Abstract
The mating system, patterns of pollen mediated gene flow and levels of genetic contamination were investigated in a planted stand of Acacia saligna subsp. saligna via paternity analysis using microsatellite markers. High levels of outcrossing were detected within the stand (t m = 0.98), and the average pollen dispersal distance was 37 m with the majority of progeny sired by paternal trees within a 50-m neighbourhood of the maternal tree. Genetic contamination from the natural background population of A. saligna subsp. lindleyi was detected in 14% of the progeny of A. saligna subsp. saligna and varied among maternal trees. Long distance inter-subspecific pollen dispersal was detected for distances of over 1,500 m. The results provide information for use in the breeding and domestication programme aimed at developing A. saligna as an agroforestry crop for the low rainfall areas of southern Australia.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tree species tend to be highly outcrossing and exhibit extensive levels of pollen mediated gene flow over large distances especially when populations are fragmented (Bacles et al. 2005; Byrne et al. 2007; Dick 2001; Nason and Hamrick 1997; White et al. 2002). Understanding the mating system and patterns of pollen dispersal in tree species is important for achieving conservation and natural resource management aims as well as for success in breeding programmes for improvement and domestication.
In the context of breeding and domestication programmes, knowledge of mating systems, patterns of pollen mediated gene flow and levels of genetic contamination are vital for the efficient management of production populations such as seed orchards (El-Kassaby 2000). Seed orchards are cultivated for the abundant production of easily harvestable seed from superior trees and are established by setting out clones or seedling progeny of trees selected for desired characteristics. Most tree seed orchards are open pollinated and aim to produce seed crops of high genetic value capturing an adequate amount of genetic diversity (Kang et al. 2001).
The genetic efficiency of seed orchards can be adversely affected by a number of factors including a high proportion of selfing events, departures from random mating because of factors such as unequal contribution of individuals to seed crops and genetic contamination with pollen from trees located outside the orchard (Friedman and Adams 1985). To implement suitable orchard design and management options and achieve predicted genetic gains, it is useful to have knowledge of outcrossing rates, patterns of pollen dispersal and maternal and paternal reproductive contributions. It is also important to determine the levels of long distance gene flow that may lead to genetic contamination from outside the orchard (Adams and Burczyk 2000).
Acacia saligna is native to southwestern Australia and has a long history of utilisation across Australia and worldwide. It is planted in many temperate and semi-arid countries for erosion control and sand dune stabilization and as a source of firewood and fodder for sheep and goats (Crompton 1992; Midgley and Turnbull 2003). The species is now a priority for further development as a short-rotation phase or coppice agroforestry crop for the low rainfall agricultural areas of southern Australia (Hobbs et al. 2006; Olsen et al. 2004). A breeding and domestication programme for A. saligna commenced in 2004 when extensive germplasm collections were made and intensive field performance and progeny testing will select for improved growth rate, form, biomass production and feedstock characteristics (Olsen et al. 2004).
While the ecological characteristics of the A. saligna species complex are well described (Fox 1995), little is known about the mating system and patterns of gene flow via pollen dispersal that can be expected within and between populations. Mating systems and pollen dispersal parameters can be directly estimated through paternity analysis using genetic markers. Microsatellite markers or short sequence repeats are suitable for studies of pollen dispersal as they are highly polymorphic, co-dominant, considered to be selectively neutral and can be amplified from small amounts of genomic deoxyribonucleic acid (DNA). As part of a larger effort in the breeding and domestication programme, we developed and screened microsatellite markers in A. saligna and used them to investigate the mating system and pollen dispersal via paternity analysis for a planted stand of A. saligna subsp. saligna. The specific aims of the study were to determine (1) the level of outcrossing, (2) patterns of pollen dispersal, (3) relative paternal contributions to seed crops within the stand and (4) the extent of genetic contamination via pollen dispersal from the background population of A. saligna subsp. lindleyi. Seed production areas have not been established for A. saligna, and these data will be used to design efficient seed orchards as part of the breeding and domestication programme.
Materials and methods
Study species
A. saligna is a bushy shrub or tree that grows to 2–10 m at maturity and lives for up to 20 years under favourable conditions (Fox 1995). It is a leguminous species that is allogamous and insect pollinated. Its natural distribution in southwestern Australia has a Mediterranean climate with annual rainfall of between 300 and 1,200 mm mainly in the winter months. Trees bear hermaphroditic, many-flowered, globular inflorescences in late winter to early spring (Maslin 2001). A. saligna has a widespread native range where it occupies a variety of habitats. Morphological and genetic studies have revealed that A. saligna is a species complex that exhibits a large degree of variation (Byrne and Millar 2004; George 2005) and consists of four main variants to be formally described as subspecies (McDonald and Maslin, unpublished). A. saligna subsp. saligna is native to the coastal areas around Perth and on the south coast and shows the greatest biomass and floral fecundity of the four subspecies. This subspecies has been commonly used as an ornamental and in re-vegetation programmes and is likely to be the subspecies most commonly utilised for agroforestry. A. saligna subsp. lindleyi is the most widespread of the subspecies and occurs across the inland ‘wheat and sheep belt’ area of Western Australia, which is the target area for re-vegetation.
Study site and sample collection
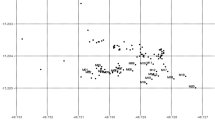
The study site consists of a planted stand of A. saligna subsp. saligna within the natural distribution of subsp. lindleyi and is located south east of Toodyay, Western Australia. The planted stand occupies an area of approximately 0.55 ha (Fig. 1) and occurs on a grassed paddock at a corner of the intersection of two roads. Tree spacing is uneven probably because of variable planting and survival. Remnant roadside trees of subsp. lindleyi occur at varying distances from the planted stand. The largest patch of remnant subsp. lindleyi trees occurs directly across a road at distances of up to 130 m away from the A. saligna stand, and other trees occur in two patches roughly 900 m and 1,500 m away. All trees within the planted stand and 29 selected trees of subsp. lindleyi were mapped using a differential global-positioning satellite system.
Mature phyllode material was collected from all 107 trees within the stand of subsp. saligna and from 29 selected trees of subsp. lindleyi in December 2004. Open-pollinated seed was sampled from ten trees of A. saligna subsp. saligna chosen at random from within the stand in December 2005. The seed was sterilised in 1.0% sodium hypochlorite solution for 1 h and rinsed thoroughly in sterile water. Boiling water was poured over the seeds, and they were left to imbibe overnight. No seed germinated for one of the maternal trees so seed crops were analysed for nine maternal trees.
Microsatellite marker development
A microsatellite library was developed using a hybridisation selection method. Genomic DNA was extracted from adult phyllodes using a modified cetyltrimethyl ammonium bromide extraction method (Byrne et al. 1993) with the addition of 0.1 M sodium sulphite to the extraction and wash buffers (Byrne et al. 2001). Genomic DNA (10 μg) was digested with Sau3A and ligated into 2 μg of pUC18 plasmid vector then transformed into competent Escherichia coli DH5α cells (Gibco) according to the manufacturer’s protocol. The resultant colonies were screened by colony lift hybridisation onto Hybond-N+ membranes using γ-32P–deoxyadenosine triphosphate (PerkinElmer Life Sciences) end-labelled dinucleotides (AC/AG). Hybridisation was carried out with equal concentration (100 ng/30 ml) of the nucleotide probes in 6× sodium chloride–sodium phosphate–ethylenediamine tetraacetic acid buffer, 5 × Denhardts and 1% sodium dodecyl sulphate (SDS) at 65°C overnight. After three post-hybridisation washes in 0.5× sodium chloride–sodium citrate and 0.1% SDS, the membranes were exposed to radiography film. Positive colonies were picked and gridded onto new membranes then re-screened by hybridisation with the probes.
Positive colonies were grown in Luria–Bertani media overnight at 37°C. Plasmid preparations were carried out with UltraClean Mini Plasmid Prep Kit (MoBio Laboratories) following the manufacturer’s instructions. Plasmid inserts were sequenced using BigDye Terminator chemistry, and 12 of 16 clones (75%) contained a microsatellite sequence of eight or more repeats. Sequences flanking the microsatellite repeat motifs were analysed, and compatible primer sequences were identified for nine sequences using PRIMER 3 (Rozen and Skaletsky 2000). Primer pairs were tested for mispriming and primer dimers with AMPLIFY 1.2 (Engels 1993).
Microsatellite loci were amplified in a total volume of 15 μl per reaction, containing 30 ng template DNA, 50 mM KCl, 20 mM Tris–HCl pH 8.4, 0.2 mM each deoxyribonucleotide phosphate, 0.3 μM of each primer, 0.5 U of Taq DNA polymerase and variable MgCl2 concentration depending on the locus (see Table 1). Amplification conditions used either protocol 1: 96°C for 2 min, 30 cycles of 30 s at 95°C, 30 s at 56°C, 30 s at 72°C followed by 5 min at 72°C, or protocol 2: 94°C for 2 min, 30 cycles of 30 s at 94°C, 30 s at 65°C with a step down of 0.3°C per cycle to 56.3°C, 5 s at 72°C, three cycles of 30 s at 94°C, 30 s at 55°C, 5 s at 72°C followed by 5 min at 72°C. All reactions were completed in a Touchdown Thermal Cycler (Hybaid). The amplification products were subjected to electrophoresis on 8% polyacrylamide gels and stained with ethidium bromide. Suitable primers were selected based on the production of amplification products that could be interpreted as single locus.
In addition, microsatellite markers developed for A. mangium (Butcher et al. 2000) and the hybrid A. mangium × A. auriculiformis (Ng et al. 2005) were tested for cross amplification in A. saligna. A range of amplification conditions was tested, and loci were considered suitable for use in A. saligna when sharp bands were produced that showed polymorphism that could be interpreted as a single locus.
DNA extraction and genotyping
Genomic DNA was extracted from phyllodes of the adult trees as described above. DNA was extracted from planted seedlings that had grown to a height of approximately 5 cm, using a modified Doyle and Doyle procedure (Doyle and Doyle 1990). Genotypes of all A. saligna subsp. saligna trees in the planted stand, progeny arrays for nine families consisting of 11–22 seedlings (186 in total), and 29 A. saligna subspecies lindleyi trees were determined for five microsatellite loci as described above except that the size of PCR products was determined by automated fluorescent scanning detection using an Applied Biosystems 3730 DNA Analyser (Applied Biosystems, Foster City, CA). Random sets of samples were re-run to check for repeatability of genotyping, and segregation of maternal alleles in the progeny was checked.
Data analysis
Genetic diversity parameters
Genetic diversity parameters were computed for the adult trees in the study stand, for the progeny alone and for the adults and progeny combined using the CERVUS 2.0 programme (Marshall et al. 1998). Differences between observed and expected heterozygosity of adults and progenies combined and as separate cohorts were assessed using the chi-squared (χ 2) test.
To quantify the genetic heterogeneity of pollen pools among maternal trees of A. saligna subsp. saligna, analysis of molecular variance (Excoffier et al. 1992) based on pollen haplotypes was performed using TWOGENER (Smouse et al. 2001). This provides a ‘global’ estimate of spatial genetic variation of pollen pools over all maternal trees, Φ ft. Pollen haplotypes were derived by subtracting maternal haplotype components from the progeny genotypes. Homogeneity of gene frequencies in the progeny of maternal trees was tested using chi-squared and likelihood ratio (G 2) tests implemented in POPGENE (Yeh and Boyle 1997).
Mating system
Estimates of mating system parameters were produced using the multi-locus estimator programme MLTR version 3.0 (Ritland 2002). Analysis used the expectation–maximisation (EM) method for determining maternal and pollen allele frequencies and the Newton Raphson method for joint maximum likelihood estimation of the multi-locus outcrossing rate (t m) and single-locus outcrossing estimate averaged over all loci (t s). Estimates of bi-parental inbreeding (t m − t s), correlation of paternity (r p) and the correlation of t among loci (r s) were also obtained. Standard errors were computed based on 500 bootstraps. The correlation of paternity was translated to provide estimates of the effective number of pollen donors using the equation N ep = 1/r p (Ritland 1989). Standard error was used to determine whether mating parameters were significantly lower than one or greater than zero. Pollen and ovule frequencies were estimated for each locus using the EM method.
Paternity assignment
Maximum likelihood-based paternity assignment was conducted using CERVUS 2.0 (Marshall et al. 1998). The total size of the candidate male breeding population used in paternity assignment included the 107 trees of A. saligna subsp. saligna in the study stand and an estimated 50 candidate male parents from the nearby patch of A. saligna subsp. lindleyi. Cryptic gene flow (Devlin and Ellstrand 1990) was estimated using FAMOZ software (Gerber et al. 2003) for a simulated population developed using data for the mother trees and progenies of A. saligna subsp. saligna. Parameters for estimates of type I (α) and type II (β) errors included 500 simulated progenies, 20 mothers and an error rate of 0.008.
Spatial analysis
Pollen dispersal patterns within the planted stand were assessed by spatial analysis. The geographic distribution patterns of pollen donors were plotted for nine maternal trees to visually assess any bias in direction for pollen dispersal. Average pollen dispersal distances were assessed using the Kruskall–Wallace test for non-normal data to determine whether pollination distances varied between maternal trees. Paternal contributions of trees in the A. saligna subsp. saligna stand were plotted as a function of the number of progeny fathered. Male mating success as a function of distance from maternal trees was analysed by sub-dividing the neighbourhood of each mother tree into increasing classes of 10-m increments and plotting the frequency distribution of all successful pollination events for paternal trees in each distance class.
A dispersal curve was fitted to the observed pollen dispersal distances over all maternal trees within the stand where f(x) is the contribution (in relative frequency) of paternal trees as a function of distance to maternal trees. A number of models suitable for describing the pollen dispersal kernel were tested including normal, exponential, exponential power, Weibull, geometric and 2D t curves.
To take into account the differing number of potential male parents in each distance class for each maternal tree, the proportion of observed male parents (ratio of the number of observed successful male parents located in the distance class over the total number of observed successful male parents over all classes within the stand) was compared with the proportion of potential male parents in each distance class (ratio of the number of possible male parents in the class over the total number of possible male parents over all classes within the stand). In this analysis, observed successful male parents were counted no more than once (even when male parents were assigned to multiple progenies of a single maternal tree) because the probability of siring multiple progenies was considered equal among pollen donors of subsp. saligna. All trees of subsp. saligna were fecund showing high levels of floral production. Significant deviation between the number of potential and observed male parents for near (<50 m) and far (>50 m) distances, averaged over all maternal trees, was tested using a chi-squared contingency test.
Results
Microsatellite marker development and utility
Only two of the nine primers developed from the constructed microsatellite library produced amplifiable products interpretable as single locus. The remaining primers appeared to amplify multiple loci, which could not be resolved even under high stringency conditions. Of the 25 loci tested for cross-species amplification, three from A. mangium, Am012 Am030 and Am367, showed clear amplification in A. saligna. No primers developed for the A. mangium × A. auriculiformis hybrid produced suitable amplification that could be interpreted as single locus in A. saligna.
The five microsatellite loci were highly variable in A. saligna. The total number of alleles for the five loci was 47, and the number of alleles per locus (A) ranged from 6 to 16 and averaged 9.40 (Table 2). Observed heterozygosity (H o) ranged from 0.323 to 0.783 and averaged 0.518. No significant differences were found in observed or expected heterozygosity for parents and progenies combined (χ 2 = 7.65, df = 4, p value = 0.105) or for parents (χ 2 = 0.0105, df = 4, p value = 0.99) and progeny (χ 2 = 0.054, df = 4, p value = 0.99) analysed as separate cohorts. Null allele frequency averaged 0.0263 for trees of subsp. saligna. As mismatches generated by null alleles are treated as typing errors in paternity assignment, null alleles with low frequencies do not have a significant effect. The polymorphic information content of the loci was high, and the total exclusion probability over the five loci was 0.953 for the second parent.
Mating system
Multi-locus outcrossing within the study stand was high (t m = 0.98) and showed little variation among maternal trees (from 0.90 to 1.0, Table 3). This indicates a mixed mating system with small amounts of inbreeding. Correlation of selfing within progeny arrays, an indication of the fraction of inbreeding because of uniparental inbreeding or true selfing, was low (r s = 0.125). The correlation of paternity is a measure of the probability that pollination events produced full sib relatives and was high (r p = 0.234). This estimate then provided a low value for the effective number of pollen donors (N ep = 4.27 donors per maternal tree).
Paternity assignment
Using the five loci, 93.5% of analysed progeny were assigned a most likely father from either within the subsp. saligna stand or from the sampled trees in the background population of subsp. lindleyi. The majority of progeny (86%) were sired by pollen from within the stand of subsp. saligna. Fourteen progenies (7.5%) were assigned a father from the sampled subsp. lindleyi trees. The paternity of 12 progenies (6.5%) was unassigned, and they were assumed to be sired by unsampled trees of the background population of subsp. lindleyi. The rate of pollen contamination into the subsp. saligna stand is the percentage of progeny whose paternity was not assigned to subsp. saligna trees and was 14%. Three selfing events were detected, one each for maternal trees TC062, TC088 and TC100. Outcrossing rates determined by paternity assignment ranged from 0.90 to 1.0 for maternal trees and averaged 0.98. The estimate of outcrossing obtained via direct paternity assignment was the same as that determined by the mating system assessment using the mixed mating model.
Over half (59.8%) of the subsp. saligna trees within the study stand contributed to the pollen pool, siring at least one offspring (Fig. 2). One paternal tree (TC050) sired nine offspring, representing 5.2% of the progeny analysed. There were 25 instances of correlated paternity detected within the stand with up to six progenies of a single maternal tree sired by the same father. The average number of progeny sired by paternal trees within the stand was 2.3. Half of the paternal trees that contributed to the effective pollen pool sired only one progeny.
Spatial analysis
Analysis of patterns of pollination within the planted stand showed individual observed pollen dispersal distances ranging from 5 to 96 m. The average pollination distance for each individual mother tree ranged from 24 to 50 m and was not significantly different among mother trees (χ 2 = 12.50, df = 8, p = 0.130). The average dispersal distance was 37 m and was similar to the average geographic distance between the maternal trees and all other trees of 40 m. Two pollination events were detected where pollen travelled a distance of 96 m. This is the maximum distance possible at this site and was observed for mother tree TC100, which is situated at the northeastern edge of the stand and hence had the opportunity for such long-distance pollination events.
Analysis of pollination events within the stand plotted over dispersal distance classes showed that for most maternal trees, the majority of pollination events were due to pollen from paternal trees within a 50-m neighbourhood (Fig. 3). For maternal tree TC100, pollination events from near (<50 m) and far (>50 m) paternal trees were equal, and for one maternal tree, TC088, the majority of pollen came from paternal trees greater than 50 m away. Of the dispersal curves tested, the negative exponential model gave the best fit and explained the highest percentage of observed mating events (R 2 = 0.746).
Analysis of dispersal distance class in Fig. 3 does not take into account the variable number of potential pollen donors in each distance class because of unequal spacing and the position of the maternal trees. To determine any bias in pollination distances the distribution of the potential and observed male parents was plotted as a function of distance to maternal trees (Fig. 4). The deviation between the potential and observed parents depicts the departure from random mating caused by the effect of distance between parental trees on male mating success. The number of observed compared to potential male parents shows a significant excess of near (<50 m) matings (χ 2 = 10.47, df = 1, p = 0.001) when averaged over the nine maternal trees, and the majority of successful paternal trees (74%) were located at distances of less than 50 m from maternal trees.
Although it was not empirically tested, pollination events detected by parentage assignment within the stand indicate a random dispersal pattern. No strong bias in direction of pollen travel because of prevailing winds was observed, and the distribution of pollen donors for each maternal tree was independent of their position in the stand (data not shown). However, a moderate level of genetic heterogeneity among pollen pools of maternal trees was evident in a global Φ ft of 0.1108. Heterogeneity in pollen clouds across maternal trees was also detected at each locus using tests for homogeneity (p < 0.01; Table 4). G 2 values are reported as they have a more accurate approximation to the log likelihood ratios of the theoretical χ 2 distribution.
Genetic contamination
Genetic contamination via pollen from A. saligna subsp. lindleyi trees into the planted stand of subsp. saligna was 14%. The level of genetic contamination can be adjusted for type I errors or the failure to assign a father to a progeny when the true father has been genotyped and for type II errors that occur when a progeny is assigned a father from the sampled trees when its true father lies in the unsampled background population. Allowing for these errors the number of progeny assigned an A. saligna subsp. saligna father that was actually sired by ungenotyped subsp. lindleyi trees may have been underestimated by 11.5%.
Genetic contamination rates varied among maternal trees within the stand and ranged from 0 to 32% (Table 3). Of the 29 trees of A. saligna subsp. lindleyi that were sampled, ten were assigned as fathers of one or more progeny in the sampled seed crops from the planted stand. Inter-subspecific pollination events because of pollen sources from the background population of subsp. lindleyi occurred over a range of distances from 5 to 1,566 m with an average pollen dispersal distance of 666 m. The level of pollen contamination (57%) from trees of subsp. lindleyi that were close to the planted stand (<130 m away) was similar to the genetic contamination (43%) that occurred via pollen from the two outlying patches of trees of subsp. lindleyi (located 930 and 1,566 m away, respectively).
Discussion
The design and genetic efficiency of seed orchards are affected by biological aspects of taxa including patterns of pollen mediated gene flow, levels of outcrossing and random mating and genetic contamination from outside the orchard. This study assessed these factors for a planted stand of A. saligna subsp. saligna and found high outcrossing with generally random mating and pollen dispersal within the stand and a low level of genetic contamination from nearby natural remnant populations of subsp. lindleyi.
The multi-locus outcrossing rate within the study stand was high and varied little for maternal trees. The high level of outcrossing is similar to levels found in a previous study using allozyme markers for several natural and one planted population of A. saligna (George 2005).
High outcrossing rates (t m > 0.9) have also been found for other Acacia species including A. auriculiformis A. Cunn. Ex Benth. A. crassicarpa A. Cunn. Ex Benth. (Moran et al. 1989), plantations of the important tannin-producing species A. mearnsii (Kenrick and Knox 1989) and some populations of A. mangium Willd. (Butcher et al. 2000). The outcrossing rate for A. saligna is much higher than that found in two other species that are native to Western Australia, A. sciophanes and A. anfractuosa (Coates et al. 2006).
The pattern of mating within the stand was essentially random. Although a significant excess of matings was detected at distances of less than 50 m, the average distance of pollen dispersal was very similar to the average distance between maternal trees and all potential pollen donors, and pollen dispersal was observed across the maximum distance of the stand. There was no evidence of directionality in pollen dispersal and no evidence of a majority of nearest-neighbour mating. There was evidence of heterogeneity in the genetic composition of pollen clouds experienced by maternal trees indicating that they were not sampling a single pollen cloud. However, this may be due to the high genetic diversity of the stand and high heterozygosity in the trees because of a diverse source of germplasm. Such high heterozygosity would lead to patchy distributions of localised allele frequencies in the pollen pool and would be expected to result in heterogeneity in paternal contributions when assessed for a sub-set of progeny. The levels of heterogeneity in pollen clouds found for the study stand therefore also do not necessarily imply that maternal trees are sampling only highly localised or restricted pollen pools.
Although there was essentially a pattern of random mating within the stand, there was some evidence of correlated paternity. Correlated paternity is generally indicative of genetic structuring within natural populations, but the values found here are more likely to be due to the nature of Acacia floral biology. In A. saligna, flowers are grouped into complex inflorescences, and pollen from the same paternal tree may regularly fertilise more than one flower within a cluster. In addition, pollen grains are compound in nature occurring as polyads of 16 grains, and there is correlation between the number of ovules within a flower and the number of pollen grains per polyad across Acacia species (Kenrick and Knox 1982). Hence, one polyad may be sufficient to sire all progeny within a pod. These aspects of floral biology have been shown to produce high levels of correlated paternity in another Acacia species (Muona et al. 1991). Correlated paternity because of this aspect of the floral morphology is not related to variable fecundity in trees of subsp. saligna, and all trees have equal probability of contributing to the correlated paternity of the seed crop; thus, the potential for random mating is maintained.
Random mating within seed orchards will be promoted by random pollen dispersal patterns, synchronised floral phenologies, equal compatibility of individuals and equality in individual reproductive success. The observations of high outcrossing and random pollen dispersal found here for the subsp. saligna stand indicate that conditions approaching panmixia may be expected for seed orchards established in this species. Thus, seed orchards are an effective means of producing high-value seed for deployment of A. saligna in agroforestry plantings.
One factor that may require active management in seed orchards of A. saligna subsp. saligna is the level of genetic contamination from outside the orchards. Inter-subspecific pollen dispersal from subsp. lindleyi trees in the nearby natural population was detected over large dispersal distances of up to 1,566 m. In the case of seed orchards, pollen dispersal from divergent populations should be minimised to maintain genetic diversity in subsequent seed crops and prevent the dilution of advanced breeding genotypes (Adams and Burczyk 2000; Friedman and Adams 1985). The magnitude of genetic contamination is difficult to predict for given taxa, but even low levels may be detrimental to the efficiency of seed orchards. Levels of genetic contamination are affected by the relative amounts of pollen produced in pollen source and sink populations and the distance between populations (Ellstrand 1992). The planted stand of subsp. saligna studied here (sink) is smaller than most commercial seed orchards, and large numbers of subsp. lindleyi trees (source) are located nearby. However, the majority of subsp. lindleyi trees were not at reproductive maturity at the time of sampling, and floral fecundity of subsp. lindleyi was much lower than that of subsp. saligna. Hence, the total amount of pollen produced by subsp. lindleyi trees was probably less than that of the subsp. saligna stand. Disparity in overall pollen production may be responsible for the relatively low level of genetic contamination found here despite the proximity of subsp. lindleyi trees to the subsp. saligna stand. However, the large distance over which contamination occurred indicates that distance itself is not a strong barrier to pollen dispersal in this species.
The level of genetic contamination found for A. saligna indicates that management options that reduce the amount of genetic contamination into seed orchards will be required. The long-distance intra-specific gene flow found in this study gives an indication that minimum isolation distances required would be greater than 1,500 m. In addition, subsp. lindleyi flowers slightly later than the peak flowering time of subsp. saligna; therefore, selective thinning of late flowering individuals may reduce the amount of genetic contamination within seed orchards, as these trees would be more susceptible to pollination from subsp. lindleyi.
References
Adams W, Burczyk J (2000) Magnitude and implications of gene flow in gene conservation reserves. In: Young A, Boshier D, Boyle T (eds) Forest conservation genetics: principles and practice. CSIRO, Collingwood, Victoria, pp 215–226
Bacles CFE, Burczyk J, Lowe AJ, Ennos RA (2005) Historical and contemporary mating patterns in remnant populations of the forest tree Fraxinus Excelsior L. Evolution 59:979–990
Bartle J, Cooper D, Olsen G, Carslake J (2002) Acacia species as large-scale crop plants in the Western Australian Wheatbelt. Conserv Sci W Aust 4:96–108
Butcher P, Decroocq S, Gray Y, Moran G (2000) Development, inheritance and cross-species amplification of microsatellite markers from Acacia mangium. Theor Appl Genet 101:1282–1290
Byrne M, Millar MA (2004) Genetic systems and issues in the development of woody perennials for revegetation. In: Ridley A, Feikma P, Bennet S, Ropgers MJ, Wilkinson R, Hirth J (eds) Proceedings of the conference salinity solutions: working with science and society, Bendigo, Victoria, 2–5 August 2004. pp 1–54
Byrne M, Moran G, Tibbits W (1993) Restriction map and maternal inheritance of chloroplast DNA in Eucalyptus nitens. Heredity 84:218–220
Byrne M, Macdonald B, Francki M (2001) Incorporation of sodium sulfite into extraction protocol minimizes degradation of Acacia DNA. BioTechniques 30:742–748
Byrne M, Elliott CP, Yates C, Coates DJ (2007) Maintenance of high pollen dispersal in Eucalyptus wandoo, a dominant tree of the fragmented agricultural region in Western Australia. Conserv Genet, doi:10.1007/s10592-007-9311-5
Coates D, Tischler G, McComb JA (2006) Genetic variation and the mating system in the rare Acacia sciophanes compared with its common sister species Acacia anfractuosa (Mimosaceae). Conserv Genet 7:931–944
Crompton H (1992) Acacia saligna—for dryland fodder and soil stabilisation. NFT Highlights, Nitrogen Fixing Tree Association, Waimanalo, HI
Devlin B, Ellstrand NC (1990) The development and application of refined methods for estimating gene flow from angiosperm paternity analysis. Evolution 44:248–259
Dick C (2001) Genetic rescue of remnant tropical trees by an alien pollinator. Proc R Soc Lond B Biol Sci 268:2391–2396
Doyle J, Doyle J (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
El-Kassaby Y (2000) Effect of forest tree domestication on gene pools. In: Young A, Boshier D, Boyle T (eds) Forest conservation genetics: principles and practice. CSIRO, Collingwood, Victoria, pp 197–213
Ellstrand N (1992) Gene flow among seed plant populations. New For 6:241–246
Engels W (1993) Contributing software to the Internet: the Amplify program. Trends Biochem Sci 18:448–450
Excoffier L, Smouse P, Quattro J (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Fox JED (1995) A review of the ecological characteristics of Acacia saligna (Labill.) H.Wendl. Mulga Res Cent J 12:65–71
Friedman S, Adams W (1985) Estimation of gene flow into two seed orchards of loblolly pine (Pinus taeda L.). Theor Appl Genet 69:609–615
George N (2005) Koojong (Acacia saligna), a species with potential as a perennial forage for dryland salinity management—genetic variation, feed quality and reproductive biology. Ph.D. thesis, University of Western Australia
Gerber S, Chabrier P, Kremer A (2003) FAMOZ: a software for parentage analysis using dominant, codominant and uniparentally inherited markers. Mol Ecol Notes 3:479–481
Hobbs T, Bennell M, Huxtable D, Bartle J, Neumann C, George N, O’Sullivan W (2006) FloraSearch Species Profiles – Low rainfall farm forestry options for southern Australia. RIRDC Report, Rural Industries Research and Development, Adelaide
Kang K, Lindgren D, Mullion T (2001) Prediction of genetic gain and gene diversity in seed orchard crops under alternative management strategies. Theor Appl Genet 103:1099–1107
Kenrick J, Knox B (1982) Function of the polyad in reproduction of Acacia. Ann Bot (Lond) 50:721–727
Kenrick J, Knox B (1989) Quantitative analysis of self incompatibility in trees of seven species of Acacia. J Hered 80:240–245
Marshall T, Slate J, Kruuk L, Pemberton J (1998) Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol 7:639–655
Maslin B (2001) Acacia. In: Orchard AE, Wilson AJG (eds) Flora of Australia: Mimosaceae, Acacia part 1. 11B. ABRS/CSIRO, Melbourne, pp 3–13
Midgley SJ, Turnbull JW (2003) Domestication and use of Australian acacias: case studies of five important species. Aust Syst Bot 16:89–102
Moran G, Muona O, Bell J (1989) Breeding systems and genetic diversity in Acacia auriculiformis and A. crassicarpa. Biotropica 21:250–256
Muona O, Moran G, Bell J (1991) Hierarchical patterns of correlated mating in Acacia melanoxylon. Genetics 127:619–626
Nason J, Hamrick J (1997) Reproductive and genetic consequences of forest fragmentation: two case studies of neotropical canopy trees. J Hered 88:264–276
Ng C, Koh S, Lee S, Ng K, Mark A, Norwati M, Wickneswari R (2005) Isolation of 15 polymorphic microsatellite loci in Acacia hybrid (Acacia mangium x Acacia auriculiformis). Mol Ecol Notes 5:572–575
Olsen G, Cooper D, Carslake J, Huxtable D, Bartle J (2004) Search Project Report. Final report for NHT Project 973849, National Heritage Trust, Western Australian Department of Conservation and Land Management, Perth Western Australia
Ritland K (1989) Correlated mating in the partial selfer, Mimulus guttatus. Evolution 43:848–859
Ritland K (2002) Extensions of models for the estimation of mating systems using n independent loci. Heredity 88:221–228
Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz SSM (ed) Bioinformatics methods and protocols: methods in molecular biology. Humana, Totowa, NJ, pp 365–386
Scheltema M (1992) Direct seeding of trees and shrubs. Greening Western Australia, Perth
Smouse P, Dyer R, Westfall R, Sork V (2001) Two generation analysis of pollen flow across a landscape. I. Male gamete heterogeneity among females. Evolution 55:260–271
White G, Boshier D, Powell W (2002) Increased pollen flow counteracts fragmentation in a tropical dry forest: an example from Swietenia humilis Zuccarini. Proc Natl Acad Sci USA 99:2038–2042
Yeh F, Boyle T (1997) Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg J Bot 129:127
Acknowledgments
We thank Bronwyn Macdonald and Lyn Wong for assistance in the laboratory, Wayne O’Sullivan for assistance with morphological identification at the study site and Mathew Williams for assistance with statistical procedures.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by O. Savolainen
Rights and permissions
About this article
Cite this article
Millar, M.A., Byrne, M., Nuberg, I. et al. High outcrossing and random pollen dispersal in a planted stand of Acacia saligna subsp. saligna revealed by paternity analysis using microsatellites. Tree Genetics & Genomes 4, 367–377 (2008). https://doi.org/10.1007/s11295-007-0115-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-007-0115-z