Abstract
Understanding the present-day distribution of molecular variation requires knowledge about the history of the species. Past colonization routes and locations of refugia of Scots pine (Pinus sylvestris) were inferred from variation in mitochondrial DNA in material collected from 37 populations located in countries within, and immediately adjacent to the continent of Europe. Two mitochondrial regions, nad1 intron (exon B/C) and nad7 intron 1, were included in the study. Differentiation in maternally inherited mitochondria was high (G ST′ = 0.824). Two new haplotypes were found at the nad7 intron 1. The occurrence of a 5-bp indel variant was restricted to the Turkish Kalabak population and a 32 bp only found in Central, Eastern, and Northern Europe. The complete absence of the 32-bp indel from the Mediterranean peninsulas supports the view that coniferous forests existed outside these areas during the last glacial maximum, and these populations contributed to the subsequent colonization of the northern parts of Europe. P. sylvestris shares features of its glacial and postglacial history with two other northern, cold-tolerant tree species, Picea abies and Betula sp. These three species differ from many other European trees for which pollen core and molecular evidence indicate colonization from southern refugia after the last glacial period.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pinus sylvestris (Scots pine) is the most widely distributed Eurasian conifer. It is found in a range of environments, soils, and climates from arid, mountainous areas of Spain and Turkey to subarctic forests of northern Scandinavia and Siberia. A large part of its current distribution was covered by the continental ice or was otherwise uninhabitable during the last glacial maximum (LGM) which occurred about 20,000 years ago (Willis et al. 1998; Svendsen et al. 1999). During the LGM, Scots pine was present in the ice-free regions of Southern and Central Europe, but it is also possible that it survived in Siberia and in more eastern parts of Europe (Willis and van Andel 2004). After the LGM, Scots pine expanded to new areas as the habitats became more suitable for it. Based on the European pollen records of the genus Pinus, its distribution started to expand about 16,000 years ago in Southern European regions, and it reached northernmost Scandinavia 7,800 years ago (Willis et al. 1998). However, little is known about the origin of the populations that colonized the northern parts of Europe.
The European pine pollen fossil records are exhaustive (Huntley and Birks 1983), but for the following reasons, they only provide an approximation of the species distribution. First, pollen disperses long distances, and second, pollen production tends to be low in severe environments (Sarvas 1962). Furthermore, P. sylvestris pollen cannot be distinguished morphologically from other Diploxylon pollen. In areas where several species of this same group exist or have existed, conclusions based on pollen occurrence are thus uncertain. Macro- and megafossil data, which are considered to be more reliable than pollen data, suggest that P. sylvestris survived in Hungary and the Czech Republic more than 30,000 years ago (reviewed by Willis and van Andel 2004). Based on genetic data, it is highly probable that there were some isolated populations in the Iberian and Italian Peninsulas during the LGM (Sinclair et al. 1999; Soranzo et al. 2000; Cheddadi et al. 2006). Little is known about the history of Scots pine east of Central Europe. The possibility that more eastern populations have also contributed to postglacial colonization of Europe cannot be ignored.
Scots pine occurs at present in very diverse habitats and exhibits genetic adaptation to local conditions in quantitative traits such as timing of growth (Hurme et al. 1997 and citations therein). However, nuclear genetic markers have not shown clear signs of genetic differentiation (Wang et al. 1991; Szmidt and Wang 1993; Karhu et al. 1996; Dvornyk 2001), mainly due to effective pollen-mediated gene flow (Muona and Harju 1989; Dvornyk et al. 2002). Some of the modern adaptive variations could have been present already in ancestral populations. Present differences in adaptive traits may just be due to these precolonization differences. The other option is that response to local conditions, for example climate–day length combination, has emerged during and after the colonization. To resolve the relative importance of these two processes resulting in adaptation, it is important to know the colonization history.
The history of colonization of forest trees also helps to deduce the climate and habitat conditions in different parts of Eurasia during the Ice Age. The extensive study of Petit et al. (2003) on the postglacial history of 25 European trees and shrubs clarified the history of European forests. It has been concluded that even if the refugia for most species were located in the same regions, the routes and dynamics of recolonization differ from species to species (Ferris et al. 1998; Petit et al. 2002). Further, more cold-tolerant species such as Picea abies and Betula sp. are likely to have more northern and eastern origins (Lagercrantz and Ryman 1990; Palme et al. 2003). Therefore, the history of Scots pine, the most widespread Pinus species, must be resolved from species-specific data.
In Pinaceae, mitochondrial DNA (mtDNA) is maternally inherited (Neale and Sederoff 1989 and citations therein; see Wagner et al. 1991 for exception; Wang et al. 1996). A lower effective population size, a clonal mode of evolution, and different means of gene flow of the organellar genomes have contrasting effects on the level of differentiation compared to the nuclear genome (Petit et al. 1993; Ennos 1994), especially in wind pollinated species such as P. sylvestris. In this study, we use data on mitochondrial markers to distinguish between the alternative locations of glacial refugia and postglacial colonization routes of Scots pine by examining the distribution of mitochondrial haplotypes with special emphasis on resolving the descent of Northern European populations.
Materials and methods
Sampling
We sampled 37 populations (containing 3–35 trees) covering most of the present Scots pine distribution in Europe and its neighboring countries (Table 1, Fig. 1). The total sample size was 714 trees. Material was provided by the Finnish Forest Research Institute, except for the Spanish samples, which were provided by SIA (Servicio de Investigación Agroalimentaria de la Diputación de Aragón) in Spain, and Northern Swedish and Russian samples, which were kindly provided by The Forestry Research Institute of Sweden and the Russian Academy of Sciences, Komi Science Center.
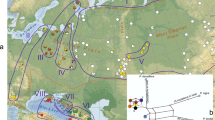
a Pie diagrams describe the distribution of mitochondrial haplotypes in P. sylvestris populations. Patterns in diagrams are equivalent to the patterns in haplotype network. For sample sizes, see Table 1. b Sequences of all three variants in the polymorphic region of mitochondrial nad7 intron 1. c Mitochondrial haplotype network with one indel mutation between the neighboring haplotypes
DNA extraction, polymerase chain reaction, sequencing, and polymerase chain reaction-restriction fragment length polymorphism
DNA was extracted from needles (Lapinjärvi, Lithuania, some of Kolari, Puebla de Lillo) with the DNeasy Plant Mini Kit (Qiagen) or from germinating seeds by FastDNA® Kit (Q-BIO Gene). Several mitochondrial regions were sequenced from a screening set consisting of at least 12 individuals from eight geographical regions to find polymorphisms. Both strands were sequenced using the Big Dye Terminator Kit v3.1 (Applied Biosystems). Multiscreen plates (Millipore) with Sephadex matrix (Sigma-Aldrich) were used for purification of sequencing products. Sequencing was conducted with ABI PRISM 377 and 3730 DNA Analyzer (Applied Biosystems).
The mitochondrial regions nad5a (intron 1), nad3-rps12, and cox2 (exon 1; Wu et al. 1998) were monomorphic in our primary screening set. Polymorphisms were found in regions nad7 intron 1. nad1 intron (exon B/C) was already known to be polymorphic in Europe (Soranzo et al. 2000). Primers for the polymorphic nad7 intron 1 region were originally developed for Picea mariana (Jaramillo-Correa et al. 2004), and the region includes a variable minisatellite motif in Pinus banksiana (Godbout et al. 2005). For amplifying a known polymorphic indel in nad1, diagnostic primers nad1 H and nad1 I (Soranzo et al. 2000) were used. Sequences have been deposited to GenBank (Accession numbers EU072470–EU072475).
The polymerase chain reaction (PCR) for nad1 had concentration 2.5 mM of MgCl2, 0.2 μM of each primer, 0.2 mM of each deoxyribonucleotide triphosphate (dNTP), included 1 U of Taq polymerase (Invitrogen), and 20 ng of template DNA in 15 μl volume of 1X Taq buffer. The PCR reaction for nad7 had concentration 2.5 mM of MgCl2, 0.5 μM of each primer, 0.3 mM of each dNTP, included 0.5 U of EXT polymerase (Finnzymes), and 20 ng of template DNA in 25 μl volume of 1× EXT buffer. Amplification protocol for nad1 was 4 min at 94°; followed by 35 cycles of 45 s at 94°, 1 min at 53°, 1 min 30 s at 72°; and finally one cycle for 10 min at 72°, and for nad7, 3 min at 94°; followed by 38 cycles of 1 min at 94°, 1 min 30 s at 59°, 2 min 30 s at 72°; and finally one cycle 20 min at 72°.
The 31-bp indel in nad1 intron (exon B/C) was scored by electrophoresis of the PCR product through 2% agarose gel. The polymorphisms in nad7 were initially scored by sequencing 90 individuals from various populations, but as no additional polymorphisms were found, the rest of the samples were genotyped by digesting 3 μl of PCR products with 0.5 U of DraII and electrophoresing through 2% agarose gel.
In this study, mitochondrial haplotypes a and b are combinations of nad1 types a and b in Soranzo et al. (2000) and the most common type in nad7. Haplotypes c and d are combinations of rare types in nad7 with a of Soranzo et al. (2000) at nad1.
Data analysis
Sequence contigs were assembled with SequencherTM 4.0.5 (Gene Codes Corporation) and aligned with Clustal X (1.83; Thompson et al. 1997). GeneDoc (2.6.002; Nicholas et al. 1997) was used for editing the alignment manually. A standardized measure of G ST, G ST′ was calculated according to Hedrick (2005) to remove the effect of level of polymorphism on the estimate.
Mitochondrial gene diversities (which in the case of haploid data is the probability of two randomly chosen alleles being different in the sample) and their standard deviations (Nei 1987) for each population were calculated with Arlequin version 2000 (Schneider et al. 2000). Average population diversity, total diversity, and estimates of differentiation parameter for unordered (H S, H T, G ST) and ordered (V S, V T, N ST) haplotypes were obtained with the software PERMUT (http://www.pierroton.inra.fr/genetics/labo/Software/Permut/). PERMUT was also used for testing whether N ST is higher than G ST for the mitochondrial data by permuting rows and columns of distance matrices between alleles (Pons and Petit 1996). G ST is an F ST estimate that takes into account only the allele frequencies, whereas N ST makes use of the distance between different alleles. A higher N ST than G ST indicates phylogeographic structure within populations (Pons and Petit 1996).
The software package SPAGeDi 2.1 (Hardy and Vekemans 2002) was used to analyze the relationship between geographic and genetic distance. In a regression analysis, genetic distance was defined as F ST/(1 − F ST) and geographic distance as the natural logarithm of distance as kilometers (Rousset 1997). A pattern of isolation by distance is detected as significant positive regression coefficient. Significance was tested by permuting population locations among populations (Mantel’s test). To examine whether the pattern of isolation by distance was especially strong in some specific distance class, estimates of genetic differentiation were binned by geographical distance. The level of differentiation was scored in six distinct classes for every 1,000 km from 1,000 to 6,000 km.
Results
Patterns and amount of genetic diversity
We found altogether four mitochondrial haplotypes (Fig. 1). None of the 90 samples sequenced or 624 samples genotyped by PCR-restriction fragment length polymorphism (RFLP) in this study showed signs of heteroplasmy as expected, given the maternal inheritance of mtDNA. New polymorphisms were found in the nad7 region, where two indels (32 bp and 5 bp) resulted in three different types in this region (Fig. 1b). There was one common mitochondrial haplotype (a), and the other three haplotypes (b, c, and d) were all within one mutational (insertion or deletion) step from it (Fig. 1c). The 32-bp indel is not the same as found by Godbout et al. (2005) in P. banksiana.
The mitochondrial gene diversities varied from 0 to 0.6 (Table 1). Total gene diversity (H T) was 0.583, average gene diversity (H S) 0.201, and between population component of diversity, G ST 0.655. The standardized genetic differentiation, G ST′, was 0.824. The ordered and unordered differentiation statistics were quite similar: N ST was 0.700 and slightly but not significantly higher than G ST when tested with 1,000 permutations. There was a pattern of isolation by distance between pairwise mitochondrial genetic differentiation and geographic distance. The regression coefficient between F ST/(1 − F ST) and natural logarithm of distance as kilometers between 37 populations was significantly greater than zero (P = 0.0012, one-sided test, population locations permuted among all populations). However, the distance explained only 3% of the variation in genetic distance (r 2 = 0.029). In the analysis of genetic differentiation by distance classes, the isolation by distance was significant up to distance class of 4,000 km (Fig. 2).
Distribution of mitochondrial haplotypes
The most common haplotype, a, was present in all geographical areas and is probably the ancestral type (Fig. 1a). The indel in the nad1 (haplotype b) is restricted to the Iberian Peninsula. Two haplotypes which have not been previously found, c and d, had their own characteristic distributions. Haplotype d was restricted to the Kalabak population in Turkey only. Haplotype c was found in central, northern, and eastern parts of Europe but was absent from Western Europe, the Iberian Peninsula, Turkey, and Scotland. Three easternmost Russian populations also had only the mitochondrial type a. Two Finnish populations, Kälviä and Utsjoki, were monomorphic for type c. Type c is the most common haplotype in the northern Finnish populations and the Moscow and St. Petersburg region. The frequency of type c gradually diminishes towards west and east from these regions. There is a large difference in frequency of type c between northern Finland and northern Sweden (0.53 in Kolari vs 0.1 in Leiporova). The observation is supported by data set of 18 additional single trees sampled from different locations in the northern Sweden (data not presented in Table 1 and Fig. 1), where only one tree was of type c.
Discussion
Geographical patterns of genetic diversity
The level of mitochondrial gene diversity outside the Iberian Peninsula was considerably higher than was previously detected due to the discovery of the previously undescribed variation in nad7. The indel variant of nad1 was restricted to Spain and has been detected in earlier studies (Sinclair et al. 1999; Soranzo et al. 2000; Cheddadi et al. 2006). In earlier studies, all continental populations outside the Iberian Peninsula were fixed for a single mitochondrial haplotype (Sinclair et al. 1999), with few exceptions (Soranzo et al. 2000; Cheddadi et al. 2006). The higher level of variation results both from geographically more extensive sampling and the better resolution in mitochondrial markers used in this paper. Nonetheless, some of the individual populations exhibit no mitochondrial variation at all. Many natural Western European populations are fixed for type a which is predominant also in the easternmost part of the distribution. This is probably due to lack of resolution in our markers rather than a common origin of Eastern Russian and Western European populations. It is likely that the addition of more markers would result in higher levels of mitochondrial haplotype diversity. However, mtDNA has very low variability, as screening of many regions did not find much variation.
In P. sylvestris, the amount of differentiation in mtDNA was very much higher than what was previously documented in nuclear markers, as expected. The lower effective population size and more restricted dispersal in seeds vs pollen result in definite genetic structure in maternally inherited DNA (Ennos 1994). Similar relationships have been observed in other pine species, e.g., in Pinus contorta (Dong and Wagner 1993, 1994) and Pinus flexilis (Latta and Mitton 1997; Mitton et al. 2000).
P. sylvestris seeds are wind dispersed, and the species has mostly continuous distribution, although there are some isolated populations at the margins of distribution. We thus expect a pattern of isolation by distance in genetic differentiation (Wright 1943). Although the number of haplotypes was low, isolation by distance was observed. It extended at least up to distance class of 4,000 km. Mitochondrial types separated by two indels were never found in the same population, which indicates some degree of phylogeographical pattern. However, N ST and G ST did not differ significantly. This is probably due to low power of the test, as there were only four haplotypes.
Recolonization of Scots pine from different refugia since the last glacial maximum
The distribution of the haplotypes supports the view that populations in the Iberian Peninsula are relicts that have not contributed to the recolonization of the rest of Europe as already concluded by Soranzo et al. (2000) and Sinclair et al. (1999). The distribution of haplotypes a and b in our study is consistent with the results of Soranzo et al. (2000) and Cheddadi et al. (2006), although in the current study, haplotype b was not detected outside the Iberian Peninsula. The Scots pine population in Turkey probably did not have a role in colonization because it is isolated from the main distribution. The private Turkish haplotype has not been found from any population earlier. It is probable that there was a glacial refugium in Asia Minor, and populations in this region may be more connected to Asian than European Scots pine populations. To verify the role of Asia Minor in colonization history of P. sylvestris, more populations from Asia Minor and adjacent regions need to be studied.
The previously undetected mitotype c was present in Central Europe, Scandinavia, and Eastern Europe. The origin of this type is not clear, but most probably it is neither from the Iberian Peninsula, Italian Peninsula, nor Turkey. Turkey and the Iberian Peninsula can be ruled out based on our results and those of Soranzo et al. (2000) and Cheddadi et al. (2006). The Italian Peninsula is one of the main hypothetical locations of glacial refugia for many species (Taberlet et al. 1998). Our study did not address the question, but the recent results of Cheddadi et al. (2006) show that Scots pine populations from Italian Peninsula have not contributed to postglacial colonization of Central and Northern Europe. For P. abies, another cold-tolerant European conifer, the Carpathians have been suggested as refugial area (Gugerli et al. 2001). However, the role of the Carpathians and Balkan Peninsula in colonization history of P. sylvestris remains unresolved, as our sampling does not cover these areas adequately.
According to comparative studies based on phylogeography of ten species, two main postglacial colonization routes meet in Central Sweden (Taberlet et al. 1998). One is from the northeast, and the other one from the southwest. Haplotype c has a low frequency in northern Sweden in contrast with higher frequencies in northern Finnish populations. Sinclair et al. (1999) suggested dual migration of P. sylvestris to northern Scandinavia and Finland from the northeast and the south. The difference in the frequency of haplotype c between northern Scandinavia and Finland are concordant with that hypothesis. However, type c is quite common in southern Sweden, and it is possible that the observed differences in haplotype frequencies between Scandinavia and Finland result from drift. They might have the same origin, but drift could have resulted in a high frequency of haplotype a in Sweden and haplotype c in Finland.
The northern cold-tolerant species, P. sylvestris, Betula, and P. abies are widely distributed in Eurasia. The distribution of chloroplastic and mitochondrial haplotypes of P. abies and chloroplastic haplotypes of Betula pendula in eastern Russia and in Northern and Central Europe (Sperisen et al. 1998; Vendramin et al. 2000; Palme et al. 2003) resemble the distribution of haplotypes in Scots pine. Our results are in accordance with the hypothesis that there was a refugium near Hungary and the Czech Republic, as suggested by macrofossil data from Central and Eastern Europe (reviewed in Willis and van Andel 2004). However, the highest frequencies of type c are relatively far north: northern Finland, St. Petersburg, and Moscow, which would suggest more eastern and northern origin of type c. Genetic data imply that one of the origins of current European P. abies is in the region of Moscow (Lagercrantz and Ryman 1990), as suggested by pollen analyses (Huntley and Birks 1983). To resolve whether Northern European P. sylvestris has an eastern origin as P. abies or a past refugium in Central Europe, denser sampling in central, northern, and eastern parts of Europe and preferably also more mitochondrial markers are needed.
These results are consistent with the view that during the last glacial maximum, there have been populations of cold-tolerant tree species such as P. sylvestris, P. abies, and Betula outside the Mediterranean peninsulas. The two northern conifers and Betula thus have a very different glacial and postglacial history compared to many European angiosperm tree species.
References
Cheddadi R, Vendramin GG, Litt T, Fraçois L, Kageyama M, Lorentz S, Laurent JM, de Beaulieu JL, Sadori L, Jost A, Lunt D (2006) Imprints of glacial refugia in the modern genetic diversity of Pinus sylvestris. Glob Ecol Biogeogr 15:271–282
Dong J, Wagner DB (1993) Taxonomic and population differentiation of mitochondrial diversity in Pinus banksiana and Pinus contorta. Theor Appl Genet 86:573–578
Dong JS, Wagner DB (1994) Paternally inherited chloroplast polymorphism in Pinus—estimation of diversity and population subdivision, and tests of disequilibrium with a maternally inherited mitochondrial polymorphism. Genetics 136:1187–1194
Dvornyk V (2001) Genetic variability and differentiation of geographically marginal Scots pine populations from Ukraine. Silvae Genet 50:64–69
Dvornyk V, Sirviö A, Mikkonen M, Savolainen O (2002) Low nucleotide diversity at the pal1 locus in the widely distributed Pinus sylvestris. Mol Biol Evol 19:179–188
Ennos RA (1994) Estimating the relative rates of pollen and seed migration among plant populations. Heredity 72:250–259
Ferris C, King RA, Väinölä R, Hewitt GM (1998) Chloroplast DNA recognizes three refugial sources of European oaks and suggests independent eastern and western immigrations to Finland. Heredity 80:584–593
Godbout J, Jaramillo-Correa JP, Beaulieu J, Bousquet J (2005) A mitochondrial DNA minisatellite reveals the postglacial history of jack pine (Pinus banksiana), a broad-range North American conifer. Mol Ecol 14:3497–3512
Gugerli F, Sperisen C, Buchler U, Magni F, Geburek T, Jeandroz S, Senn J (2001) Haplotype variation in a mitochondrial tandem repeat of Norway spruce (Picea abies) populations suggests a serious founder effect during postglacial re-colonization of the western Alps. Mol Ecol 10:1255–1263
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Hedrick P (2005) A standardized genetic differentiation measure. Evolution 59:1633–1638
Huntley B, Birks HJB (1983) An atlas of past and present pollen maps for Europe 0–13,000 years ago. Cambridge University Press, Cambridge
Hurme P, Repo T, Savolainen O, Pääkkönen T (1997) Climatic adaptation of bud set and frost hardiness in Scots pine (Pinus sylvestris). Can J For Res-Rev Can Rech For 27:716–723
Jaramillo-Correa JP, Beaulieu J, Bousquet J (2004) Variation in mitochondrial DNA reveals multiple distant glacial refugia in black spruce (Picea mariana), a transcontinental North American conifer. Mol Ecol 13:2735–2747
Karhu A, Hurme P, Karjalainen M, Karvonen P, Kärkkäinen K, Neale D, Savolainen O (1996) Do molecular markers reflect patterns of differentiation in adaptive traits of conifers? Theor Appl Genet 93:215–221
Lagercrantz U, Ryman N (1990) Genetic structure of Norway spruce (Picea abies)—concordance of morphological and allozymic variation. Evolution 44:38–53
Latta RG, Mitton JB (1997) A comparison of population differentiation across four classes of gene marker in limber pine (Pinus flexilis James). Genetics 146:1153–1163
Mitton JB, Kreiser BR, Latta RG (2000) Glacial refugia of limber pine (Pinus flexilis James) inferred from the population structure of mitochondrial DNA. Mol Ecol 9:91–97
Muona O, Harju A (1989) Effective population sizes, genetic variability, and mating system in natural stands and seed orchards of Pinus sylvestris. Silvae Genet 38:221–228
Neale DB, Sederoff RR (1989) Paternal inheritance of chloroplast DNA and maternal inheritance of mitochondrial DNA in loblolly pine. Theor Appl Genet 77:212–216
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York, NY, USA
Nicholas KB, Nicholas HB Jr, Deerfield DW II (1997) GeneDoc: Analysis and visualization of genetic variation. EMBNEW.NEWS 4:14
Palmé AE, Su Q, Rautenberg A, Manni F, Lascoux M (2003) Postglacial recolonization and cpDNA variation of silver birch, Betula pendula. Mol Ecol 12:201–212
Petit RJ, Kremer A, Wagner DB (1993) Finite island model for organelle and nuclear genes in plants. Heredity 71:630–641
Petit RJ, Brewer S, Bordács S, Burg K, Cheddadi R, Coart E, Cottrell J, Csaikl UM, van Dam B, Deans JD, Espinel S, Fineschi S, Finkeldey R, Glaz I, Goicoechea PG, Jensen JS, König AO, Lowe AJ, Madsen SF, Mátyás G, Munro RC, Popescu F, Slade D, Tabbener H, de Vries SGM, Ziegenhagen B, de Beaulieu JL, Kremer A (2002) Identification of refugia and post-glacial colonisation routes of European white oaks based on chloroplast DNA and fossil pollen evidence. For Ecol Manag 156:49–74
Petit RJ, Aguinagalde I, de Beaulieu JL, Bittkau C, Brewer S, Cheddadi R, Ennos R, Fineschi S, Grivet D, Lascoux M, Mohanty A, Müller-Starck GM, Demesure-Musch B, Palmé A, Martin JP, Rendell S, Vendramin GG (2003) Glacial refugia: Hotspots but not melting pots of genetic diversity. Science 300:1563–1565
Pons O, Petit RJ (1996) Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144:1237–1245
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145:1219–1228
Sarvas R (1962) Investigations on the flowering and seed crop of Pinus sylvestris. Valtion painatuskeskus, Helsinki
Schneider S, Roessli D, Excoffier L (2000) Arlequin: A software for population genetics data analysis. Ver 2.000. Genetics and Biometry Lab, Dept. of Anthropology, University of Geneva
Sinclair WT, Morman JD, Ennos RA (1999) The postglacial history of Scots pine (Pinus sylvestris L.) in Western Europe: evidence from mitochondrial DNA variation. Mol Ecol 8:83–88
Soranzo N, Alia R, Provan J, Powell W (2000) Patterns of variation at a mitochondrial sequence-tagged-site locus provides new insights into the postglacial history of European Pinus sylvestris populations. Mol Ecol 9:1205–1211
Sperisen C, Buchler U, Mátyás G (1998) Genetic variation of mitochondrial DNA reveals subdivision of Norway Spruce (Picea abies (L.) Karst.). In: Ingram DS (ed) Molecular tools for screening biodiversity: Plants and animals, Chapman & Hall, London, pp 413–417
Svendsen JI, Astakhov VI, Bolshiyanov DY, Demidov I, Dowdeswell JA, Gataullin V, Hjort C, Hubberten HW, Larsen E, Mangerud J, Melles M, Möller P, Saarnisto M, Siegert MJ (1999) Maximum extent of the Eurasian ice sheets in the Barents and Kara Sea region during the Weichselian. Boreas 28:234–242
Szmidt AE, Wang X (1993) Molecular systematics and genetic differentiation of Pinus sylvestris (L.) and P. densiflora (Sieb. et Zucc.). Theor Appl Genet 86:159–165
Taberlet P, Fumagalli L, Wust-Saucy AG, Cosson JF (1998) Comparative phylogeography and postglacial colonization routes in Europe. Mol Ecol 7:453–464
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Vendramin GG, Anzidei M, Madaghiele A, Sperisen C, Bucci G (2000) Chloroplast microsatellite analysis reveals the presence of population subdivision in Norway spruce (Picea abies K.). Genome 43:68–78
Wagner DB, Dong J, Carlson MR, Yanchuk AD (1991) Paternal leakage of mitochondrial DNA in Pinus. Theor Appl Genet 82:510–514
Wang X, Szmidt AE, Lindgren D (1991) Allozyme differentiation among populations of Pinus sylvestris (L.) from Sweden and China. Hereditas 114:219–226
Wang X, Szmidt AE, Lu M (1996) Genetic evidence for the presence of cytoplasmic DNA in pollen and megagametophytes and maternal inheritance of mitochondrial DNA in Pinus. For Genet 3:37–44
Willis KJ, van Andel TH (2004) Trees or no trees? The environments of central and Eastern Europe during the Last Glaciation. Quat Sci Rev 23:2369–2387
Willis KJ, Bennett KD, Birks JB (1998) The late Quaternary dynamics of pines in Europe. In: Richardson DM (ed) Ecology and biogeography of Pinus, Cambridge University Press, pp 107–121
Wright SI (1943) Isolation by distance. Genetics 28:114–138
Wu JY, Krutovskii KV, Strauss SH (1998) Abundant mitochondrial genome diversity, population differentiation and convergent evolution in pines. Genetics 150:1605–1614
Acknowledgments
We thank Katri Kärkkäinen, Leena Yrjänä, and Jukka Lehtonen from Finnish Forest Research Institute, Torgny Persson from The Forestry Research Institute of Sweden, Eduardo Notivol from SIA, and Aleksei Fedorkov from Russian Academy of Sciences, who have kindly provided us with the material. We thank Anna Palmé and Rosario García-Gil for their contribution and discussion on the manuscript. This research was supported by the National Graduate School for Population Genetics to TP, by the Thule Institute to MS and by the University of Oulu, and the Research Council for Biosciences and Environment to OS.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Kremer
Rights and permissions
About this article
Cite this article
Pyhäjärvi, T., Salmela, M.J. & Savolainen, O. Colonization routes of Pinus sylvestris inferred from distribution of mitochondrial DNA variation. Tree Genetics & Genomes 4, 247–254 (2008). https://doi.org/10.1007/s11295-007-0105-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-007-0105-1