-Hernandez JJ, Lopez-Upton J, Borja-de la Rosa A (2005) Genetic parameters for growth and wood density in juvenile Eucalyptus urophylla S. T. Blake. Agrociencia 39:469–479
Jordan G, Potts BM, Wiltshire RJ (1999) Strong independent quantitative genetic control of the timing of vegetative phase change and first flowering in Eucalyptus globulus ssp. globulus (Tasmanian Blue Gum). Heredity 83:179–187
Jordan GJ, Potts BM, Chalmers P, Wiltshire RE (2000) Quantitative genetic evidence that the timing of vegetative phase change in Eucalyptus globulus ssp. globulus is an adaptive trait. Aust J Bot 48:561–567
Kien ND, Jansson G, Harwood CE, Almqvist C, Thinh HH (2008) Genetic variation in wood basic density and Pilodyn penetration and their relationships with growth, stem straightness, and branch size for Eucalyptus urophylla in Northern Vietnam. NZ J For Sci 38:160–174
Klein T, DeFries J, Finkbeiner C (1973) Heritability and genetic correlation: standard errors of estimates and sample size. Behav Genet 3:355–364
Kube P, Raymond CA, Banham P (2001) Breeding Eucalyptus nitens to improve wood quality and profitability. In: Developing the eucalypt of the future, IUFRO, Valdivia, Chile, 10–15 September 2001
Li Y, Dutkowski GW, Apiolaza L, Pilbeam D, Costa e Silva J, Potts BM (2007) The genetic architecture of a Eucalyptus globulus full-sib breeding population in Australia. For Genet 12:167–179
Loo J, Tauer C, van Buijtenen J (1984) Juvenile–mature relationships and heritability estimates of several traits in loblolly pine (Pinus taeda). Can J For Res 14:822–825
Lopez GA, Potts BM, Dutkowski GW, Apiolaza LA, Gelid PE (2002) Genetic variation and inter-trait correlations in Eucalyptus globulus base population trials in Argentina. For Genet 9:217–231
Lopez GA, Potts BM, Vaillancourt RE, Apiolaza LA (2003) Maternal and carryover effects on early growth of Eucalyptus globulus. Can J For Res 33:2108–2115
MacDonald AC, Borralho NMG, Potts BM (1997) Genetic variation for growth and wood density in Eucalyptus globulus ssp. globulus in Tasmania (Australia). Silvae Genet 46:236–241
Magnussen S (1989) Effects and adjustments of competition bias in progeny trials with single-tree plots. For Sci 35:532–547
Matheson AC, Raymond CA (1984) Effects of thinning in progeny tests on estimates of genetic parameters in Pinus radiata. Silvae Genet 33:125–128
McGowen MH (2007) Genetic control of reproductive traits in Eucalyptus globulus. Ph.D. thesis, University of Tasmania
McRae T, Apiolaza LA, Dutkowski GW, Kerr R, Pilbeam D, Powell M, Tier B (2003) Treeplan—a genetic evaluation system for forest trees. In: 27th Bienial Southern Forest Tree Improvement Conference, Stillwater, Oklahoma, USA, 24–27 June 2003
Milgate A, Potts BM, Joyce K, Mohammed C, Vaillancourt RE (2005) Genetic variation in Eucalyptus globulus for susceptibility to Mycosphaerella nubilosa and its association with growth rate. Aust Plant Pathol 34:11–18
Miranda I, Almeida MH, Pereira H (2001) Provenance and site variation of wood density in Eucalyptus globulus Labill., at harvest age and its relation to a non-destructive early assessment. For Ecol Manage 149:235–240
Muneri A, Raymond CA (2000) Genetic parameters and genotype-by-environment interactions for basic density, pilodyn penetration and stem diameter in Eucalyptus globulus. Forest Genetics 7:317–328
Olsen M (2007) Wood ontogeny as a model for studying heterochrony with an example of paedomorphosis in Moringa (Moringaceae). Syst Biodivers 5:145–158
O’Reilly-Wapstra JM, McArthur C, Potts BM (2002) Genetic variation in resistance of Eucalyptus globulus to marsupial browsers. Oecologia 130:289–296
Osorio LF, White TL, Huber DA (2001) Age trends of heritabilities and genotype-by-environment interactions for growth traits and wood density from clonal trials of Eucalyptus grandis Hill ex Maiden. Silvae Genet 50:108–117
Osorio LF, White TL, Huber DA (2003) Age–age and trait–trait correlations for Eucalyptus grandis Hill ex Maiden and their implications for optimal selection age and design of clonal trials. Theor Appl Genet 106:735–743
Potts BM, Vaillancourt RE et al (2004) Exploration of the Eucalyptus globulus gene pool. In: Eucalyptus in a changing world, IUFRO, Aviero, Portugal, 11–15 October 2004
Rapley LP, Allen GR, Potts BM (2004) Genetic variation in Eucalyptus globulus in relation to susceptibility from attack by the southern eucalypt leaf beetle, Chrysophtharta agricola. Aust J Bot 52:747–756
Raymond CA (2002) Genetics of Eucalyptus wood properties. Ann For Sci 59:525–531
Raymond CA, Muneri A (2001) Non-destructive sampling of Eucalyptus globulus and E. nitens for wood properties. I. Basic density. Wood Sci Technol 35:27–39
Raymond CA, Schimleck LR, Muneri A, Michell AJ (2001) Genetic parameters and genotype-by-environment interactions for pulp yield predicted using near infrared reflectance analysis and pulp productivity in Eucalyptus globulus. For Genet 8:213–224
Retief E, Stanger T, Galloway G (2001) Early results from a trial to test the effect of plot design on Eucalyptus hybrid clonal ranking in coastal Zululand, South Africa. In: Developing the eucalypt of the future, IUFRO, Valdivia, Chile, 10–15 September 2001
Sanhueza RP, White TL, Huber DA, Griffin AR (2002) Genetic parameters estimates, selection indices and predicted genetic gains from selection of Eucalyptus globulus in Chile. For Genet 9:19–29
SAS Institute Inc (2002) SAS/STAT users guide, version 9. SAS Institute, Cary
Schneeberger M, Barwick SA, Crow GH, Hammond K (1992) Economic indices using breeding values predicted by BLUP. J Anim Breed Genet 109:180–187
Self SG, Liang KY (1987) Asymptotic properties of maximum likelihood estimators and likelihood ratio tests under nonstandard conditions. J Am Stat Assoc 82:605–610
Sgro C, Hoffman A (2004) Genetic correlations, tradeoffs and environmental variation. Heredity 93:241–248
Steane DA, Conod N, Jones RC, Vaillancourt RE, Potts BM (2006) A comparative analysis of population structure of a forest tree, Eucalyptus globulus (Myrtaceae), using microsatellite markers and quantitative traits. Tree Genet Genom 2:30–38
Stram DO, Lee JW (1994) Variance components testing in the longitudinal mixed effects model. Biometrics 50:1171–1177
Suitor S, Potts BM, Brown PH, Gracie AJ, Gore PL (2008) Post-pollination capsule development in Eucalyptus globulus seed orchards. Aust J Bot 56:51–58
TAPPI (1989) Basic density and moisture content of pulpwood T258 om-98. Technical Association of the Pulp and Paper Industry, Norcross
Tibbits W, White T, Hodge G, Borralho N (2006) Genetic variability in freezing tolerance of Eucalyptus globulus ssp globulus assessed by artificial freezing in winter. Aust J Bot 54:521–529
Tibbits WN, Boomsma DB, Jarvis S (1997) Distribution, biology, genetics and improvement programs for Eucalyptus globulus and E. nitens around the world. In: 24th Biennial Southern Forest Tree Improvement Conference, University of Florida, Gainesville USA 9-12 June 1997
van Wyk G, Pierce BT, Verryn SD (1991) Two year results from a site by clone interaction trial series of Eucalyptus grandis. In: Intensive forestry: the role of eucalypts, P2.02-01 productivity of eucalypts, Southern African Institute of Forestry, Durban, South Africa, 2–6 September 1991
Volker PW (2002) Quantitative genetics of Eucalyptus globulus, E. nitens and their F1 hybrid. Ph.D. thesis, University of Tasmania
Volker PW, Potts BM, Borralho NMG (2008) Genetic parameters of intra- and inter-specific hybrids of Eucalyptus globulus and E. nitens. Tree Genet Genom 4:445–460
Watkins T (2001) A quantitative genetic test of adaptive decoupling across metamorphosis for locomotor and life-history traits in the Pacific tree frog Hyla regilla. Evolution 55:1668–1677
Wei X, Borralho NMG (1997) Genetic control of wood basic density and bark thickness and their relationships with growth traits of Eucalyptus urophylla in south east China. Silvae Genet 46:245–250
Wei X, Borralho NMG (1998) Use of individual tree mixed models to account for mortality and selective thinning when estimating base population genetic parameters. For Sci 44:246–253
White TL, Adams WT, Neale DB (2007) Forest genetics. CABI, Wallingford
Wiseman D, Smethurst P, Pinkard L, Wardlaw T, Beadle C, Hall M, Baillie C, Mohammed C (2006) Pruning and fertiliser effects on branch size and decay in two Eucalyptus nitens plantations. For Ecol Manag 225:123-133
Xie C, Mosjidis JA (1999) Influence of sample size on precision of genetic correlations in red clover. Crop Sci 39:863–867
Zamudio F (1995) On the genotype-by-time interaction: growth increments, stability over time and their effect on genetic gain. Ph.D. thesis, North Carolina State University
Zamudio F, Rozenberg P, Baettig R, Vergara A, Yanez M, Gantz C (2005) Genetic variation of wood density components in a radiata pine progeny test located in the south of Chile. Ann For Sci 62:105–114
Zar JH (1974) Biostatistical analysis. Prentice Hall, Englewood Cliffs
Acknowledgments
We thank Kelsey Joyce, Mark Reynolds, Linda Ballard, and Paul Tilyard for their assistance; the CRC for Forestry and the Australian Research Council and partners on Linkage grant LP0453704 for support; and Gunns Ltd for access to the field trial.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by D. Grattapaglia
Rights and permissions
About this article
Cite this article
Stackpole, D.J., Vaillancourt, R.E., de Aguigar, M. et al. Age trends in genetic parameters for growth and wood density in Eucalyptus globulus . Tree Genetics & Genomes 6, 179–193 (2010). https://doi.org/10.1007/s11295-009-0239-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-009-0239-4