Abstract
To evaluate the microbial diversity of Fushan forest soils, the variation of soil properties, microbial populations, and soil DNA with soil depth in three sites of different altitude were analyzed. Microbial population, moisture content, total organic carbon (Corg), and total nitrogen (Ntot) decreased with increasing soil depth. The valley site had the lowest microbial populations among the three tested sites due to the low organic matter content. Bacterial population was the highest among the microbial populations. The ratios of cellulolytic microbes to the total bacteria in organic layers were high, implying their roles in the carbon cycle. The microbial biomass carbon (Cmic) and nitrogen (Nmic) contents ranged from 130.5 to 564.1 μg g−1 and from 16.7 to 95.4 μg g−1, respectively. The valley had the lowest Cmic and Nmic. The organic layer had the highest Cmic and Nmic and decreased with soil depth. Analysis using denaturing gradient gel electrophoresis (DGGE) of polymerase chain reaction (PCR) amplicons of 16S rDNA showed that the bacterial diversity of the three sites were very similar to each other in the major bands, and the variation was in the minor bands. However, the patterns in PCR-DGGE profile through gradient horizons were different, indicating the prevalence of specific microbes at different horizons. These results suggest that the microbial diversity in the deeper horizons is not simply the diluted analogs of the surface soils and that some microbes dominate only in the deeper horizons. Topography influenced the quantity and diversity of microbial populations.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Natural forests have gained recognition as sites of high biodiversity, where complex relationships among fauna, flora, and microflora are maintained due to the structural richness of the habitat. Soil microbes are essential components of the biotic community in natural forests, and they are largely responsible for ecosystem functioning because they participate in most nutrient transformations (Hackl et al. 2004). Usually, the microbial composition of the surface horizon has been studied because it is supposed to be the most active one, whereas little attention has been paid to the deeper horizons of the soil profile (Agnelli et al. 2004). Few papers take into account the subsurface soil layers (Bundt et al. 2001; Ekelund et al. 2001; Fierer et al. 2003), and fewer, as far as we know, consider the pedogenetic horizons in assessing soil-inhabiting microorganisms (Fritze et al. 2000; Agnelli et al. 2001). Microbes in these deeper horizons play an important role in ecosystem biogeochemistry, soil formation, and maintenance of groundwater quality (Konopka and Turco 1991). However, it is not clear whether the subsurface microbial community is closely tied to the surface-soil microbial community or is an independent ecosystem with a distinct assemblage of microorganisms.
The number of culturable microbes in soil is often only about 1% of the total number of microbe presents (Schoenborn et al. 2004), which hinders microbial diversity analysis. To overcome this limitation, a number of approaches, such as DNA reassociation (Torsvik et al. 1990), DNA–DNA and mRNA–DNA hybridization (Cho and Tiedje 2001), analysis of bacterial 16S rDNA library (Axelrood et al. 2002), and other polymerase chain reaction (PCR)-based methods of denaturing gradient gel electrophoresis (DGGE), temperature gradient gel electrophoresis (TGGE) (Muyzer 1999), and ribosomal intergenic spacer analysis (Fisher and Triplett 1999), have been developed to study molecular microbial diversity (Kirk et al. 2004). DGGE has the advantages of being reliable, reproducible, rapid, and inexpensive. Multiple samples can also be analyzed concurrently, making it possible to follow changes in the microbial populations (Muyzer 1999). PCR-DGGE is based on the variation in base composition and secondary structure of the 16S rDNA molecule fragment, and DGGE can separate the DNA with one base-pair difference. Application of DNA fingerprinting methods enables detection of soil microbial community diversity, even including those microbes not yet cultivated.
Fushan Mountain is located in the northern part of Taiwan and is a typical low-altitude, ecosystem-protected area. Fushan forest is identified as one of the four natural forest sites by Taiwan Long Term Ecological Research Network (TERN) to study ecosystem response to environmental disturbances such as typhoons and acidic deposition (Lin et al. 1997, 2000, 2003b; Liu et al. 2004). Although, few studies were conducted in the Fushan forest, such as nitrogen (N) mineralization and nitrification rates (Owen et al. 2003) and fluvial transportation and sedimentation (Jen et al. 2006), the microbial ecology of this forest is not characterized. Hence, in the present study, microbial populations, microbial biomass carbon (Cmic) and nitrogen (Nmic) were determined, and DGGE of PCR-amplified ribosomal RNA (rDNA) genes was performed to evaluate whether the subsurface bacterial community is closely tied to the surface soil bacterial community or is a distinct assemblage of bacteria through different soil profiles from three altitude locations.
Materials and methods
Site description
Fushan forest in northern Taiwan (24°34′N, 121°34′E) has an elevation from 400 m to 1,400 m (King et al. 2003). Fushan is a moist, subtropical, mixed evergreen/hardwood broadleaf forest with a flora of more than 500 species (Mabry et al. 1998). Members of Lauraceae, Fagaceae, and Theaceae are dominant in this area. Mean annual precipitation is about 3,990 mm (Hsia and Hwong 1999). The soil is classified into Hapludults, Dystrochrepts, Udipsamments, and Udorthents according to the Keys of Soil Taxonomy (Lin et al. 1996). The ridge is the richest in vegetation, with the highest species diversity and density. Diversity decreases gradually through the middle slope, and the valley habitats are the least diverse. The ridge also has the greatest elevation, effective soil depth, and mean canopy height; the middle-slope area has the highest slope; and the valley has the greatest canopy gaps and the shallowest soil formation due to erosion. Topography was significantly correlated with soil disturbance, which induced differentiation of microenvironment and species compositions.
Sampling
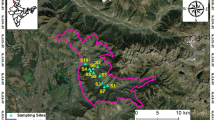
During September 2005, soil samples were collected from three locations—valley, middle slope, and ridge—at OA (0–15 cm), A (16–35 cm), BW1 (36–55 cm), BW2 (56–75 cm), and BC (76–95 cm) horizons, which differed from each other in altitude, characteristic plant species, and slope (Table 1). Soil profiles were classified into horizons according to the International Soil Classification System (Soil Survey Staff 2003). Three replicate samples were taken at each horizon for plate count, chemical analysis, and DNA extraction. The samples were sieved to 2 mm, and DNA extraction was carried out immediately after reaching the laboratory.
Culture media and conditions
For plate count, 1 g soil was used to prepare the serial dilution, and 1 ml from selected dilutions was pour plated. Bacteria were counted at 25°C after 5 days on nutrient agar (Merck, Darmstadt, Germany), and actinomycetes were cultivated at 25°C for 7 days on glycerol–yeast extract medium composed of (g l−1) glycerol 5.0, yeast extract 2.0, K2HPO4 1.0, and agar 15.0 at pH 7.0 ± 0.1. Streptomycin and cycloheximide were added to inhibit the growth of bacteria and fungi at a final concentration of 10 μg ml−1 (Yang and Yang 2001). Fungi were grown at 25°C for 5 days on Rose Bengal medium containing (g l−1) glucose 10.0, peptone 5.0, K2HPO4 1.0, MgSO4·7H2O 0.5, Rose Bengal 0.033, and agar 15.0 at pH 6.8 ± 0.1. Cellulolytic microbes were assayed at 25°C after 7 days’ incubation on Mandels–Reese medium (Mandels and Reese 1957) with modifications, containing (g l−1) carboxymethylcellulose (CMC, Sigma) 10.0, peptone 1.0, urea 0.3, (NH4)2SO4 1.4, KH2PO4 2.0, CaCl2·H2O 0.3, MgSO4·7H2O 0.3, FeSO4·7H2O 0.005, ZnSO4·7H2O 0.0014, MnSO4·4H2O 0.0016, CoCl2·7H2O 0.002, and agar 20.0 at pH 6.0 ± 0.1. The plates were sprayed with Congo red to show a clear zone around the colonies. Phosphate-solubilizing microbes were measured at 25°C after 5 days on rock phosphate medium by the clear zone around the colonies (Chang et al. 2001). Nitrogen-fixing microbes were counted after incubation for 7 days at 25°C on nitrogen-free mannitol medium (Yang et al. 2003). All experiments were carried out in triplicate.
Microbial biomass carbon and nitrogen
Cmic and Nmic were determined by the chloroform fumigation extraction method (Brookes et al. 1985; Vance et al. 1987). Fresh soil samples (25 g) at 40% water-holding capacity were placed in a 50-ml beaker and kept in a vacuum desiccator containing a 100-ml beaker with 50 ml chloroform (ethanol removed). The desiccator was evacuated using a vacuum pump until the chloroform boiled rapidly. After 72-h incubation, the soil samples were extracted with 0.5 M K2SO4 (100 ml) by shaking at 250 rpm for 30 min, and filtered through Whatman no. 42 filter paper. A nonfumigated incubation (without chloroform) was conducted as the control. For the Cmic analysis, 8 ml of soil extract was mixed with 0.066 M K2Cr2O7 2 ml, HgO 70 mg, concentrated H2SO4 10 ml, and 85% H3PO4 5 ml; digested at 150°C for 30 min; and titrated against 0.033 M ferrous (II) ammonium sulfate using 1,10-phenanthroline–ferrous sulfate mixture as indicator. Nmic was determined by a modified Kjeldahl method (Yang et al. 1991). The Cmic was calculated using the formula Cmic = E C/K EC, where E C is the difference between extractable C from fumigated and nonfumigated samples, and K EC = 0.45 (Wu et al. 1990). The Nmic was calculated as Nmic = E N/K EN, where E N is the difference between extractable N from fumigated and nonfumigated samples and K EN = 0.54 (Brookes et al. 1985).
DNA extraction
Genomic DNA of the soil samples was extracted following a modified protocol of Krsek and Wellington (1999). Two grams of wet soil was used to extract DNA, and the yields were expressed in dry-weight basis. Soil DNA was extracted with Crombach buffer (33 mM pH 8.0 Tris–HCl, 1 mM pH 8.0 EDTA) containing lysozyme (5 mg ml−1) and sodium dodecyl sulfate (1%). After centrifugation, supernatants were subjected to potassium acetate and polyethylene glycol precipitation, phenol/chloroform/iso-amylalcohol purification, isopropanol precipitation, and spermine-HCl precipitation. The crude DNA was purified by Gene-Spin 1-4-3 DNA Extraction Kit (Protech, Protech Technology Enterprise Ltd., Taiwan) according to the manufacturer’s recommendations. After extraction, absorbances at 230, 260, and 280 nm were measured to evaluate the levels of protein (A 260/A 280) and humic acid (A 260/A 230) contamination. DNA was quantified by measuring the absorbance at 260 nm, and the concentration was determined from a λ phage DNA standard curve (Promega, Madison, WI, USA).
PCR amplification
Total bacterial 16S rDNA was amplified using the primer set GC−968f (5′-CGC CCG GGG CGC GCC CCG GGC GGG GCG GGG GCA CGG GGG GAA CGC GAA GAA CCT TA-3′) and 1,401r (5′-GCG TGT GTA CAA GAC CC-3′), as described by Felske et al. (1997). The DNA template was amplified with 5 U μl−1 of Pro Taq DNA polymerase (Protech), 10 mM of each primer, 10 mM of each dNTP (Protech), and reaction buffer 1× (Protech, with MgCl2) in a final reaction volume of 50 μl. The PCR was performed with an Applied Biosystems 2720 Thermal Cycler (Foster City, CA, USA) with the following reaction conditions: 94°C for 90 s, followed by 33 cycles at 95°C for 20 s, 56°C for 30 s, 72°C for 45 s, and a final extension step at 72°C for 10 min. The PCR product, 5 μl, was analyzed by electrophoresis on 2% (w/v) agarose gel with 100 bp DNA ladder (Promega) to conform the size and the approximate quantity of generated amplicons.
Denaturing gradient gel electrophoresis
The PCR products, 20 μl, were separated in a vertical denaturing gradient gel using Dcode Universal Mutation Detection System (Bio-Rad Laboratories, Hercules, CA, USA) at 60°C. The polyacrylamide (6%) gels with gradients of 45–65% denaturants (where 100% denaturants contains 7 M urea and 40% formamide) (Muyzer et al. 1996) and a running time of 6 h at 150 V were selected, as these conditions optimally separated a maximal number of bands. After electrophoresis, gels were stained with ethidium bromide and photographed under ultraviolet (UV) light.
Chemical analysis
Moisture contents were determined by drying the sample at 105°C overnight to a constant weight; pH was measured in 1:5 soil:water extracts. Air and soil temperatures were measured directly and at 5 cm depth of soil, respectively, with a thermometer. Total nitrogen (Ntot) was determined by a modified Kjeldahl method (Yang et al. 1991), and total organic carbon (Corg) was estimated using a modified Walkley–Black method, as described by Nelson and Sommers (1982).
Statistical analysis
Analyses were carried out using triplicate samples, and the mean values with standard error (SE) are presented after statistical analyses. All results were reported on a dry-weight basis. Data analysis was performed using analysis of variance (ANOVA) and Tukey’s multiple range tests (P = 0.05) using the Statistical Analysis System (SAS Institute 2002). For DGGE analysis, samples were run to get at least three identical profiles, and a representative figure was presented. DGGE banding patterns were analyzed by ImageJ software (National Institutes of Health, Bethesda, MD, USA), which used band intensity. Band-matching data and relative band densities were exported via Excel (Microsoft Corp.) for cluster analysis using the BioDiversity Pro ver.2 software (http://www.sams.ac.uk/dml/projects/benthic/bdpro/downloads.htm). The similarity matrices were generated using the Bray–Curtis cluster analysis, and dendrograms were created using group average link.
Results
Soil properties and environmental conditions
Soil properties of the valley, middle slope, and ridge are presented in Fig. 1. Fushan soil is acidic (pH 4.39–4.97), and the pH of deeper horizons was higher than that of the organic layer, but the differences were not statistically significant. The pH of the valley soils was the highest, and the ridge soil was the lowest, although there were no significant differences among them. The Corg and Ntot contents of the ridge soils were the highest, while the valley samples were the lowest. Further, Corg, Ntot, and the C/N ratio were high in the organic layer and low in the deeper layer. Air and soil temperatures ranged from 12.0°C to 26.6°C and from 11.3°C to 23.3°C, respectively.
Microbial populations
Microbial populations of three tested sites are presented in Fig. 2. The valley had the lowest microbial population among the tested sites. The organic layer contained the highest populations, and the populations decreased gradually through deeper layers. The bacterial, actinomycete, fungal, cellulolytic, phosphate-solubilizing, and nitrogen-fixing populations at the BC horizon were 1.7–1.9, 4.7–10.0, 3.1–6.2, 0.3–2.0, 0.3–7.2, and 4.9–9.0% of those at the OA horizon, respectively. The ratios of cellulolytic, phosphate-solubilizing, and nitrogen-fixing microbes to the total bacteria in organic layers were 85.8–95.6, 1.4–6.6, and 1.0–2.5%, respectively.
Microbial biomass carbon and nitrogen
Cmic and Nmic in the three topographically different sites are presented in Fig. 3. The Cmic and Nmic contents in the Fushan forest soils ranged from 130.5 to 564.1 μg g−1 and from 16.7 to 95.4 μg g−1, respectively (Fig. 3a, b). The valley had low Cmic and Nmic in all tested sites, and the differences in Cmic were significant in the upper layers (OA, A, and BW1), whereas the Nmic values were not significantly different among all sites. For all profiles, the organic layer had the highest Cmic and Nmic, which decreased with soil depth. However, below 55 cm depth in the three tested profiles, Cmic and Nmic contents remained fairly constant. The proportion of organic carbon present as Cmic/Corg (Fig. 3c) and Cmic/Nmic ratios (Fig. 3e) were high in deeper horizons, and the highest values were in the BC horizon, while all values of Nmic/Ntot were not significantly different (Fig. 3d).
PCR-DGGE diversity profiles of different soil layers
The molecular profiles of bacterial communities at different topographies and different layers are shown in Fig. 4. The yield of total genomic DNA through the soil profiles decreased from 36.0–43.3 μg g−1 in the organic layer to 3.0–10.4 μg g−1 in the BC horizon (75- to 95-cm depth), and the valley had the lowest DNA yield among the three tested sites (Fig. 3f). The ratio of A 260/A 230 greater than 2.0 and the ratio of A 260/A 280 larger than 1.7 indicate the yield of high-purity DNA. Our soil DNA extraction method resulted in high yield of DNA and low contamination with humic acid and protein, as revealed by A 260/A 230 and A 260/A 280 ratios (1.79–2.03 and 1.63–1.83, respectively). Total number of bands in the DGGE profile varied from 14 to 17 in the valley, 16 to 18 in the middle slope, and 20–25 in the ridge sites (Fig. 4). Four strong bands were common to all the profiles, and the total number of strong bands was five to six in the valley and the middle slope and five to eight in the ridge site. DGGE profiles showed that bacterial diversity is almost similar when comparing the strong bands, which may represent the numerically dominant bacteria. However, the soil microbial biomass had a gradient through different layers. Together with these strong bands, a greater number of fainter, well-resolved bands appeared in the profiles, which were all considered when the clustering method was applied. A striking diversity was observed in DGGE patterns due to numerous faint bands, indicating that the structure of microbial communities was rather complex. Profiles of replicates were generally highly repetitive (data not shown). The Bray–Curtis cluster analysis was used to cluster data based upon band intensity. The similarity dendrogram (Fig. 5) clustered into two groups: one group comprising the ridge and valley sites and other one from middle-slope, except the BC layer of the middle slope.
Dendrogram indicating the relationships among soil samples according to polymerase chain reaction—denaturing gradient gel electrophoresis (PCR-DGGE) bacterial diversity profiles of soil samples from the valley, middle slope, and ridge sites. Soil sites were compared using Biodiversity Pro software. The tree was constructed using Bray–Curtis distance equation and group average linkage method. V valley, M middle-slope, R ridge
Correlation analysis
Soil depth had a significant positive correlation with soil pH (R > 0.94) and a negative correlation with DNA yield (R > −0.95), Cmic (R > −0.97), Nmic (R > −0.99), Corg (R > −0.91), Ntot (R > −0.99), and C/N ratio (R > −0.94) in the three tested profiles (Table 2). There was significant positive correlation between DNA yield and microbial biomass (Cmic and Nmic) (R > 0.93) and DNA yield and microbial population (R > 0.86). The correlation between moisture content and DNA yield was significant in the ridge (R = 0.95) and valley (R = 0.88) sites but not in the middle slope. A high correlation was found between microbial biomass and Corg (R > 0.86) and between microbial biomass and Ntot contents (R > 0.96) (Table 2). Further, Cmic had high correlation with Nmic in all tested sites (R > 0.95).
Discussion
Corg and Ntot were high in the organic layer and decreased through the deeper horizons, which indicated that the major nutrient pool in Fushan forest was the organic layer. The ridge and the middle-slope soils had higher tree density and above-ground tree biomass than those of the valley (Lin et al. 2003a). The woody debris biomass of the ridge and the middle slope (36.1 Mg ha−1) and the valley (8.5 Mg ha−1) in Fushan forest (Lin et al. 2003a) were lower than those in the temperate forests (58–381 Mg ha−1) (Carmona et al. 2002), and was similar to those in tropical forests (1.6–51.5 Mg ha−1) (Delaney et al. 1998). There were large areas of canopy gaps in the valley patches, and the effective soil was shallow due to erosion, which led to reduce leaf litter and higher pH in the valley soils when compared with the middle slope and ridge soils. Diaz-Ravina et al. (1995) reported that lack of water seemed to limit the microbial biomass more than temperature did, since lower microbial biomass contents were observed in the dry period than in the wet period. Several studies reported a close relationship between soil moisture and microbial biomass (Chen et al. 2005; Devi and Yadava 2006). The moisture content had a positive correlation with microbial populations especially in the valley due to the topographic effect (Table 2). Low leaf littering in the valley site makes the soil vulnerable to erosion, and the moisture content also is less when compared with the other two sites, which also results in lower Corg and Ntot content in the valley soils. Moreover, the soil type is stony lithosols in the valley site, and stones of <2 mm size can also account for the comparatively lower microbial population and DNA recovery from this site. Since the correlation among different microbial populations and abiotic factors (Corg and Ntot) are consistent in the valley site, we recommend that this site would be appropriate to monitor environmental effects such as acid rain in the Fushan forest ecosystem. Further, if the forest ecosystem needs to be monitored by microbial populations as an indicator, the topographic variations should be appropriately considered.
Microbial populations were significantly high in the organic layers and decreased gradually through the deeper layers. Similar patterns in microbial abundance had been reported in the profiles of agricultural fields (Taylor et al. 2002) and the spruce profile of Tatachia Mountain (Yang et al. 2003). There is much leaf deposit on the surface of Fushan forest soil. The soil pH increased slightly with increasing depth from the OA horizon (0–15 cm) to the BC horizon (75–95 cm) due to abundant leaf litter on the surface, incomplete decomposition of the litter, and accumulation of organic acids (Yang et al. 2003). Berg et al. (1998) also reported that Wekerom forest soil in the Netherlands had a pH of 3.8 due to organic matter content. The differences in chemical composition of litter and root exudates would be expected to influence the availability of different carbon sources, which subsequently influenced the prevalence of microbes. Corg and Ntot also decreased with increasing depth—from 13.25–17.62% and 0.38–0.57% in the OA horizon to 0.51–0.82% and 0.08–0.15% in the BC horizon, respectively. The differences among Corg contents of different layers are statistically significant, where as such, a clear distinction was not observed in Ntot.
The microbial populations of the BC horizon of the valley, middle slope and ridge sites were less than 10% of those in the OA horizon. Many studies using different methods—such as conventional microbial counts (Priha et al. 2001), respiration and ergosterol content (Imberger and Chiu 2001), microbial activity, respiration, biomass, and adenosine triphosphate (ATP) content (Lavahun et al. 1996)—have reported that the microbial populations in the organic layer were higher than those in the surface or subsurface layers. The low microbial populations in the BC horizon may be attributed to the limitation of organic matter, Ntot, and oxygen contents. The valley had the lowest microbial populations because of the lowest amount of organic matter and Ntot contents among the three tested locations. However, the differences in microbial populations of the valley site in comparison with other sites are not significant, except at the organic layer. Among these abiotic factors, the Corg correlates significantly with the microbial population, indicating its importance on that population.
The Cmic/Nmic ratio is often used to describe the structure and state of the microbial community. A high Cmic/Nmic ratio indicates that the microbial biomass contains a high proportion of fungi, whereas a low value suggests that bacteria predominate in the microbial population (Joergensen et al. 1995). Paul and Clark (1996) also indicated that bacteria had a C/N ratio as low as 3.5 and fungi had the values from 10 to 15. In this study, the Cmic/Nmic ratio of the valley, middle slope, and ridge were 5.6–8.2, 4.5–7.8, and 5.8–6.8, respectively. The differences in Cmic/Nmic ratios among different sites are not significant (P < 0.05), with a few exceptions that are not consistent. The bacterial population was significantly higher than the fungal population, which resulted in low Cmic/Nmic ratio, as reported by Joergensen et al. (1995). Moreover, the bacteria-to-fungi ratios of the valley, middle slope, and ridge were 46–143, 155–495, and 170–555, respectively. It showed that the valley had the lowest bacteria-to-fungi ratio, which resulted in high Cmic/Nmic ratio. Further, Joergensen et al. (1995) suggested that forest soil had comparatively low C and N availability and Cmic/Nmic ratio, which was lower than the optimum value (5–8). Similar results were shown in the valley, middle slope, and ridge sites of Fushan forest soils.
Cmic and Nmic also decreased with increasing soil depth and showed significant positive correlation with Corg and Ntot, respectively. Agnelli et al. (2004) reported that the Cmic/Corg ratio increased with soil depth and had the highest value in the BCb2 horizon. The same phenomenon was found in this study, which might be due to the low organic matter in the deep soil layer. Moore et al. (2000) reported that the Nmic showed more pronounced fluctuations than those of Cmic because the microorganisms differed much more in their nitrogen contents than in their carbon contents. Therefore, small shifts in the structure of the microbial community can result the large changes in Nmic. The Cmic/Nmic ratio was affected by soil properties such as moisture content, texture, and pH (Moore et al. 2000). In this study, the Cmic/Nmic ratio increased with soil depth (Fig. 3) in the valley and ridge sites, whereas a fluctuation was observed in the middle slope. The gradual increase of the Cmic/Nmic ratio may be due to the presence of microbes, which has low nitrogen content. The Cmic and Nmic of Fushan forest soils (130.5–564.1 and 16.7–95.4 μg g−1, respectively) were similar to the tropical forest soil in India (341–872 and 38–78 μg g−1, respectively) (Raghubanshi 1991) but lower than the spruce (438–2,018 and 43–210 μg g−1, respectively) and hemlock (100–1,582 and 11–147 μg g−1, respectively) soils in Tatachia Mountain of Taiwan (Yang et al. 2003, 2006) and the forest soil in Germany (317–2,116 and 30–347 μg g−1, respectively) (Joergensen et al. 1995). The low value could be due to the relatively warm climate, which enhanced decomposition of soil organic matter and retained less microbial biomass.
Cmic/Corg ratios of the valley (0.3–2.6%), middle slope (0.3–2.7%), and ridge (0.3–2.2%) sites were lower than that of the tropical forest (1.5–5.3%) (Luizao et al. 1992) and the tropical wet evergreen forest (4–6%) (Barbhuiya et al. 2005) and similar to the subtropical humid forests (0.7–1.7%) (Maithani et al. 1996). However, Nmic/Ntot ratios of the valley (2.0–2.6%), middle slope (1.8–2.1%), and ridge (1.5–2.0%) sites were also lower than that of the forest soils (3.4–5.9%) (Martikainen and Palojarvi 1990) and the forest regrowths (7.3–8.3%) (Maithani et al. 1996) and similar to the tropical wet evergreen forest (1.3–1.7%) (Barbhuiya et al. 2005). This indicates low C and N immobilization as microbial biomass in Fushan forest soils. The dynamic nature of C and N circulation of forest floor and microbial populations is very important in nutrient conservation and regeneration in Fushan forest. Soil moisture had a strong influence on soil properties, and the dynamics of Cmic and Nmic. Similar relationship, i.e., a positive correlation (R > 0.74 and significant at P < 0.001) between moisture content and Cmic and Nmic was reported from the subtropical humid forest of northeast India (Arunachalam and Arunachalam 2000).
The 16S rDNA-DGGE analysis can be used to study changes in the structure of the microbial community (Fritze et al. 2000). The 16S rDNA-DGGE analysis showed the structure of the microbial community changed with increasing soil depth (Fig. 4). Comparatively more bands were observed in ridge soil DGGE profiles, implying higher diversity of the bacterial population. DGGE analysis had very complex banding patterns in the upper horizons, indicating a high diversity and density of microorganisms in contrast to less diversity in the deeper horizons. Numerically dominant bacterial types in the upper layers (bands with high intensity) gradually decrease through the deeper horizons, and a few bacterial types prefer the deeper regions and exist in large numbers (high-intensity bands appeared in the BW2 and BC horizons of the middle slope), which may not be numerically dominant in the upper layers (Fig. 4). Cluster analysis of bacterial DGGE banding patterns in the three tested sites resulted in two groups: (1) middle slope and (2) ridge and valley samples (Fig. 5). Within each group, different layers, especially OA-A, formed different groups, suggesting that the microbial community in the deeper horizons (BW1-BW2-BC horizons) was not simply the diluted analogs of the surface soils and that some microbes dominate only in the deeper horizons. These results indicate that topography influenced both microbial populations and bacterial diversity.
In conclusion, the valley site had the lowest microbial populations (microbial biomass) among the three tested sites due to the low organic matter content, and these populations decreased with soil depth. The PCR-DGGE community profiles of three tested sites were very similar to each other in the major bands, and the variation was in the minor bands only. However, the pattern through gradient profiles was different, indicating the prevalence of specific microbes at different horizons. Topography influenced the quantity and diversity of microbial populations.
References
Agnelli A, Ugolini FC, Corti G, Pietramellara G (2001) Microbial biomass-C and basal respiration of fine earth and highly altered rock fragments of two forest soils. Soil Biol Biochem 33:613–620
Agnelli A, Asche J, Corti G, Ceccherini MT, Nannipieri P, Pietramellara G (2004) Distribution of microbial communities in a forest soil profile investigated by microbial biomass, soil respiration and DGGE of total and extracellular DNA. Soil Biol Biochem 36:859–868
Arunachalam A, Arunachalam K (2000) Influence of gap size and soil properties on microbial biomass in a subtropical humid forest of Northeast India. Plant Soil 223:185–193
Axelrood PE, Chow ML, Radomski CC, McDermott JM, Davies J (2002) Molecular characterization of bacterial diversity from British Columbia forest soils subjected to disturbance. Can J Microbiol 48:655–674
Barbhuiya AR, Arunachalam A, Pandey HN, Arunachalam K, Khan ML, Nath PC (2005) Dynamics of soil microbial biomass C, N and P in disturbed and undisturbed stands of a tropical wet-evergreen forest. Eur J Soil Biol 40:113–121
Berg MP, Kniese JP, Verhoef HA (1998) Dynamic and stratification of bacteria and fungi in the organic layers of a Scots pine forest soil. Biol Fertil Soils 26:313–322
Brookes PC, Landman A, Pruden G, Jenkinson DS (1985) Chloroform fumigation and the release of soil nitrogen: a rapid direct extraction method to measure microbial biomass nitrogen in soil. Soil Biol Biochem 17:837–842
Bundt M, Widmer F, Pesaro M, Zeyer J, Blaser P (2001) Preferential flow paths: biological hot spots in soils. Soil Biol Biochem 33:729–738
Carmona MR, Armesto JJ, Aravena JC, Pérez CA (2002) Coarse woody debris biomass in successional and primary temperate forests in Chiloé Island, Chile. For Ecol Manage 164:265–275
Chang CH, Hsieh CY, Yang SS (2001) Effect of cultural media on the phosphate-solubilizing activity of thermo-tolerant microbes. J Biomass Energy Soc China 20:79–90
Chen TH, Chiu CY, Tian G (2005) Seasonal dynamics of soil microbial biomass in coastal sand dune forest. Pedobiologia 49:645–653
Cho J-C, Tiedje JM (2001) Bacterial species determination from DNA–DNA hybridization by using genome fragments and DNA microarrays. Appl Environ Microbiol 67:3677–3682
Delaney M, Brown S, Lugo AE, Torres-Lezama A, Quintero NB (1998) The quantity and turnover of dead wood in permanent forest plots in six life zones of Venezuela. Biotropica 30:2–11
Devi NB, Yadava PS (2006) Seasonal dynamics in soil microbial biomass C, N and P in a mixed-oak forest ecosystem of Manipur, Northeast India. Appl Soil Ecol 31:220–227
Diaz-Ravina M, Acea MJ, Carballas T (1995) Seasonal changes in microbial biomass and nutrient flush in forest soils. Biol Fertil Soils 19:220–226
Ekelund F, Rønn R, Christensen S (2001) Distribution with depth of protozoa, bacteria and fungi in soil profiles from three Danish forest sites. Soil Biol Biochem 33:475–481
Felske A, Rheims H, Wolterink A, Stackebrandt E, Akkermans ADL (1997) Ribosome analysis reveals prominent activity of an uncultured member of the class Actinobacteria in grassland soils. Microbiology 143:2983–2989
Fierer N, Schimel JP, Holden PA (2003) Variations in microbial community composition through two soil depth profiles. Soil Biol Biochem 35:167–176
Fisher MM, Triplett EW (1999) Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl Environ Microbiol 65:4630–4636
Fritze H, Pietikainen J, Pennanen T (2000) Distribution of microbial biomass and phospholipid fatty acids in Podzol profiles under coniferous forest. Eur J Soil Sci 51:565–573
Hackl E, Zechmeister-Boltenstern S, Bodrossy L, Sessitsch A (2004) Comparison of diversities and compositions of bacterial populations inhabiting natural forest soils. Appl Environ Microbiol 70:5057–5065
Hsia YJ, Hwong JL (1999) Hydrological characteristics of Fushan experimental forest. Q J Chin For 32:39–51
Imberger KT, Chiu CY (2001) Spatial changes of soil fungal and bacterial biomass from a sub-alpine coniferous forest to grassland in a humid, sub-tropical region. Biol Fertil Soils 33:105–110
Jen CH, Lin JC, Hsu ML, Petley DN (2006) Fluvial transportation and sedimentation of the Fu-shan small experimental catchments. Quat Int 147:34–43
Joergensen RG, Anderson TH, Wolters T (1995) Carbon and nitrogen relationships in the microbial biomass of soils in beech (Fagus sylvatica) forests. Biol Fertil Soils 19:141–147
King HB, Liu CP, Hsia YJ, Hwong JL (2003) Interactons of the Fushan hardwood forest ecosystem and the water chemistry of precipitation. Taiwan J For Sci 18:367–373
Kirk JL, Beaudette LA, Hart M, Moutoglis P, Klironomos JN, Lee H, Trevors JT (2004) Methods of studying soil microbial diversity. J Microbiol Methods 58:169–188
Konopka A, Turco R (1991) Biodegradation of organic compounds in vadose zone and aquifer sediments. Appl Environ Microbiol 57:2260–2268
Krsek M, Wellington EMH (1999) Comparison of different methods for the isolation and purification of total community DNA from soil. J Microbiol Methods 39:1–16
Lavahun MFE, Joergensen RG, Meyer B (1996) Activity and biomass of soil microorganisms at different depths. Biol Fertil Soils 23:38–42
Lin KC, Horng FW, Cheng WE, Chiang HC, Chang UC (1996) Soil survey and classification of the Fushan experimental forest. Taiwan J For Sci 11:159–174
Lin KC, Duh CT, Ma FC, Wang HH (2003a) Biomass and nutrient content of woody debris in the Fushan subtropical broadleaf forest of northeastern Taiwan. Taiwan J For Sci 8:235–244
Lin KC, Hamburg SP, Tang S, Hsia YJ, Lin TC (2003b) Typhoon effects on litterfall in a subtropical forest. Can J For Res 33:2184–2192
Lin NH, Lee CT, Chan CC, Hsu WC, Chang MB, Lin WL, Hong CA, King HB, Hsia YJ (2000) A preliminary analysis of chemical characteristics of atmospheric pollutants and their deposition budget on the Fu-Shan forest in Taiwan. Terr Atmos Ocean Sci 11:481–500
Lin TC, Hamburg SP, King HB, Hsia YJ (1997) Spatial variability of throughfall in a subtropical rain forest in Taiwan. J Environ Qual 26:172–180
Liu CP, King HB, Wang MK, Hsia YJ, Hwong JL (2004) Water chemistry and temporal variation of nutrients in stemflow of three dominant tree species in the subtropics of the Fu-shan forest. Water Air Soil Pollut 155:239–249
Luizao RCC, Bonde TA, Rosswall T (1992) Seasonal variation of soil microbial biomass—the effect of clear felling in a tropical rain forest and establishment of pasture in the Central Amazon. Soil Biol Biochem 24:805–813
Mabry CM, Hamburg SP, Lin TC, Horng FW, King HB, Hsia YJ (1998) Typhoon disturbance and stand level damage patterns at a subtropical forest in Taiwan. Biotropica 30:238–250
Maithani K, Tripathi RS, Arunachalam A, Pandey HN (1996) Seasonal dynamics of microbial biomass C, N and P during regrowth of a disturbed subtropical humid forest in northeast India. Appl Soil Ecol 4:31–37
Mandels M, Reese ET (1957) Induction of cellulase in Trichoderma viride as influenced by carbon sources and metals. J Bacteriol 73:269–278
Martikainen PJ, Palojarvi A (1990) Evaluation of the fumigation extraction method for determination of microbial C and N in a range of forest soils. Soil Biol Biochem 27:797–802
Moore JM, Klose S, Tabatabai MA (2000) Soil microbial biomass carbon and nitrogen as affected by cropping systems. Biol Fertil Soils 31:200–210
Muyzer G (1999) DGGE/TGGE: a method for identifying genes from natural ecosystems. Curr Opin Microbiol 2:317–322
Muyzer G, Hottenträger S, Teske A, Wawer C (1996) Denaturing gradient gel electrophoresis of PCR-amplified 16S rRNA: a new molecular approach to analyse the genetic diversity of mixed microbial communities. In: Akkermans ADL, van Elsas JD, de Bruijn FJ (eds) Molecular microbial ecology manual. Kluwer, Dordrecht, pp 1–27
Nelson DW, Sommers LE (1982) Total carbon, organic carbon and organic matter. In: Page AL (ed) Methods of soil analysis. Part 2. Chemical and microbiological properties, 2nd edn. The American Society of Agronomy, Wisconsin, pp 539–580
Owen JS, Wang MK, Wang CH, King HB, Sun HL (2003) Net N mineralization and nitrification rates in a forested ecosystem in northeastern Taiwan. For Ecol Manage 176:519–530
Paul EA, Clark FE (1996) Soil microbiology and biochemistry, 2nd edn. Academic, London, pp 129–155
Priha O, Grayston SJ, Hiukka R, Pennanen T, Smolander A (2001) Microbial community structure and characteristics of the organic matter in soils under Pinus sylvestris, Picea abies and Betula pendula at two forest sites. Biol Fertil Soils 33:17–24
Raghubanshi AS (1991) Dynamics of soil biomass C, N, and P in a dry tropical forest in India. Biol Fertil Soils 12:55–59
SAS Institute (2002) SAS/STAT user’s guide, Release 6.03, SAS Institute, Cary, North Carolina
Schoenborn L, Yates PS, Grinton BE, Hugenholtz P, Janssen PH (2004) Liquid serial dilution is inferior to solid media for isolation of cultures representative of the phylum-level diversity of soil bacteria. Appl Environ Microbiol 70:4363–4366
Soil Survey Staff (2003) Keys to soil taxonomy, 9th edn. United States Department of Agriculture, Soil Conservation Service, Blacksburg, http://www.soils.usda.gov/technical/classification/tax_keys/
Taylor JP, Wilson B, Mills MS, Burns RG (2002) Comparison of microbial numbers and enzymatic activities in surface soils and subsoils using various techniques. Soil Biol Biochem 34:387–401
Torsvik V, Salte K, Soerheim R, Goksoeyr J (1990) Comparison of phenotypic diversity and DNA heterogeneity in a population of soil bacteria. Appl Environ Microbiol 56:776–781
Vance ED, Brookes PC, Jenkinson DS (1987) Microbial biomass measurements in forest soils: the use of the chloroform fumigation-incubation method in strongly acid soils. Soil Biol Biochem 19:697–702
Wu J, Joergensen RG, Pommerening B, Chaussod R, Brookes PC (1990) Measurement of soil microbial biomass C by fumigation-extraction—an autoclaved procedure. Soil Biol Biochem 22:1167–1169
Yang CK, Yang SS (2001) Microbial ecology of soils surrounding nuclear and thermal power plants in Taiwan. Environ Int 26:315–322
Yang SS, Chang HL, Wei CB, Lin HC (1991) Reduce waste production with modified Kjeldahl method for nitrogen measurement. J Biomass Energy Soc China 10:147–155
Yang SS, Fan HY, Yang CK, Lin IC (2003) Microbial population of spruce soil in Tatachia Mountain of Taiwan. Chemosphere 52:1489–1498
Yang SS, Fan HY, Yang CK, Hung WL, Cho ST (2006) Microbial ecology population of hemlock soil in Tatachia Mountain of Taiwan. J Microb Immunol Infect 39:195–205
Acknowledgments
The authors thank Professors H. P. King and K. C. Lin for their helpful assistance, and the National Science Council of Taiwan for financial support (NSC 92–2621-B002-007 and NSC 93-2621-B002-005).
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Tsai, SH., Selvam, A. & Yang, SS. Microbial diversity of topographical gradient profiles in Fushan forest soils of Taiwan. Ecol Res 22, 814–824 (2007). https://doi.org/10.1007/s11284-006-0323-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11284-006-0323-2