Abstract
Knowledge of pistachio genetic diversity is necessary for the formulation of appropriate management strategies for the conservation of these species. We analysed amplified fragment length polymorphisms in a total of 216 pistachio accessions, which included seven populations from three wild species (Pistacia vera, Pistacia khinjuk and Pistacia atlantica subsp. kurdica) and most of the important cultivars from Iran, together with some foreign cultivars. High levels of genetic diversity were detected within the Iranian cultivars, and they showed a clear separation from foreign cultivars, as revealed by unweighted pair group method with arithmetic averaging and supported by analysis of molecular variance. The lowest amount of polymorphism was observed in P. atlantica subsp. kurdica, which showed the lowest number of total bands as compared to the other species. This revealed strong genetic erosion of P. atlantica subsp. kurdica, which reflected a severe decline in habitat and over-exploitation. Based on these findings, strategies are proposed for the genetic conservation and management of pistachio species and cultivars.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Iran is considered to be one of the main centres of genetic diversity for pistachio due to the fact that a large number of cultivated pistachio accessions, in addition to wild species, are found in this region. In total, these species form the major part of Iranian forests. The deciduous, dioecious and wind-pollinated tree species Pistacia vera L. is a member of the Anacardiaceae family. P. vera is the only cultivated and commercially grown species in the genus (Zohary 1996). Pistachio cultivation dates back to ancient times and probably started in an area that is close to the wild stands of pistachio, in the low mountains and foothills of the semi-desert region of south-central Asia. This range extends from northeastern Iran and northern Afghanistan to western Tien-Shan and the Karatau mountains, through Turkmenistan, Uzbekistan, Tajikistan, Kazakhstan and Kyrgyzstan (Kayimov et al. 2001). From its presumed centre of origin, pistachio cultivation spread first within the ancient Persian Empire and then moved gradually westward. In fact, according to Joret (1976), the name pistachio appears to originate from the word ‘pista-pistak’ in the ancient Persian language ‘Avestan’ (Hormaza and Wünsch 2007). Today, Iran, which has 350,000 ha that are devoted to growing pistachios and which produces more than 300,000 tons/year, is the main pistachio producer in the world (FAO 2006).
The Iranian pistachio industry relies upon a limited number of cultivars, which are speculated to have a narrow genetic base (Maggs 1973; Tous and Ferguson 1996). Therefore, they are vulnerable to attack by pests or diseases, which means that the well-being of thousands of people who depend on this crop as a source of livelihood is at risk. It has been suggested that the reason for this low genetic diversity is due to the long juvenile period of the tree (10–12 years) and its long life span (Whitehouse and Stone 1941). Today, even in areas where the greatest diversity is found, traditional cultivars are being replaced by a small number of improved varieties. The destruction of natural habitat is also contributing to the loss of wild species. However, the majority of the current research efforts are being directed towards crop breeding (Parfitt 1995) rather than the sustainable exploitation of existing diversity within wild or cultivated species (Bahsa et al. 2007).
The genus Pistacia includes 11 species and, in addition to the cultivated pistachio (P. vera L.), three wild species of Pistacia, P. vera, Pistacia khinjuk and Pistacia atlantica, are found in Iran. P. atlantica has three subspecies: P. atlantica subsp. mutica, P. atlantica subsp. kurdica and P. atlantica subsp. cabulica (Katamsaz 1988). Wild pistachio forests are distributed throughout most parts of Iran and cover approximately 2,400,000 ha. Wild species are important in forest regions because they provide protection from wind and soil erosion, which contributes to soil stability, and they also provide resin, fruit and oil. In addition, they are important for the development of new pistachio varieties because they can be a source of resistance to biotic and abiotic stresses, including drought tolerance and growth under poor soil conditions.
Molecular methods are being used increasingly to study diversity in crops. The use of molecular techniques to assess, at the DNA level, the genetic distance and variation between accessions is widespread. The amplified fragment length polymorphism (AFLP) technique (Vos et al. 1995) provides a reliable tool for estimation of the degree of genetic relatedness among a number of members of the plant kingdom (Hartl and Seefelder 1998; Manubens et al. 1999; Angiolillo et al. 1999). It is also useful for diversity studies, phylogeny, genomic linkage mapping and the identification of different varieties (Sensi et al. 1996; Maughan et al. 1996; Wang et al. 1997; Xu et al. 2000; Katsiotis et al. 2003; Golan-Goldhirsh et al. 2004; Kafkas 2006). AFLPs have also been recognised as a more reliable DNA marker system than random amplification of polymorphic DNA or microsatellite markers (Powell et al. 1996; Winfield et al. 1998; Russell et al. 1999).
In this study, AFLP markers were used (1) to investigate the extent of diversity among cultivated varieties of P. vera L. that were gathered from across Iran and to assess the level of diversity that is maintained on farms and (2) to provide basic information that can be used for gene conservation and the management of wild pistachio forests, by studying the genetic diversity within and among populations of the different species.
Materials and methods
Plant material
A total of 216 pistachio (Pistacia spp.) genotypes were analysed: 43 local cultivars from the main pistachio collection in Iran (Rafsanjan); 23 foreign cultivars (ten male and 13 female) from the Centro di Ricerca per la Frutticoltura (CRA); and 151 individuals, which represented one, two and four populations of P. khinjuk, P. atlantica subsp. kurdica and P. vera (wild type), respectively (Tables 1 and 2). Young leaf samples were collected and immersed immediately in liquid nitrogen. DNA extraction was carried out using the DNeasy Plant Mini Kit (Qiagen).
AFLP analysis
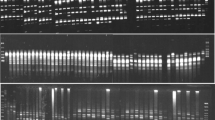
AFLP analysis was performed based on the method described by Vos et al. (1995) using the restriction enzymes EcoRI and MseI. The following seven primer combinations were chosen for the final selective amplification: EcoRI + TAT/MseI + ATA, EcoRI + TAT/MseI + CAT, EcoRI + GTT/MseI + CAA, EcoRI + GTT/MseI + CAT, EcoRI + GTT/MseI + GAG, EcoRI + GTT/MseI + ATA and EcoRI + ACT/MseI + CAT (see Table 3 for primer sequences). The amplified fragments were separated by 6% denaturing polyacrylamide gel electrophoresis on a Sequencing gel apparatus (Bio-Rad). The AFLP analysis of some samples was repeated twice in order to check the reproducibility of the data.
Data analysis
The amplified bands for all the individuals were scored as either 1 (present) or 0 (absent), and only unambiguous bands were considered. GenAlEx 6 software (Peakal and Smouse 2006) was used for the analysis of molecular variance (AMOVA; Schneider et al. 2000) in order to partition the genetic variation among species, among populations within species and among individuals within populations. The significance of each variance component was tested with permutation tests (Excoffier et al. 1992). Genetic distances were estimated according to Nei (1987), and principal coordinate analysis (PCO) (Gower 1966) and neighbour joining (NJ) analysis were performed. The NJ dendrogram was constructed with the Mega 4 software (Tamura et al. 2007). Pairwise genetic similarity was estimated using Jacard of the software package NTSYS-pc (Rohlf 2004), according to Nei and Li (1979). The similarity matrix was used to construct a dendrogram for all 65 cultivated accessions (Iranian and foreign cultivars) using the SHAN module of NTSYS-pc (Rohlf 2004), which is based on the unweighted pair group method with arithmetic averaging (UPGMA). The COPH option in NTSYS-pc was used to generate a matrix of cophenetic values, which was used in turn with the MXCOMP option to calculate the correlation between the cophenetic matrix and the original SIMQUAL matrix. This analysis measured goodness-of-fit under 1,000 permutations and provided a cophenetic correlation value (r).
Results
AFLP variation
The seven combinations of AFLP primer pairs yielded a total of 151 distinct polymorphic bands. The number of polymorphic loci that were amplified by each primer combination varied from eight to 32, with an average of 21.6 bands per primer pair. The percentage of polymorphic loci was 53% on average. The highest degree of polymorphism in the wild species was observed among wild populations of P. vera, whereas the lowest was in P. atlantica subsp. kurdica (Table 4). The level of polymorphism among the Iranian cultivars was twice that seen in the foreign cultivars. Two population-specific bands were observed in the former data set; however, no population-specific bands were found in the latter. The number of alleles per locus (Na) with a frequency of ≥5% was slightly higher in the wild species than in the cultivated accessions of P. vera.
Assuming Hardy–Weinberg equilibrium, the values of Nei’s genetic diversity (He) ranged from 0.08 (foreign cultivars) to 0.22 (wild-type populations of P. vera). The He values for the wild accessions of P. vera (0.22) and P. khinjuk (0.20) were rather similar, whereas the value for the populations of P. atlantica subsp. kurdica was lower (0.11). At the population level, four wild populations of P. vera and two populations of P. atlantica subsp. kurdica had rather similar He values.
Genetic relationships
Estimates of the genetic distances between the analysed datasets of the pistachios (wild species and pooled Iranian and foreign cultivars) ranged from 0.035 (P. vera Kh1–P. vera S) to 0.296 (P. atlantica subsp. kurdica–P. khinjuk; Table 5). As expected, the Iranian cultivars showed the most genetic similarity to P. vera (wild type) (0.065), whereas the foreign cultivars were more similar to P. atlantica subsp. kurdica (0.062) than to P. vera (0.144). Both the foreign and Iranian cultivars showed the greatest genetic distance from P. khinjuk (0.280 in both cases).
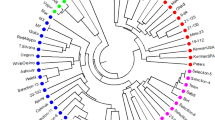
The NJ phenogram was constructed using Nei’s genetic distance (Fig. 1). The NJ tree had long terminal branches, which suggested that the populations/cultivars/species were well differentiated and that the relationships among the pistachio cultivars/species were well resolved. The branching order and the grouping of species and populations/cultivars (clades) were consistent and supported the phenetic analysis. All the populations in each species clustered together. The branching of P. khinjuk with wild-type populations of P. vera suggested that a direct relationship existed between them. P. atlantica subsp. kurdica was found to be related closely to the foreign cultivars. PCO was also used (Fig. 2). The first two coordinates separated the species well and accounted for 89.4% of the total variation (Fig. 2a). The Iranian cultivars were found to be related more closely to the wild-type populations of P. vera, as compared to the foreign cultivars, which were at a greater distance. The separation of the female and male individuals of foreign origin into two groups (Fig. 2b) or the inclusion of only the female individuals (Fig. 2c) did not change considerably the pattern of the genetic relationships among the wild species and pooled Iranian and foreign cultivars.
AMOVA was used to estimate and partition the total molecular variance among species, populations within species and within populations, as well as to test the significance of partitioned variance components using a permutation procedure. Variation among species accounted for 19% of the total variation, among populations within species 14% and within populations 67% (Table 6). The variation for all three sources was significant (P = 0.01). Although there were large morphological differences between species, the proportion of the total variation that was due to variation between species (19%) was smaller than that due to variation within populations (67%) but larger than that due to variation between populations within each species (14%).
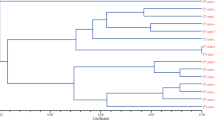
The UPGMA dendrogram, which was based on Jaccard’s similarity matrix, clustered the 65 genotypes in two main groups (Fig. 3). Group A included all the cultivars of Iranian origin, which were further divided into subgroups, whereas group B included the cultivars of foreign origin. The cophenetic correlation between the dendrogram and the similarity matrix revealed a good degree of fit (r = 0.9). Inclusion of either the male accessions of foreign origin (Fig. 4) or the female accessions (Fig. 5) with the Iranian cultivars resulted in UPGMA dendrograms that again contained two main groups, similar to those in Fig. 3. PCO pattern (Fig. 6) was comparable to the clustering of the accessions in the dendrogram. The Iranian cultivars were located apart from the foreign ones.
Three-dimensional scatter plot of PCO of 65 Iranian and foreign cultivars. The numbers refer to the codes of the pistachio genotypes as shown in Table 1
Discussion
Molecular markers have been used previously to estimate genetic relationships among different species and cultivars of pistachio. In particular, AFLP technology has been shown in earlier studies to be useful for discriminating between species and cultivars [Float1](Katsiotis et al. 2003; Kafkas 2006; Kafkas et al. 2006a,b). In the present study, we used AFLP markers to evaluate the genetic diversity among wild pistachio species and cultivars from Iran in comparison with foreign cultivars. In our investigation, we found that seven combinations of primer pairs produced a relatively high number of polymorphic bands (151) among 219 accessions, which indicates that a high level of genetic diversity exists within the germplasm of the wild species and cultivars of pistachio.
We showed that all the cultivars that were analysed could be separated into two distinct groups, as revealed by both UPGMA and PCO analyses and further justified by AMOVA. Due to the fact that almost half of the foreign cultivars that were tested were male pollinators, which, by their nature (non-commercially harvested), can accommodate much greater levels of interspecific gene introgression, we compared the UPGMA dendrograms that were derived for Iranian/foreign cultivars, Iranian/male foreign individuals and Iranian/female foreign individuals. Interestingly, in all three UPGMA dendrograms, the existence of two distinct groups (Iranian and foreign) was obvious. These results are in agreement with those of Hormaza et al. (1994) and Kafkas and Perl-Treves (2001), who screened hundreds of primers in Pistacia in order to identify sex-specific polymorphisms but found only a few putative sex-related bands[Float1]. They have suggested that the low frequency of sex-linked bands may indicate that the number of DNA segments that is involved in sex determination is small and probably involves a single or very few genes. Such a clear-cut differentiation between the Iranian and foreign-cultivated germplasm was in agreement with the results of previous studies (Hormaza et al. 1994; Kafkas et al. 2006a). In both these studies, Iranian cultivars were separated distinctly from cultivars that originated from other countries (within the Mediterranean region). Therefore, our study, which examined a larger set of Iranian pistachio germplasms, supported the previous reports. Knowledge of the genetic variation and genetic relationship among pistachio genotypes is important for the efficient utilisation of the available germplasm resources.
Native Iranian cultivars demonstrated a wider degree of diversity than the foreign cultivars that were examined. This fact confirms our speculation that the cultivation of pistachio has led increasingly to a reduction in genetic variation due to the utilisation of improved varieties and to the availability of private- or public-grafted seedling nurseries for pistachio. In addition, changing livelihood conditions may have had an effect; for example, in the past, exchange of local cultivars among farmers was very popular along with the consumption of nuts from wild species, which were also object of exchange based on a 50% ratio with cultivated material. Recently, the method of pistachio cultivation has changed, and this has also led to an increased reduction of diversity in the cultivars that are used on farms. In the past (until approximately 25 years ago), a high level of pistachio diversity in the field was maintained through a number of cultivation practices, such as the use of male varieties that were derived from seed, the use of wild Pistacia species to boost pollination and hence fruit setting and the use of natural populations of wild Pistacia (P. atlantica) as rootstock because of their well-known resistance to stony and calcareous soils (Bahsa et al. 2007).
These results suggest that the Iranian cultivars represent an interesting and unique genetic resource that needs to be conserved and maintained. Although the history of pistachio domestication and cultivation in Iran is not yet clear, archaeological findings indicate that pistachio cultivation spread first within the ancient Persian Empire and then expanded gradually westward (Hormaza and Wünsch 2007). Therefore, the clear-cut separation that is observed between the total Iranian- and foreign-cultivated germplasm may be due to diverse anthropogenic processes throughout history.
The domestication of P. vera and the spread of pistachio cultivation far beyond the natural range of its wild progenitor brought the crop into contact with several different Pistacia species. Zohary (1996) states that, in traditional areas of pistachio cultivation, contact between the cultivated clones and wild species is quite common and has existed for hundreds or even thousands of years. Therefore, cross-pollination between these sympatric species takes place and may have led, over the centuries, to the development of various interspecific hybrids with different backgrounds (Maggs 1973; Kafkas and Perl-Treves 2001). Interspecies introgression in Turkey is so prevalent that Kafkas and Perl-Treves (2001) have reported that, in this region, it is sometimes difficult to obtain uncontaminated wild material and to identify a Pistacia tree correctly on the basis of its morphology. In the present study, interspecies introgression may explain the closer genetic similarity of foreign cultivars to accessions of P. atlantica subsp. kurdica.
At the interspecific level, molecular DNA markers have been used in Pistacia to analyse the phylogenetic and genetic similarity among the species of the genus (Kafkas and Perl-Treves 2001, 2002; Katsiotis et al. 2003; Golan-Goldhirsh et al. 2004; Kafkas 2006). Zohary (1952), on the basis of morphological observations, has suggested that P. vera and P. khinjuk are the most primitive species of Pistacia, and this has been confirmed by the previously mentioned studies. According to the results of the present study, P. atlantica subsp. kurdica, which was identified taxonomically by Khatamsaz (1988), originated directly from P. vera (Fig. 1). Furthermore, P. khinjuk is also connected directly to P. vera. The shorter distance between P. vera and P. atlantica subsp. kurdica, as compared to P. khinjuk, suggests that the former diverged from P. vera more recently.
Considerations for conservation
The primary objective in nature conservation is to preserve the evolutionary potential of species through maintaining as much genetic diversity as possible. Thus, knowledge of the genetic variation between and within different populations of plant species plays a significant role in the formulation of appropriate strategies for their conservation (Miligan et al. 1994). According to our results, the genetic diversity of P. atlantica subsp. kurdica, as compared to the other species that were examined, is relatively low. This may be related to a severe decline in habitat, in addition to the over-collection and over-exploitation of P. atlantica (Fig. 7). The fruits of P. atlantica are used as edible nuts and for oil extraction (56% of the kernel and 30% of the total weight is composed of oil). In addition, physical and internal seed dormancy, low seed germination and low seedling vigor in the early stages make regeneration of this species difficult. Moreover, this species has experienced strong selection for higher resin production, which has resulted in genetic erosion (Esmail-pour 2001). Although further study is necessary, including that of the other two P. atlantica subsp., P. atlantica subsp. cabolica and P. atlantica subsp. mutica, the present analysis provides information that policy makers and scientists can use to improve the conservation and sustainable use of the major part of wild pistachio forests. The low genetic diversity of P. atlantica subsp. kurdica in comparison with that of other species reveals its susceptibility to anthropogenic processes, environmental change and the presence of biotic stresses. Protection by in situ conservation and the management of seed and resin exploitation to prevent the loss of genetic diversity is required urgently.
The present analysis revealed that Iranian-cultivated germplasm is highly variable and genetically distinct from foreign accessions. This can be attributed to different local genetic backgrounds, particular breeding pressures and the limited interchange of genetic material. The apparently unique nature of the Iranian pistachio germplasm, which was revealed by our results, supports the case for the implementation of more intense characterisation and conservation strategies.
References
Angiolillo A, Mencuccini M, Baldoni L (1999) Olive genetic diversity assessed using amplified fragment length polymorphisms. Theor Appl Genet 98:411–421
Bahsa AI, Padulosi S, Chabane K (2007) Genetic diversity of Syrian pistachio (Pistacia vera L.) varieties evaluated by AFLP markers. Gene Resour Crop Evol 54:1807–1816, doi:10.1007/s10722-006-9202-5
Esmail-pour A (2001) Distribution, use and conservation of pistachio in Iran. In: Padulosi S, Hadj-Hassan A (eds) In towards a comprehensive documentation and use of pistacia genetic diversity in central and West Asia, North Africa and Europe. Report of the IPGRI workshop, 14–17 December 1998, Ibrid, Jordan. IPGRI, Rome, Italy
Excoffier L, Smouse P, Quattro J (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
FAO (2006) FAOSTAT database. http://apps.fao.org/page/form?collection= Production.Crops.Primary&Domain=Production&servlet=1&language=EN&hostname=apps.fao.org&version=default
Golan-Goldhirsh A, Barazani O, Wang ZS, Khadkal DK, Saunders JA, Kostiukovsky V, Rowland LJ (2004) Genetic relationships among Mediterranean Pistacia species evaluated by RAPD and AFLP markers. Plant Systemat Evol 246:9–18
Gower JC (1966) Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 53:325–338
Joret C (1976) Les Plantes dansL'antiquité et au moyen âge; histoire, usages et symbolisme. Slatkine Reprints, Genève. Reprinted from the book first published in 1897–1904
Hartl L, Seefleder S (1998) Diversity of selected Hop cultivars detected by fluorescent AFLPs. Theo Appl Gene 96:112–116
Hormaza JI, Wünsch A (2007) Pistachio. In: Kole C (ed) Genome mapping and molecular breeding in plants, fruits and nuts, vol 4. Springer, New York, USA, pp 243–251
Hormaza JI, Dollo L, Polito VS (1994) Determination of relatedness and geographic movements of Pistacia vera (Pistachio; Anacardiaceae) germplasm by RAPD analysis. Econ Bot 48:349–358
Kafkas S (2006) Phylogenetic analysis of the genus Pistacia by AFLP markers. Plant Syst Evol 262:113–124
Kafkas S, Perl-Treves R (2001) Morphological and molecular phylogeny of Pistacia species in Turkey. Theo Appl Gene 102:908–915
Kafkas S, Perl-Treves R (2002) Interspecific relationships in Pistacia based on RAPD fingerprinting. Hort Sci 37:168–171
Kafkas S, Kaska A, Wassimi AN, Padulosi S (2006a) Molecular characterisation of Afghan pistachio accessions by amplified fragment length polymorphisms (AFLPs). J Hort Sci Biotechnol 81:864–868
Kafkas S, Ozkan H, Erol Ak B, Acar I, Alti HS (2006b) Detecting DNA polymorphism and genetic diversity in a wide pistachio germplasm: comparison of AFLP, ISSR, and RAPD marker. J Am Soc Hort Sci 131:522–529
Katsiotis A, Hagidimitriou M, Drossoul A, Pontikis C, Loukas M (2003) Genetic relationships among species and cultivars of Pistacia using PAPDs and AFLPs. Euphytica 132:279–286
Kayimov AK, Sultanov RA, Chernova GM (2001) Pistacia in Central Asia. In: Padulosi S, Hadj-Hassan A (eds) Project on underutilizes Mediterranean species. Pistacia: towards a comprehensive documentation of distribution and use of its genetic diversity in Central & West Asia, North Africa and Mediterranean Europe. IPGRI, Rome, Italy
Khatamsaz M (1988) Flora of Iran No. 30: Anacardiaceae. Research Institute of Forests and Rangelands, Tehran, Iran
Maggs DH (1973) Genetic resources in pistachio. Plant Gene Resour Newsle 29:7–15
Manubens A, Lobos S, Jadue Y, Toro M, Messina R, Lladser M, Seelenfrund D (1999) DNA isolation and AFLP fingerprinting of nectarine and peach varieties (Prunus persica). Mol Biol Rep 17:255–267
Maughan PJ, Saghai Maroof MA Buss GR, Huestis GM (1996) Amplified fragment length polymorphism (AFLP) in soybean: species diversity, inheritance, and neaisogenic line analysis. Theo Appl Gene 93:392–401
Milligan BG, Leebens-Mack J, Strand AE (1994) Conservation genetics: beyond the maintenance of marker diversity. Mol Ecol 12:844–855
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Nei M, Li W (1979) Mathematical model for study genetic variation in terms of restriction endonucleases. Proc Nati Acad Sci USA 74:5267–5273
Parfitt DE (1995) Pistachio cultivars. In: Ferguson L (ed) Pistachio production. University of California, Davis, pp 43–46
Peakal R, Smouse PE (2006) GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (Microsatellites) markers for germplasm analysis. Mol Breed 2:225–235
Rohlf JF (2004) NTSYS-pc: numerical taxonomy and multivariate analysis system, version 2.11. Exeter, Setauket, NY
Russell JR, Weber JC, Booth A, Powell W, Sotelo-Montes C, Dawson IK (1999) Genetic variation of Calycophyllum spruceanum in the Peruvian Amazon basin, revealed by amplified fragment length polymorphism (AFLP) analysis. Mol Ecol 8:199–204
Schneider S, Roessli D, Excoffier L (2000) Arlequin: A software for population genetics data analysis version 2.000 genetics and biometry laboratory, dept. of anthropology. University of Geneva, Switzerland
Sensi E, Vignani R, Rhode W, Biricolti S (1996) Characterization of genetic biodiversity with Vitis vinifera L. sangiovese and colorino genotypes by AFLP and ISTR DNA marker technology. Vitis 35:183–188
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tous J, Fergusen L (1996) Mediterranean fruits. In: Janick J (ed) Progress in new crops. ASHS, Arlington, VA
Vos P, Hogers R, Bleeker M, Reijans M, van der Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Wang YH, Thomas CE, Dean RA (1997) A genetic map of melon (Cucumis melo L.) based on amplified fragment length polymorphism (AFLP) Markers. Theo Appl Gene 95:791–798
Whitehouse WE, Stone CL (1941) Some aspects of dichogamy and pollination in pistachio. Am Soc Hort Sci 39:95–100
Winfield MO, Arnold GM, Cooper F, Le Ray M, White J, Karp A, Edwards KJ (1998) A study of genetic diversity in Populus nigra subsp. betulifolia in The upper seven area of The UK using AFLP markers. Mol Ecol 7:3–10
Xu RQ, Tomooka N, Vaughan DA (2000) AFLP markers for characterizing the Azuki Bean complex. Crop Sci 40:808–815
Zohary M (1952) A monographical study of the genus Pistacia. Palest J Bot Jerus Ser 5:187–228
Zohary D (1996) The genus Pistacia L. In: Padulosi S, Caruso T, Barone E (eds) Taxonomy, distribution, conservation and uses of Pistacia genetic resources. IPGRI, Palermo, Italy, pp 1–11
Acknowledgments
This study was supported by the Agricultural Biotechnology Research Institute of Iran. The authors would like to thank Dr. Hasan Maddah Arefy from the Research Institute of Forests and Rangelands of Iran for the photographs of pistachio. They would also like to thank Dr A. A. Javanshah (Director General of the Iranian Pistachio Research Institute), in particular, for his contribution to the collection of plant material.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shanjani, P.S., Mardi, M., Pazouki, L. et al. Analysis of the molecular variation between and within cultivated and wild Pistacia species using AFLPs. Tree Genetics & Genomes 5, 447–458 (2009). https://doi.org/10.1007/s11295-008-0198-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-008-0198-1