Abstract
Precocious trifoliate orange (Poncirus trifoliata [L.] Raf), an extremely early flowering mutant of P. trifoliata, is an attractive model for functional genomics research in Citrus. A procedure for efficient regeneration and transformation of this genotype was developed by using green fluorescent protein (GFP) gene as visual marker and etiolated stem segments as explants. In vivo monitoring of GFP expression permitted a rapid and easy discrimination of transgenic shoots and escapes. Transformation efficiency was 20.7% and the transformants were identified by polymerase chain reaction (PCR) and Southern blot analysis. Moreover, the transgenic lines expressed variable amounts of the GFP gene as revealed by real-time PCR analysis. Fifteen transgenic plants flowered 18 months after transfer to the greenhouse and six of them set fruits. GFP expression was also observed in the transgenic flowers and fruits. To test the utility of this system for functional genomics studies, an Arabidopsis thaliana MAC12.2 gene with the potential to produce seedless fruits was introduced into this genotype, and the traits of the transgenic fruits were characterized. The successful transformation of this perennial woody genotype with extremely short juvenility will allow us to test the function of cloned genes in citrus, the improvement of which is hindered by a long juvenility period.
Similar content being viewed by others

Avoid common mistakes on your manuscript.
Introduction
Citrus is a major fruit crop in the world. The market is demanding citrus fruits with improved quality, e.g., seedlessness, easy peeling with attractive size and color, etc. (Deng 2005). The improvement of fruit quality by traditional breeding techniques has been constrained by complex biology of citrus, especially the long juvenile period. Genetic transformation provides the means for adding a single agronomic trait in perennial plant cultivars without altering their phenotype (Deng and Duan 2006). Until now, various genes have been introduced into citrus species and its relatives to improve agronomical traits, including enhancing abiotic stress tolerance (Cervera et al. 2000; Fagoaga et al. 2007) and disease resistance (Ananthakrishnan et al. 2007; Gonzalez-Ramos et al. 2005; Omar et al. 2007; Zanek et al. 2008), shortening the juvenile phase (Duan et al. 2007a; Endo et al. 2005; Peña et al. 2001), rootstock (Gentile et al. 2004), and fruit improvement (Costa et al. 2002; Guo et al. 2005; Koltunow et al. 2000; Li et al. 2002; Wong et al. 2001). Among these, fruit improvement is of particular importance as it can be very attractive for consumers. However, the characteristics of transgenic fruits were not analyzed in these reports due to the long juvenile period.
Juvenile phase in citrus ranges between 2 and 20 years and has been one of the limiting factors for citrus genetic improvement. An alternative to resolve the problem of the long juvenility in genetic improvement of citrus fruits is to use mature tissues as the explants. This method has been proved successful for the transformation of adult sweet orange (Cervera et al. 1998, 2005), sour orange (Ghorbel et al. 2000), and some mandarin genotypes (Cervera et al. 2008). But the procedure has limited application because mature tissues are recalcitrant to Agrobacterium tumefaciens infection and transformation. Another method is to use a model genotype to test gene function prior to the application in genetic improvement of commercial citrus fruits. Arabidopsis and tobacco as test systems were successfully used to pretest genes to be used in citrus in efforts to obtain seedless fruits (Koltunow et al. 1998). Tomato can also be considered as a model plant to confirm the function of genes cloned from citrus. However, these systems have limitations due to the different genetic background, expression pattern, and regulating mechanism between citrus and the model species. The ideal strategy is to use a short juvenility citrus genotype as model transformation system. The efficient transformation system of West Indian lime (Citrus aurantifolia) and kumquat (Fortunella sp.), which have a short juvenility period, has been successfully established (Koltunow et al. 2000; Yang et al. 2007).
Precocious trifoliate orange (Poncirus trifoliata [L.] Raf), an extremely early flowering mutant from P. trifoliata, has a short juvenile period of 1–2 years. The mutant has significant differences from its wild-type: (1) high sprouting rate, short juvenile period, and early fruit set; (2) fast formation of floral bud; (3) flowering and bearing fruits several times in 1 year; (4) high fruit setting; (5) dwarf and short internode (Liang et al. 1999). These special traits make it a useful model plant for the evaluation of gene function related to the characteristics of flowers and fruits in citrus. So far, reporter genes have been widely used in citrus genetic transformation (Almeida et al. 2003; Cervera et al. 2008; Dominguez et al. 2004; Duan et al. 2007b; Ghorbel et al. 1999; Grosser et al. 2000; Guo et al. 2005; Omar and Grosser 2008; Zheng et al. 2006). The green fluorescent protein (GFP) from Aequorea victoria as a vital marker has several advantages over other visual reporter genes and has been proved extremely useful for reporting gene expression in transformed plants. In this study, we intend to establish an efficient regeneration and transformation procedure of precocious trifoliate orange by introducing a modified GFP gene (mgfp5-ER) using stem segments as explants for potential functional genomics studies in citrus specifically and long juvenile perennial woody plants generally. Furthermore, an Arabidopsis thaliana MAC12.2 gene (Payne et al. 2004) with the potential to produce seedless fruits was introduced into this genotype to test the utility of this system for functional genomics studies.
Materials and methods
Plant materials and culture media
Etiolated seedlings of precocious trifoliate orange were used as explant sources. Well-developed precocious trifoliate orange seeds freshly removed from the fruits were treated in 1 M NaOH for 10 min and rinsed with tap water, followed by a 2% solution of sodium hypochlorite treatment for 15 min. The sterilized seeds were rinsed three times with sterile distilled water. Seed coats were removed and embryos were placed in test tubes containing 25 mL of MT (Murashige and Tucker 1969) medium with 25 g/L sucrose and 7.5 g/L agar and then maintained at 26 ± 2°C in the dark for 5 weeks. Stem segments approximately 1.0-cm-long were collected. Coculture medium (CM) consisted of solid MT with 1.0 mg/L 6-benzyladenine (BA), 0.5 mg/L kinetin (KT), 0.1 mg/L α-napthaleneacetic acid (NAA), 100 mmol/L acetosyringone (AS), and 30 g/L sucrose. The primary shoot regeneration medium (SRM1) consisted of CM (without 100 mmol/L AS) with 50 mg/L kanamycin (Km) and 400 mg/L cefotaxime (Cef). The secondary shoot regeneration medium (SRM2) was mostly identical to the SRM1 with a minor modification that BA concentration was decreased to 0.5 mg/L. Shoot elongation medium (SEM) consisted of solid MT with 0.1 mg/L BA, 0.25 mg/L gibberellic acid (GA3), 0.1 mg/L indole butyric acid (IBA), and 0.2 mg/L activated charcoal with 200 mg/L Cef. Root regeneration medium (RRM) consisted of solid 1/2 MT with 0.5 mg/L NAA, 0.1 mg/L IBA, and 0.5 g/L activated charcoal with 20 mg/L Km and 200 mg/L Cef. The antibiotics and hormones were filter sterilized and added to autoclaved medium.
Transformation and regeneration
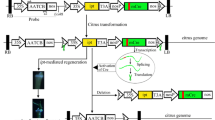
The disarmed A. tumefaciens EHA105 harboring the binary plasmid pBIN-mgfp5-ER (Haseloff et al. 1997) was used as the vector system for transformation. The pBIN-mgfp5-ER T-DNA contained the Nospro-nptII-Noster cassette as a selectable marker and the 35Spro-mgfp5-ER-35Ster cassette as a vital marker. Three experiments were performed. The transformation procedure was as described by Duan et al. (2007a, b) with several minor modifications. Briefly, the explants were blotted dry, placed horizontally on CM, and cultured at 21°C in the dark for 3 days after 20 min incubation. Stem segments were soaked in sterile water with 400 mg/L Cef for 5 min and rinsed three times with sterile distilled water. The explants were dried by sterile filter paper and transferred to SRM1. The cultures were maintained in the dark for 2 weeks at 28°C and then transferred to 16 h photoperiod (Peña et al. 1995). After 3 weeks, the explants producing shoots were transferred to the SRM2 medium and excised the latent buds in the explants.
Detection of GFP by fluorescence stereomicroscopy
Before transfer to SRM2, explants were examined in vivo periodically under a fluorescence stereomicroscope equipped with a Leica fluorescence stereomicroscope (MZFLIII) comprising a 480/40-nm exciter filter, a 505-nm LP dichromatic beam splitter, and a 510-nm LP barrier filter. The red autofluorescence from chlorophyll was not blocked with any interference filter. Photographs were taken using Nikon E4500. Transformation frequency was evaluated as the total number of green fluorescent shoots per total number of Agrobacterium-inoculated explants.
PCR and Southern blot analyses
Standard polymerase chain reaction (PCR) techniques were used to detect target gene sequences in leaf samples from regenerated transgenic plants. The GFP gene primers were: 5′-AGGACCATGTGGTCTCTCTT-3′ and 5′-TGGCCAACACTTGTCACTAC-3′, which produced a 500-bp fragment. Reactions were subjected to 35 cycles of 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C.
DNA was isolated from leaves of 15 individual PCR-positive lines (lines 1–15) and control plants in the greenhouse according to Cheng et al. (2003). For Southern blot analysis, 15 µg DNA samples were digested with HindIII, separated on 0.8% (w/v) agarose gels, blotted to nylon membranes (Hybond-N+, Amersham), and probed with P32-labeled specific fragment.
Real-time PCR quantification
Total RNA was extracted from the fresh leaves of six individual transgenic lines (lines 1, 3, 7, 10, 13, and 15) and one nontransgenic plant according to Liu et al. (2007). To monitor GFP expression in transgenic plants, a primer set was designed with the Primer Express software (Applied Biosystems, Foster City, CA, USA) for real-time PCR (forward primer: TCCAAGGAGATATAACAATGAAGACTAATC; reverse primer: AATTGGGACAACTCCAGTGAAAA). The control primer set and real-time PCR procedure were as described by Liu et al. (2007). Output data generated by the instrument onboard software Sequence Detector Version 1.3.1 (PE Applied Biosystems) were transferred to a custom-designed Microsoft Excel macro for analysis.
p12.2GUS2-1 construct, Southern blot, and characteristics of transgenic fruits
In order to evaluate the efficiency of the precocious trifoliate orange transformation system for gene function test, a MAC12.2 gene which was cloned from parthenocarpic knuckles mutant of Arabidopsis was used. The construction of p12.2GUS2-1 (kindly provided by Dr. Koltunow, Australia, CSIRO) T-DNA contained the Nospro-nptII-Noster and the KNUpro-MAC12.2:GUS-Noster cassettes as described in detail by Payne et al. (2004).
Southern blot analysis was performed to confirm the stable integration of the MAC12.2:GUS fusion gene in the transgenic plants. DNA samples (15 µg) were digested with HindIII, separated on 0.8% (w/v) agarose gels, blotted to nylon membranes (Hybond-N+, Amersham), and probed with P32-labeled specific fragment of the MAC12.2:GUS fusion gene by PCR. The MAC12.2:GUS fusion gene primers were 5′-CACACACATCCTTCACTCTTC-3′ and 5′-CATTACGCTGCGATGGATCCG-3′.
The fruits of MAC12.2 lines and control were picked after full maturation and their polar and equatorial diameters were evaluated, respectively. Each fruit was cut transversely to evaluate total seed number.
Results
Optimization of transformation for precocious trifoliate orange
Stem segments from precocious trifoliate orange were cocultivated with A. tumefaciens EHA105 carrying the binary vector pBIN-mgfp5-ER. After cocultivation in the dark, the explants were examined in vivo under a stereomicroscope with 480 nm excited light. The cut end of the transformed stem segments showed green fluorescence (Fig. 2a). Then, the cultures were transferred to the SRM1 in darkness for 2 weeks and afterward under 16 h photoperiod. On this medium, a pale green and compact callus (Fig. 1a), which showed green fluorescence (Fig. 2b), appeared on the cutting end in about 10 days. And after selection for an additional 3 weeks, multiple adventitious buds differentiated on the surface of these calli through a process of indirect organogenesis. The escapes and nontransformed shoots appeared red due to the autofluorescence of the chlorophyll. The shoots which did not show green fluorescence were excised. The explants with shoots showing bright green fluorescence (Fig. 2c) were transferred to SRM2 for subsequent molecular analyses. Meanwhile, the latent buds from the leaf axil of the explants (Fig. 1b) were excised to avoid competition of nutrition with the shoots from the cut end. It is a pivotal step in the transformation of precocious trifoliate orange because of its short internodes.
Production of transformed precocious trifoliate plants. a Regenerated callus from the cut end formed after about 10 days in SRM1 (white arrow); b regenerated latent bud from the leaf axil of stem segment (white arrow); c transgenic shoots self-rooted in the SEM (white arrow); d transgenic plants flowering after 18 months in the greenhouse; e normal flower; f abnormal flower; g pistil-abortion flower; h transgenic plant bearing fruit
GFP expression in stem segments cut ends, shoots, plantlets, flower, fruitlet, and seeds (a–j). Green fluorescence in transformed stem segments after cocultivation (a) and cultured on SRM1 for 10 days (b); green fluorescence in transformed shoots (c); red fluorescence in nontransformed plantlet (d); green fluorescence in different transformed plantlets (e, f); green fluorescence in flower (g), fruitlet (h), and seeds (i) of transgenic plants. Scale bars = 1.0 mm
Defining regeneration efficiency as the percentage of stem segments with shoots obtained per stem segments inoculated and transformation efficiency as the percentage of GFP-positive shoots obtained per stem segments infected, the efficiency of regeneration and transformation was 91.8% and 20.7%, respectively (Table 1). GFP-expressing shoots (longer than 0.5 cm) were physically separated in vitro and elongated on SEM. Interestingly, several GFP-expressing shoots were rooted automatically after transfer into SEM for 3 weeks (Fig. 1c). Transgenic plantlets were recovered by rooting on RRM. When rooted shoots were approximately 3–4 cm long and had two to three well-developed roots, they were enclosed in a plastic bag, watered, and monitored. Totally, over 100 transgenic precocious trifoliate orange plantlets were obtained.
The transgenic plantlets were successfully acclimatized and hardened. Twenty-five well-developed plantlets were repotted to the greenhouse for subsequent analyses. Fifteen transgenic plants flowered 18 months after transfer to the greenhouse and showed normal growth (Fig. 1d). Normal flower (Fig. 1e), abnormal flower (petal is closed in the beginning of flowering and opened in the full bloom, as shown in Fig. 1f), and pistil-abortion flower (Fig. 1g) were observed among the 15 transgenic plants with flowers. Among the three flower phenotypes, normal flower is the main phenotype and was also observed in the control plants. Six of 25 transgenic plants were bearing fruits (Fig. 1h). Using the procedure for genetic transformation, plants with fruits were obtained within 2 years, significantly faster than with other citrus cultivars.
Different patterns of GFP expression in transgenic plantlets under the fluorescence stereomicroscope
About 80–100 days after cocultivation with Agrobacterium, the plantlets were examined in vivo under a stereomicroscope with 480 nm excited blue light. Different phenotypes over the expression of the GFP gene in regenerated transgenic plantlets were observed. Some transformants appeared full green fluorescence (Fig. 2e), others showed partial green fluorescence (Fig. 2f), and nontransformed plantlets showed red fluorescence (Fig. 2d). The fluorescence intensity varied among different parts of the plantlets, being higher in young tissues (shoot apices, young leaves) than in old ones (Fig. 2f). The roots of all transformants showed bright green fluorescence. In old tissues, low metabolism and chlorophyll accumulation partially masked the green fluorescence provided by GFP (Ghorbel et al. 1999).
The stability of GFP expression was maintained in the transgenic precocious trifoliate orange plants under greenhouse condition, in not only vegetative tissues but also reproductive organs, such as flowers (Fig. 2g) and resulting fruitlets (Fig. 2h) and seeds (Fig. 2i). GFP analysis seems to be a good choice for rapid visual selection of transformed tissues, but further and final confirmation of transformation by PCR and Southern blot is required.
Molecular analyses of the GFP-expressing plants
Putative transgenic plantlets were first observed to test if green fluorescence exists and then analyzed by PCR to verify the presence of the GFP gene in their genome. The 500-bp GFP gene fragment was amplified from 15 randomly selected plants (Fig. 3a). No amplificant was detected in the DNA samples from nontransgenic regenerated control plants (Fig. 3a, lane C), which emitted red autofluorescence under blue light (Fig. 2d).
Molecular analyses of GFP-expressing plants. a PCR detection of the GFP gene in putative GFP-expressing plants. Lanes 1–15 GFP-expressing plants, lane C nontransformed plant, lane P plasmid pBIN-mgfp5-ER, lane M 100 bp DNA ladder. b Southern blot analysis of the GFP gene from PCR-positive plants. Genomic DNA was digested with HindIII, which has only one enzyme site in the plasmid. Lanes 1–15 PCR-positive plants (lines 1–15), C nontransformed plant, M λ DNA/HindIII molecular weight marker; molecular weights are indicated on the left. c Real-time PCR analysis of the relative expression levels of the GFP gene in transgenic precocious trifoliate orange lines. 1, 3, 7, 10, 13, and 15 transgenic lines of precocious trifoliate orange, C nontransgenic plant
Southern blot analyses were performed to confirm the stable integration of the GFP gene cassette. The GFP gene fragment (500 bp) was used as a probe to reveal the presence of the transgene. Different integration patterns in the transgenic plants from one to three copies at different loci were observed when HindIII was used. As shown in Fig. 3b, all 15 GFP-expressing plants contained the target gene (lanes 1–15). No hybridization signal was detected in a nontransformed control plant (lane C).
Six individual transgenic lines with full green fluorescence expression and varied copy numbers as confirmed by Southern blot were selected for real-time PCR analysis. The results indicated that the GFP transgenes expressed in the leaves of the transgenic precocious trifoliate orange plants with variable transcript accumulation among individual transgenic lines. As shown in Fig. 3c, the GFP expression level of line 13 (three copies) was significantly higher than that of line 10 (three copies) and that of lines 1, 3, and 15 (one copy) were obviously different and slightly higher than line 7 (two copies). This result suggested that there was no correlation between copy number and transgene expression.
Production of transgenic lines with MAC12.2 gene
A total of 35 plant lines were obtained from cocultivated etiolated stem segments. Control and 35 putative transgenic plants were repotted into the greenhouse for subsequent analyses. Southern blot analysis performed on selected transgenic plants using a gene-specific probe confirmed the presence of MAC12.2 and GUS sequence in precocious trifoliate orange genomes (Fig. 4). The hybridized bands indicated the integration of one to four copies of the target gene in transgenic plants (lanes 1–10). Thirty-three transgenic plants flowered 15 months after being transferred to the greenhouse and nine of them set fruitlets. The nontransformed plants also flowered after being transferred to the greenhouse and set fruitlets.
Southern blot analysis of the MAC12.2:GUS fusion gene from PCR-positive plants. Genomic DNA was digested with HindIII, which has only one restriction site in the plasmid. Lanes 1–10 PCR-positive plants (lines 1–10), C nontransformed plant, M λ DNA/HindIII molecular weight marker; molecular weights are indicated on the left
Five ripe fruits from five different transgenic lines (P8, P5, P2, P16, and P6-2) were eventually obtained, as each transgenic line yielded only one fruit due to severe physiological drop. Fruit characteristics of five lines were analyzed and compared to the control. The fruit size of transgenic lines was almost the same as that of the control except for P16 (Fig. 5a–f; Table 2). Fruits of the control were very seedy and have 22.8 seeds on average (Fig. 5g), while the seed number of transgenic lines was much less and possessed only 11, seven, six, or as few as four seeds per fruit (Fig. 5h–l; Table 2). Moreover, some seeds in the transgenic lines were flattened and reduced in size.
Discussion
This study described successful Agrobacterium-mediated transformation of the precocious trifoliate orange with the plasmid pBIN-mgfp5-ER, encoding for the GFP gene. Over 100 transgenic lines containing the GFP gene were obtained and 15 transgenic plants were analyzed by PCR and Southern blot analyses. The GFP expression was also observed in six individual transgenic lines by real-time PCR analysis.
One advantage of precocious trifoliate orange transformation is the high regeneration efficiency (91.8%) and transformation efficiency (20.7%). In the indirect regeneration pathway, BA is essential for callus development and bud formation (Moreira-Dias et al. 2000). And as Peña et al. (2003) reported, cambial cells on the cut surfaces of the explants under specific culture conditions producing callus cells are the major target for Agrobacterium action. When explants were transferred to SRM1 (BA 1.0 mg/L) in darkness for 2 weeks, the regeneration process was similar to that described by Peña et al. (2004) and callus was observed 10 days later. SRM1 was successful in promoting the regeneration of more transgenic adventitious buds or shoots.
Transformation efficiency depends considerably on plant genotype. In comparison with published data, the efficiency of transformation reported in the present investigation is much higher than that of “Xuegan” (4.3%) (Yu et al. 2002), Ridge pineapple (2%) (Gutiérrez-E et al. 1997), and Bingtang sweet orange (10%) (Duan et al. 2007b), lower than that of Valencia (23.8%) (Boscariol et al. 2003) and similar to the Carrizo citrange (20.6%) (Peña et al. 1995). Previous studies in trifoliate orange (Endo et al. 2005; Iwanami et al. 2004; Kaneyoshi et al. 1994; Wong et al. 2001) were also very successful. But transformation efficiency is different among previous reports. Kaneyoshi et al. (1994) has shown that the frequency of shoot formation was 35.6% in the segments inoculated with pBI121 and 44.6% in pBI101-O12-p1, and histochemical GUS assay showed that 55.4% (pBI121) and 87.7% (pBI101-12-p1) of shoots expressed the GUS gene. In the present investigation, the ability of shoot regeneration in precocious trifoliate orange mutant is much higher than that of the wild trifoliate orange, while transformation efficiency is lower in the mutant. However, transformation efficiency of the wild-type was only 2.1–4.6% in another report (Iwanami et al. 2004). These differences were probably due to the factors that affect the transformation and regeneration process. However, the protocol reported in the present investigation is suitable for the transformation system of fruit-specific gene function tests.
The primary advantage of transforming precocious trifoliate orange is that transgenic plants could flower and set fruits within 1–2 years. Production of transgenic plants is fundamental to investigate plant gene function as well as to improve agronomic traits in citrus. However, the long juvenility phase is a serious limiting factor in genetic modification, especially for developmental evaluation of flowers and fruits. Direct transformation of mature tissue is ideal for early evaluation of genetically modified characteristics. Cervera et al. (1998, 2005) produced sweet orange transformants that flowered and set fruits in about 1 year after in vitro regeneration by using this method. However, direct transformation of mature material is not easily achieved because of low transformation competence and regeneration potential of mature tissues (Cervera et al. 2008). Koltunow et al. (2000) introduced the SDSL-1 gene, a decreased seed number gene, into West Indian lime which had a shorter juvenile period of 2–3 years and ultimately obtained fruits with reduced seeds. But the transformation efficiency was very low (3.2%). In addition, Yang et al. (2007) established the transformation procedure of kumquat, which is very close to Citrus and had a short juvenile period of 2 years. The final transformation efficiency was only 3.6%. Compared with West Indian lime and kumquat, transformation efficiency in precocious trifoliate orange could be up to 20.7%. Furthermore, transgenic precocious trifoliate orange plants could flower and set fruits within 18 months after transfer to the greenhouse. In other fruit crops, Petri et al. (2008) established a high transformation efficiency system in plum which could be a new tool for functional genomics studies in Prunus spp.
In summary, we showed that the transformation procedure as presented herein is a simple and highly productive system for developing transgenic precocious trifoliate orange plants. Establishment of an efficient transformation system is important for functional genomics analyses. Based on the model precocious trifoliate orange transformation system, the Arabidopsis MAC12.2 gene, which has the potential to induce fruit seedlessness in precocious trifoliate orange, was introduced to test the gene function. Thirty-five independent transgenic lines were obtained and five transgenic lines set fruits. The characteristics of the transgenic fruits were analyzed, which indicated that they have significantly less seeds than the control.
The system is ideal for the study of gene function. We expect to screen more functional genes relating to economically important traits which could be further introduced into commercial citrus cultivars for creating new transgenic lines with improved fruit quality.
Abbreviations
- AS:
-
acetosyringone
- BA:
-
6-benzyladenine
- GA3 :
-
gibberellic acid
- GFP:
-
green fluorescent protein
- IBA:
-
indole butyric acid
- KT:
-
kinetin
- MT medium:
-
Murashige and Tucker (1969)
- NAA:
-
α-napthaleneacetic acid
- NPT II:
-
neomycin phosphotransferase II
References
Almeida WAB, Filho FAAM, Mendes BMJ, Pavan A, Rodriguez APM (2003) Agrobacterium-mediated transformation of Citrus sinensis and Citrus limonia epicotyl segments. Sci Agric 60(1):23–29
Ananthakrishnan G, Orbovic V, Pasquali G, Grosser JW (2007) Transfer of CTV-derived resistance candidate sequences to four grapefruit cultivars through Agrobacterium-mediated genetic transformation. In Vitro Cell Dev Biol Plant 43:593–601
Boscariol RL, Almeida WAB, Derbyshire MTVC, Filho FAAM, Mourao FAA, Mendes BMJ (2003) The use of the PMI/mannose selection system to recover transgenic sweet orange plants (Citrus sinensis L. Osbeck). Plant Cell Rep 22:122–128
Cervera M, Juarez J, Navarro A, Pina JA, Duran-Vila N, Dominguez A, Navarro L, Peña L (1998) Genetic transformation and regeneration of mature tissues woody fruit plants bypassing the juvenile stage. Transgenic Res 7:51–59
Cervera M, Ortega C, Navarro A, Navarro M, Peña L (2000) Generation of transgenic citrus plants with tolerance-to-salinity gene HAL2 from yeast. J Hortic Sci Biotechnol 75(1):26–30
Cervera M, Juarez J, Navarro L, Peña L (2005) Genetic transformation of mature citrus plants. In: Peña L (ed) Transgenic plants, methods and protocols. Humana, Totowa, NJ, pp 177–188
Cervera M, Navarro A, Navarro L, Peña L (2008) Production of transgenic adult plants from clementine mandarin by enhancing cell competence for transformation and regeneration. Tree Physiol 28:55–66
Cheng YJ, Guo WW, Yi HL, Pang XM, Deng XX (2003) An efficient protocol for genomic DNA extraction from Citrus species. Plant Mol Biol Rep 21:177–178
Costa MGC, Otoni WC, Moore GA (2002) An evaluation of factors affecting the efficiency of Agrobacterium-mediated transformation of Citrus paradisi (Macf.) and production of transgenic plants containing carotenoid biosynthetic genes. Plant Cell Rep 21:365–373
Deng XX (2005) Advances in worldwide citrus breeding. Acta Horticulturae Sinica 32(6):1140–1146 in Chinese with English abstract
Deng XX, Duan YX (2006) Modification of perennial fruit trees. In: Fladung M, Ewald D (eds) Tree transgenesis: recent developments. Springer, Berlin, pp 48–66
Dominguez A, Cervera M, Perez RM, Romero J, Fagoaga C, Cubero J, Lopez MM, Juarez JA, Navarro L, Peña L (2004) Characterisation of regenerants obtained under selective conditions after Agrobacterium-mediated transformation of citrus explants reveals production of silenced and chimeric plants at unexpected high frequencies. Mol Breed 14:171–183
Duan YX, Guo WW, Meng HJ, Tao NG, Li DD, Deng XX (2007a) High efficient transgenic plant regeneration from embryogenic calluses of Citrus sinensis. Biol Plant 51(2):212–216
Duan YX, Liu X, Fan J, Li DL, Wu RC, Guo WW (2007b) Multiple shoot induction from seedling epicotyls and transgenic citrus plant regeneration containing the green fluorescent protein gene. Botanical Studies 48:165–171
Endo T, Shimada T, Fujii H, Kobayashi Y, Araki T, Omura M (2005) Ectopic expression of an FT homolog from Citrus confers an early flowering phenotype on trifoliate orange (Poncirus trifoliata L. Raf.). Transgenic Res 14:703–712
Fagoaga C, Tadeo FR, Iglesias DJ, Huerta L, Lliso I, Vidal AM, Talon M, Navarro L, Garcia-Martinez JL, Peña L (2007) Engineering of gibberellin levels in citrus by sense and antisense overexpression of a GA 20-oxidase gene modifies plant architecture. J Exp Bot 58(6):1407–1420
Gentile A, Deng ZN, Malfa SL, Domina F, Germana C, Tribulato E (2004) Morphological and physiological effects of rolABC genes into Citrus genome. Acta Hortic 632:235–242
Ghorbel R, Juarez J, Navarro L, Peña L (1999) Green fluorescent protein as a screenable marker to increase the efficiency of generating transgenic woody fruit plants. Theor Appl Genet 99:350–358
Ghorbel R, Dominguez A, Navarro L, Peña L (2000) High efficiency genetic transformation of sour orange (Citrus aurantium L.) and production of transgenic trees containing the coat protein gene of citrus tristeza virus. Tree Physiol 20:1183–1189
Gonzalez-Ramos J, Graham J, Mirkov TE (2005) Transformation of citrus cultivars with genes encoding potential resistance to citrus canker (Xanthomonas axonopodis pv citri). Phytopathology 95(6):S35
Grosser JW, Gmitter FG Jr, Fleming GH, Chandler JL (2000) Applications of biotechnology to citrus cultivar improvement at the Citrus Research and Education Center. Acta Hortic 535:213–220
Guo WW, Duan YX, Olivares-Fuster O, Wu RCZC, Arias CR, Burns JK, Grosser JW (2005) Protoplast transformation and regeneration of transgenic Valencia sweet orange plants containing a juice quality-related pectin methylesterase gene. Plant Cell Rep 24(8):482–486
Gutiérrez-E MA, Luth D, Moore GA (1997) Factors affecting Agrobacterium-mediated transformation in Citrus and production of sour orange (Citrus aurantium L.) plants expressing the coat protein gene of citrus tristeza virus. Plant Cell Rep 16:745–753
Haseloff J, Siemering KR, Prasher DC, Hodge S (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci U S A 94:2122–2127
Iwanami T, Shimizu T, Ito T, Hirabayashi T (2004) Tolerance to citrus mosaic virus in transgenic trifoliate orange lines harboring capsid polyprotein gene. Plant Dis 88(8):865–868
Kaneyoshi J, Kobayashi S, Nakamura Y, Shigemoto N, Doi Y (1994) A simple and efficient gene transfer system of trifoliate orange (Poncirus trifoliata Raf.). Plant Cell Rep 13:541–545
Koltunow AM, Brennan P, Bond JE, Barker SJ (1998) Evaluation of genes to reduce seed size in Arabidopsis and tobacco and their application to Citrus. Mol Breed 4:235–251
Koltunow AM, Brennan P, Protopsaltis S (2000) Regeneration of West Indian limes (Citrus aurantifolia) containing genes for decreased seed set. Acta Hortic 535:81–91
Li DD, Shi W, Deng XX (2002) Agrobacterium-mediated transformation of embryogenic calluses of Ponkan mandarin and the regeneration of plants containing the chimeric ribonuclease gene. Plant Cell Rep 21:153–156
Liang SQ, Wang XZ, Wan TX (1999) Study on biological characteristic and experiment of rootstock of precocious trifoliate orange. Zhejiang Citrus 16(3):2–4 in Chinese
Liu Q, Xu J, Liu YZ, Zhao XL, Deng XX, Guo LL, Gu JQ (2007) A novel bud mutation that confers abnormal patterns of lycopene accumulation in sweet orange fruit (Citrus sinensis L. Osbeck). J Exp Bot 58:4161–4171
Moreira-Dias JM, Molina RV, Bordon Y, Guardiola JL, Garcia-Luis A (2000) Direct and indirect shoot organogenic pathways in epicotyl cuttings of Troyer citrange differ in honmone requirements and in their response to light. Ann Bot 85:103–110
Murashige T, Tucker DPH (1969) Growth factor requirements of citrus tissue culture. Proc First Int Citrus Symp 3:1155–1161
Omar AA, Grosser JW (2008) Comparison of endoplasmic reticulum targeted and non-targeted cytoplasmic GFP as a selectable marker in citrus protoplast transformation. Plant Sci 174:131–139
Omar AA, Song WY, Grosser JW (2007) Introduction of Xa21, a Xanthomonas-resistance gene from rice, into ‘Hamlin’ sweet orange [Citrus sinensis (L.) Osbeck] using protoplast-GFP co-transformation or single plasmid transformation. J Hortic Sci Biotechnol 6:914–923
Payne T, Johnson SD, Koltunow AM (2004) KNUCKLES (KNU) encodes a C2H2 zinc-finger protein that regulates development of basal pattern elements of the Arabidopsis gynoecium. Development 131:3737–3749
Peña L, Cervera M, Juarez J, Ortega C, Pina JA, Duran-Vila N, Navarro L (1995) High efficiency Agrobacterium-mediated transformation and regeneration of citrus. Plant Sci 104:183–191
Peña L, Martin-Trillo M, Juarez J, Pina JA, Navarro L, Martinez-Zapater JM (2001) Constitutive expression of Arabidopsis LEAFY or APETALA1 genes in citrus reduces their generation time. Nat Biotechnol 19:263–267
Peña L, Cervera M, Ghorbel R, Dominguez A, Fagoaga C, Juarez J, Pina JA, Navarro L (2003) Transgenic citrus. In: Jaiwal PK, Singh RP (eds) Plant genetic engineering, vol. 3: improvement of commercial plants. SciTech, Houston, TX, pp 261–282
Peña L, Perez RM, Cervera M, Juarez JA, Navarro L (2004) Early events in Agrobacterium-mediated genetic transformation of citrus explants. Ann Bot 94:67–74
Petri C, Webb K, Hily JM, Dardick C, Scorza R (2008) High transformation efficiency in plum (Prunus domestica L.): a new tool for functional genomics studies in Prunus spp. Mol Breed 22:581–591
Wong WS, Li GG, Ning W, Xu ZF, Wendy Hsiao WL, Zhang LY, Li N (2001) Repression of chilling-induced ACC accumulation in transgenic citrus by over-production of antisense l-aminocyclopropane-l-carboxylate synthase RNA. Plant Sci 161:969–977
Yang L, Xu CJ, Hu GB, Chen KS (2007) Establishment of an Agrobacterium-mediated transformation system for Fortunella crassifolia. Biol Plantarum 51(3):541–545
Yu CH, Huang S, Chen C, Deng Z, Ling P, Gmitter FG Jr (2002) Factors affecting Agrobacterium-mediated transformation and regeneration of sweet orange and citrange. Plant Cell Tissue Organ Cult 71:147–155
Zanek MC, Reyes CA, Cervera M, Peña EJ, Velazquez K, Costa N, Plata MI, Grau O, Peña L, Garcia ML (2008) Genetic transformation of sweet orange with the coat protein gene of Citrus psorosis virus and evaluation of resistance against the virus. Plant Cell Rep 27:57–66
Zheng QF, Chen CX, Huang S, Choi YA, Gmitter FG Jr (2006) The green fluorescent protein (GFP) is a vital visual marker in citrus transgene research. Electron J Biol 2(1):1–5
Acknowledgements
This research was financially supported by the National Natural Science Foundation of China, the National 863 Project (2006AA100108, 2007AA10Z182), and the Key Project of Hubei Provincial Natural Science Foundation (no. 2008CDA069).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by F. Gmitter
Rights and permissions
About this article
Cite this article
Tan, B., Li, DL., Xu, SX. et al. Highly efficient transformation of the GFP and MAC12.2 genes into precocious trifoliate orange (Poncirus trifoliata [L.] Raf), a potential model genotype for functional genomics studies in Citrus . Tree Genetics & Genomes 5, 529–537 (2009). https://doi.org/10.1007/s11295-009-0206-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-009-0206-0