Abstract
Preparation of curd vary worldwide due to which its taste, texture and impact on human health also differ. In Assam, curd prepared from raw milk (RMC) is preferred over curd prepared from boiled milk (BMC), a tradition believed to have originated from the Mongoloid customs. Microbial diversity of raw milk (RM), boiled milk (BM), RMC and BMC collected from three farms were investigated by culture dependent and independent techniques. Additionally, metabolite profiles of RMC and BMC were studied by gas chromatography and mass spectroscopy. A total of 59 bacterial isolates were identified from the four different dairy products. In RM, lactic acid bacteria such as Lactococcus, Enterococcus, Lactobacillus and Leuconostoc were obtained along with the environmental bacteria like Bacillus, Staphylococcus, Acetobacter, Chryseobacterium, Streptococcus, Acinetobacter, Kocuria, Klebsiella and Macrococcus. Additionally, Prevotella, Oscillospira, Phascolarctobacterium and Akkermansia were also detected in BM by culture independent technique. In RMC and BMC, Lactococcus, Leuconostoc and Lactobacillus were prevalent. RM and RMC shared Enterococcus, Lactococcus, Streptococcus and Acinetobacter as common bacterial genera. However, no bacterial genus was common in BM and BMC. The correlation analysis revealed that Lactobacillus was negatively correlated to other bacterial genera. Oligotyping analysis revealed that Lactobacillus brevis and L.fermentum were abundant in RMC and BMC, respectively. In metabolomic study, ascorbic acid, dodecanoic acid and hexadecanoic acid were found to be significantly higher in RMC. Presence of different types of probiotics in these curds samples opens a new avenue to understand their effects on human health.
Graphic abstract

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Dairy products are important components of a healthy diet. Fermented dairy products are preferred worldwide and are source of health beneficial microbes too. The process of making fermented dairy products has evolved with the human civilisations and therefore, varies across the world. Naturally fermented milk (NFM), a component of the Mongolian diet is believed to have health beneficial role and offer palatability to consumers due to rich microbial diversity (Zhong et al. 2016). In the north-eastern state of Assam, India few communities who are known to be of Mongoloid origin; traditionally prefer to take curds prepared from raw milk (RMC) over those prepared from boiled milk (BMC). RMC is prepared by allowing raw milk to undergo natural fermentation without adding any inoculum, whereas BMC is prepared by back sloping technique in which an inoculum from previous batch is added to boiled milk for fermentation.
Fermented dairy products are produced by specific group of microorganisms resulting decrease in pH that leads to coagulation of milk proteins (Dewan and Tamang 2007; Kabak and Dobson 2011). The microbiota of raw milk is complex and is reported to harbour thermoresistant, thermophilic, mesophilic, adventitious and psychrotrophic bacteria whose dominance decreases in semi cooked curd (47 °C) used for the production of Fontina (an Italian cheese) (Giannino et al. 2009). Microbial composition in some of the fermented dairy products, such as curd and churpi widely preferred in Sikkim, the north-eastern state of India included different species of Lactobacillus, and Lactococcus along with Enterococcus faecium and Leuconostoc mesenteroides (Dewan and Tamang 2007; Tamang et al. 2000). However, there is no report on the microbial composition of naturally fermented curd which is highly preferred in Assam over curd prepared from boiled milk. The traditional method of isolation of microbes has limitations as it cannot simulate the exact environment for their growth due to which a majority of them remains undetected (Zhong et al. 2016). To overcome this limitation, the culture independent techniques such as next generation sequencing (NGS) analysis of 16S rRNA amplicons have been widely used to detect the microbial diversity in various dairy products such as raw milk (Li et al. 2018), cheese (Wolfe et al. 2014), Koumisses (Zhong et al. 2016), curd and churpi (Tamang et al. 2000). However, the NGS analysis should be followed by oligotyping to investigate the bacterial diversity at sub-genus level (Eren et al. 2013).
The aim of this research was to compare the bacterial diversity of raw milk, boiled milk, curd prepared from raw and boiled milk. Bacterial diversity of both milk and curd samples collected from three farms of Assam were studied by both culture dependent and independent techniques (NGS analysis of 16S rRNA amplicons). The NGS data was further explored to determine the core bacterial genera and their interacting partners. Oligotyping was performed to identify the diversity within common core bacterial genera of RMC and BMC. A gas chromatography-mass spectrometry (GC–MS) based metabolomics profiling was also carried out to compare the metabolite profiles of the two types of curd samples.
Materials and methods
Sample collection
Dairy samples including raw milk (RM), boiled milk (BM) (boiled at 100 °C for 10 min) and curd prepared from RM (RMC) and BM (BMC) were collected in triplicates from three farms located at Aanthmile, Jagiroad and Pathgaon, Assam, India. RMC was prepared by allowing raw milk to undergo natural fermentation at room temperature (25–30 °C) without adding any starter culture, while BMC was prepared from boiled milk using back sloping technique where boiled milk was allowed to ferment after adding an inoculum from a previous batch for 48 h. For isolation of culturable microbes, samples were processed within 3–4 h after collection. Samples were stored at − 80 °C for culture independent and metabolomics studies.
Bacterial isolation and characterisation
A 1 ml each of the milk and curd samples were homogenised in sterile 0.9% (w/v) NaCl solution (9 ml) for 5 min and a tenfold serial dilutions were carried out. A 100 µl of each of the dilutions of both milk (10−2 to 10−4) and curd (10−5 to 10−7) samples were spreaded onto plates containing different nutrient media. Isolation of Lactobacilli and Enterobacteria were carried out on deMan Rogosa Sharpe agar (MRSA, HiMedia, Mumbai, India) using both aerobic and anaerobic conditions (Anaerobic chamber, Don Whitely Scientific Limited, West Yorkshire, UK) at 30 °C and 37 °C, respectively for 48 h. M17 media (HiMedia, Mumbai, India) was used for isolation of Lactococci under aerobic and anaerobic conditions at 30 °C for 24 h. In addition, MRSA supplemented with 0.2% (w/v) cysteine (Merck, New Jersey, USA) was used for isolation of Bifidobacteria by incubating in an anaerobic chamber at 30 °C for 48–72 h. Pure cultures were maintained for each isolated colonies and stored at − 80 °C in 15% (v/v) glycerol. In order to obtain genomic DNA, the isolates were cultured overnight in liquid media (MRS or M17) and DNA extraction was carried out following the protocol described by Sambrook et al. (1989). The quality of DNA was assessed by 0.8% (w/v) agarose gel electrophoresis. Quantification was performed using a Nano Drop™ ND-1000 (Thermo Fisher Scientific, Wilmington, USA). All the DNA samples were stored at − 20 °C for future use. PCR amplification of the 16S rRNA region of the bacterial DNA was carried out using the primer pair, 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and L1401(5′-CGGTGTGTACAAGACCC-3′). PCR reaction was performed in a 20 µl volume in a thermal cycler. Each PCR reaction contained a final concentration of 1 × standard buffer, 1.75 mM of MgCl2, 200 µM of dNTPs, 0.2 µM of each primer, 1U of Taq DNA polymerase and 30 ng of template DNA. The PCR conditions were, initial denaturation at 94 °C for 5 min followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 60 °C for 30 s, extension at 72 °C for 30 s and a final extension at 72 °C for 7 min. The PCR products (1400 bp) were visualised in a 1.2% (w/v) agarose gel under BioDoc-It Imaging System (UVP, California, US). Sequencing of the PCR amplicons covering the 16S rRNA region was carried out with Xcelris Genomics (Gujarat, India). The DNA sequences were compared against the GenBank database of NCBI using the BLAST program (Camacho et al. 2009).
Metagenomic DNA extraction from milk and curd samples
Metagenomic DNA was extracted from milk samples (both RM & BM) by following the method earlier described by Sambrook et al. (1989). Briefly, aliquots (40 ml) of milk samples were centrifuged initially at 1000×g for 30 min and the pellets were resuspended in 1 ml of lysis buffer (Tris–EDTA buffer). A final concentration of 1 mg/ ml of proteinase K (Sigma) and 0.3% sodium dodecyl sulphate (SDS) were added to each sample and incubated at 42 °C overnight in a water bath. Similarly, the curd samples were initially centrifuged at 8000 rpm to reduce the fat content. The pellets were then resuspended in the lysis buffer (Tris–EDTA buffer) and incubated at 55 °C overnight in a water bath. Equal volume of Phenol and chloroform (1:1) was added and then DNA was precipitated by addition of sodium acetate (3 M, pH 5.2) and 100% (v/v) ethanol. The pellets were finally resuspended in 25 µl sterile Tris–EDTA buffer. Quantification was performed using a Nano Drop™ ND-1000 (Thermo Fisher Scientific, Wilmington, USA).
Next generation sequencing (NGS) analysis
Metagenomic DNA of both milk and curd samples were subjected to NGS analysis with Macrogen (Seoul, Korea). Quantification of the double stranded DNA was performed using QuantiFluor dsDNA System (Promega, Wisconsin, USA). Bacterial diversity in the sample was analyzed using V3-V4 region of 16S rRNA amplicon sequencing on Illumina MiSeq platform. A 2 × 300 bp of MiSeq amplicon library was prepared using the Nextera XT Index Kit (Illumina Inc, California, USA). Amplicons were then ligated with Illumina adaptors and were amplified and gel purified as per the standard Illumina protocols.
NGS data analysis
The NGS data analyses were performed using QIIME (version 1.9.1) pipeline (Caporaso et al. 2010) on the extracted high-quality sequences as described earlier (Zhang et al. 2016). Representative sequences were classified as an operational taxonomic unit (OTU) with a 97% threshold identity and taxonomic assignment were performed using SILVA database 128. For beta diversity, both weighted and unweighted UniFrac principal coordinate analyses (PCoA) were performed. All the sequences were uploaded in the Metagenomic RAST server (MG-RAST) and the details of each sample and IDs are tabulated in the Supplementary Table S1.
Oligotyping analysis
Oligotyping of the three core bacteria, Lactobacillus, Leuconostoc and Lactococcus obtained in both the types of curd samples was performed using the oligotyping pipeline version 2.2 (available from https://oligotyping.org) (Eren et al. 2013). Following the initial entropy analysis, oligotyping was performed considering entropy values greater than 0.5 positions (C) were different for all the three bacteria and therefore, a minimum substantive abundance criterion (M) as 50 for all the three genera was used. Oligotypes not meeting the minimum substantive abundance criterion were discarded as noise. The matrix percent count generated during analysis was used for further analysis. We performed BLAST search for dominant oligotypes in the NCBI database to assign taxonomy.
Metabolite study
Both the dairy samples (RMC & BMC) were extracted in hexane in 1:1 ratio for 12 h using a rotospin (Tarsons, Kolkata, India). After mixing, the samples were centrifuged at 7000×g for 15 min at room temperature. Supernatant was collected and were filtered with 0.45 µm syringe filter (Sartorius, Gottingen, Germany). A 200 µl of the filtrate was transferred into GC–MS vials containing inserts and further analysis was carried out using Shimadzu Plus-triple quadrupole (TP-8030) GC–MS/MS system fitted with EB-5MS column (length-30 m, thickness-0.25 μm and ID-0.25 mm). The oven program started at 80 °C, subsequently ramped at a rate of 10 °C per min to 300 °C and then held for 10 min. A 1 µl sample was injected at 250 °C using He as carrier gas (1 ml per min) in split mode (1:10). The mass spectrometer was operated at a source temperature of 230 °C and a continuous scan from 45 to 800 m/z. The peaks were identified by matching the mass spectra with the National Institute of Standards and Technology (NIST) library, USA. The noisy peaks and the column bleeds (siloxanes and silane) were removed from the metabolite list before further analysis.
Statistical analysis
The differences in microbial diversities across the milk and curd samples were investigated by Mann–Whitney U test within SPSS (SPSS Inc., Chicago IL, USA). The interactions among the microbes in both milk and curd samples were studied by bivariate correlation analysis {Spearman correlation (non-parametric) within SPSS (IBM SPSS, statistics 20)} and the networks were generated using Cytoscape (version 3.6.0) (Shannon et al. 2003) as described by Dehingia et al. (2015). Metabolite analysis was performed in MetaboAnalyst 4.0 package customized for metabolomics study (Xia et al. 2015). The metabolite data were normalized by sum method followed by log transformation and pareto scaling as described earlier (Dehingia et al. 2017; Dutta et al. 2012). Normalized data were subjected to multivariate analyses such as partial least square discriminant analysis (PLS-DA) and t-test in Metaboanalyst software.
Results
Culture dependent bacterial diversity
A total of 59 bacterial isolates were isolated from the four types of dairy products {RM (n = 19), BM (n = 15), RMC (n = 10) and BMC (n = 15)} collected from Aanthmile (AM), Jagiroad (JR) and Pathgaon (PG). The bacterial isolates were identified by 16S rRNA sequencing (Supplementary Table S2 and Table S3). The bacterial profiles of the RM, BM, RMC and BMC samples were farm specific. In the RM samples, species of Lactobacillus (ARM5, JRM8, PGRM9, ARC3, PGRC6 and PGRC20), Lactococcus (PGRM2, JRM6, PGRM10, PGRM3, ARC2, JRC2 and PGRC1) and Leuconostoc (ARM2, JRM9, ARC4, JRC1 and PGRC3) were found, which were also detected in the respective curd (RMC) samples (Supplementary Table S2). The species of Enterococcus (PGRM4 and PGBM4) were found in both the RM and BM samples of Pathgaon along with RM samples of Aanthmile (ARM3). In Aanthmile region, the species of Staphylococcus which was isolated from BM (Staphylococcus sciuri ABM4) was also obtained from its respective curd sample (BMC; Staphylococcus sciuri ABC4). In Pathgaon region, different species of Corynebacterium were isolated from BM (Corynebacterium ureicelerivorans PGBM2) and BMC (Corynebacterium nuruki PGBC1) (Supplementary Table S3). The diversity of lactic acid bacteria (LAB) such as Lactobacillus (ABC4, PGBC3, and JBC2), Lactococcus (PGBC6 and JBC8), Leuconostoc (JBC9 and PGBC5) and Acetobacter (ABC1, ABC3 and JBC6) in addition to non-lactic acid bacteria (n-LAB) such as Staphylococcus (ABC7), Corynebacterium (PGBC1), Moraxella (ABC5), Rothia (ABC6), Chryseobacterium (ABC8) were abundant in BMC samples in comparison to RMC samples (Supplementary Table S3).
Culture-independent bacterial diversity
NGS based analysis of metagenomic DNA of the dairy products
Amplicon sequencing produced a total of 1,790,112 high quality 16S rRNA gene reads. To compare the samples without biasness, the minimal read number of 1,22,614 was used as eigen value for calculation of diversity indices. Slopes of the rarefaction curves indicated sufficient coverage of bacterial diversity of the dairy samples (Supplementary Fig. S1). An unweighted Principal Coordinate Analysis (PCoA) plot based on the NGS data of 16S rRNA amplicons of the metagenomic DNA samples indicated distinct clustering of the samples RM, BM, RMC and BMC (Fig. 1).
The phylum level distributions of bacteria in the dairy products have been presented in the Fig. 2. The major bacterial phyla detected in the milk samples were Firmicutes, Proteobacteria,Bacteroidetes and Actinobacteria, while boiled milk had less Proteobacteria and more Firmicutes in comparison to raw milk. However, in the curd samples major bacterial phyla were Firmicutes and Proteobacteria (Fig. 2a–d). The major bacterial families found in raw milk of all the three regions were Micrococaceae, Staphylococcaceae, Enterococcaceae, Streptococcacea and Enterobacteriaceae. However, a region wise variation was observed in which Leuconostocaceae was specific to Jagiroad, Weeksellaceae was found in both Aanthmile and Jagiroad, while Moraxellaceae was detected in Aanthmile and Pathgaon. The bacterial family detected in the boiled milk samples of all the three regions were Staphylococcaceae and Moraxellacea, while Micrococcaceae, Enterococcaceae and Streptococcaceae were found in Aanthmile and Pathgaon and Enterobacteriaceae was found in Jagiroad and Pathgaon. There were few families which were region specific, such as Weeksellaceae, Bacillaceae, Planococcaceae and Aerococcaceae in Aanthmile, Xanthomonadaceae in Jagiroad and Comamonadaceae in Pathgaon. Curds prepared from raw milk of all the three regions had dominance of Lactobacillaceae, Leuconostocaceae, Streptococcaceae, Enterobacteriaceae and Moraxellaceae. However, dominance of Acetobacteraceae was specific to Aanthmile, Pseudomonaceae to Jagiroad and Xanthomanadaceae and Staphylococcaceae to Pathgaon. Curd prepared from boiled milk from all the three regions had dominance of Lactobacillaceae, Leuconostocaceae, Streptococcaceae and Acetobacteraceae. However, Enterobacteriaceae was specific to Aanthmile, Bifidobacteriaceae to Jagiroad and Clostridiaceae to Pathgaon (Fig. 2e).
Relative abundance of the bacterial taxa found in the dairy products. Pie charts showing phylum level distribution of bacterial relative abundance (%) in raw milk (RM) (a), boiled milk (BM) (b), raw milk curd (RMC) (c) and boiled milk curd (BMC) (d). Bar charts show relative abundance (%) of bacterial families (e) and genera (f) in the dairy samples
At the genus level, Georgenia, Xylanimicrobium, Paludibacter, Prevotella, Jeotgalicus, Dorea, Oscillospira, Phascolarctobacterium, Hylemonella, Acrobacter, Akkermansia and Sphingobacterium were found to be significantly higher in BM in comparison to RM (p < 0.05), while Escherichia was higher in RM in comparison to BM (p < 0.05) (Fig. 2f). RMC had significantly higher abundance of Corynebacterium, Kocuria, Sphingobacterium, Salinicoccus, Sphingomonas, Comamonas, Enhydrobacter and Pseudomonas in comparison to BMC (p < 0.05). The RMC samples prepared from RM had higher abundance of Sphingobacterium, Aerococcus, Lactobacillus, Pediococcus and Acetobacter in comparison to the respective RM samples (p < 0.05). However, Aerococcus, Ruminococcus and Paracoccus were significantly higher in RM in comparison to RMC (p < 0.05).
Distribution of the LAB in the dairy samples was farm specific. Lactobacillus was dominant in both RMC and BMC of Aanthmile and Lactococcus in both RMC and BMC of Jagiroad. However, Enterococcus, Leuconostoc, Streptococcus and Lactococcus were dominant in RMC, while Bacillus, Leuconostoc, Streptococcus and Acetobacteraceae were dominant in the BMC samples of Pathgaon (Fig. 2f).
Core bacterial genera in the milk and curd samples
Among the 340 bacterial genera detected, bacteria which were present in all the replicates with more than 0.1% relative abundance were considered as the core bacteria (Table 1). The core bacteria in RM were Staphylococcus, Enterococcus, Lactococcus, Streptococcus and Acinetobacter. In BM, the core bacteria were found to be Chryseobacterium, Staphylococcus, Streptococcus, Acinetobacter, Enhydrobacter, Jeotgalicus, Salinicoccus and Aerococcus. In BMC, the core bacterial genera were found to be Lactobacillus, Leuconostoc, Lactococcus and Acetobacter, while in RMC the core bacterial genera were Chryseobacterium, Enterococcus, Lactobacillus, Leuconostoc,Lactococcus, Streptococcus, Klebsiella, Acinetobacter, Pseudomonas and Enhydrobacter. The core bacteria common in both RM as well as RMC were Enterococcus, Lactococcus, Streptococcus and Acinetobacter. However, no core bacterial genus was common to BM and BMC. The core bacterial genera of RMC and BMC were found to be Lactobacillus, Leuconostoc and Lactococcus.
Networks of co-occurrence of core bacteria found in the RM, BM, RMC and BMC sample were constructed based on the significant correlations between them {r = (−)1.00 to ( + )1.00 and p < 0.01} (Fig. 4). In the RM samples, significant correlations were (i) positive correlation of Staphylococcus with Lactococcus (ii) negative correlations of Staphylococcus with Acinetobacter and Streptococcus (iii) negative correlations of Lactococcus with Streptococcus and Acinetobacter and (iv) positive correlation of Streptococcus with Acinetobacter (Fig. 3a). In BM, significant correlations were (i) positive correlations of Chryseobacterium with Jeotgalicus and Staphylococcus (ii) negative correlation of Chryseobacterium with Acinetobacter (iii) positive correlation of Jeotgalicus with Staphylococcus (iv) negative correlation of Jeotgalicus with Acinetobacter (v) negative correlation of Salinicoccus with Streptococcus (vi) negative correlation of Staphylococcus with Acinetobacter and (vii) positive correlation of Aerococcus with Enhydrobacter (Fig. 3c). In RMC, major correlations were (i) positive correlations of Chryseobacterium with Enterococcus, Streptococcus, Klebsiella and Pseudomonas (ii) negative correlation of Chryseobacterium with Lactobacillus (iii) positive correlations of Enterococcus with Streptococcus, Klebsiella and Pseudomonas (iv) negative correlation of Enterococcus with Lactobacillus (v) negative correlations of Lactobacillus with Streptococcus, Klebsiella and Pseudomonas (vi) positive correlation of Leuconostoc with Acinetobacter (vii) negative correlation of Lactococcus with Enhydrobacter (viii) positive correlations of Streptococcus with Klebsiella and Pseudomonas and (ix) positive correlation of Klebsiella with Pseudomonas (Fig. 3b). In BMC, a negative correlation of Lactobacillus was observed with Leuconostoc (Fig. 3d).
Oligotypes of the core bacterial genera
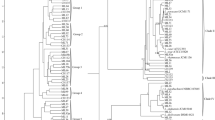
We analyzed the distribution of oligotypes of the core bacterial genera viz., Lactobacillus, Leuconostoc and Lactococcus found in the curd samples. Oligotyping of Lactobacillus revealed that the Lactobacillus delbrueckii was dominant in the samples of Aanthmile. However, Lactobacillus paracasei was found to be dominant in the RMC samples of both Jagiroad and Pathgaon along with the BMC samples of Jagiroad (Fig. 4a). Lactobacillus brevis was found to be dominant in the RMC samples of both Jagiroad and Pathgaon. Oligotyping of Leuconostoc revealed that three different strains of Leuconostoc pseudomesenteroides were dominant in both the RMC and BMC samples of all the three farms (Fig. 4b). Similarly, it was observed that four different oligotypes of Lactococcus were dominant in both the RMC and BMC samples of all the three farms (Fig. 4c). An oligotype of the member species Lactococcuslactis subsp. cremoris was the most dominant oligotype in both the RMC and BMC samples of Aanthmile, Jagiroad and Pathgaon.
Metabolomics
A GC–MS based analysis detected a total of 121 metabolites of which 88 had ≥ 0.1% peak area percentage and were selected for further analysis. Multivariate analysis and partial least squares discriminant analysis (PLS-DA) was performed based on the metabolite data among the RMC and BMC samples. PLS-DA was performed for maximizing the group separation and Leave-one-out cross validation (LOOCV) gave Q2 and R2 values which represent predictive capability and variance, respectively. The PLS-DA plot based on the curd metabolites had Q2 = 0.36 and R2 = 0.96. PLS-DA plot shows farm specific clustering of the RMC and BMC samples (Fig. 5). It was observed that few metabolites such as 10-methyl dodecanoic-5-olide, ascorbic acid and 2,2,4-Trimethyl-1,3-pentanediol disobutyrate were significantly higher in the RMC samples in comparison to the BMC samples of Aanthmile (p < 0.01). However, dodecanoic acid and glycerol 2-acetate were significantly higher in the BMC samples in comparison to the RMC samples of Aanthmile (Fig. 6a). It was observed that n-hexadecanoic acid was significantly higher in the RMC samples of Jagiroad in comparison to the BMC samples where glycerol -2-acetate was higher (p < 0.01) (Fig. 6b). 6-octadecanoic acid was found to be significantly higher in the RMC samples of Pathgaon in comparison to the BMC samples (p < 0.01) (Fig. 6c).
Discussions
In this study, a combination of both culture dependent and independent approaches were applied to explore the microbial communities in RM, BM, RMC and BMC coupled with metabolomic study to reveal their metabolite profiles. Previous reports suggested that the NFM products are vital components of regular diets in ethnic communities of the Mongolian population (Zhong et al. 2016). Many ethnic populations of Assam are of southern Mongolian origin who are referred to as Kiratas in the Sanskrit literature (Bose 1989). Similar to the Mongolians, many ethnic communities in the north-east of India still practice and prefer curd prepared from raw milk (RMC) by natural fermentation. Microbial diversities of the RM, BM, RMC and BMC collected from the three farms of Assam were found to be highly diverse indicating the crucial role of environmental factors along with indigenous milk bacteria in shaping the bacterial composition.
In culture dependent studies, the most important LAB detected in the RM samples of the three farms were Lactococcus, Enterococcus, Lactobacillus and Leuconostoc. These are the most common LAB genera found in bovine milk prior to pasteurisation (Quigley et al. 2013b). Fermentation is facilitated by the autochthonous microorganisms present in raw milk (Motato et al. 2017) due to which few bacterial genera such as Lactococcus, Leuconostoc, Lactobacillus were found to be common in both RM and RMC. Additionally, Bacillus, Staphylococcus, Acteobacter, Chryseobacterium, Streptococcus, Acinetobacter, Kocuria, Klebsiella and Macrococcus were also observed in the RM samples which were earlier reported to have originated from the environment, teat surface and milking equipment (Vacheyrou et al. 2011). All these environmental bacteria, Enterococcus and Corynebacterium were detected in BM. It was reported that Lactococcus have low tolerance to heat in comparison to Enterococcus which therefore was not detected in BM (Delgado et al. 2013). The BMC samples had abundance of LAB (Lactococcus, Lactobacillus and Leuconostocs) along with few n-LAB (Corynebacterium and Staphylococcus) genera which were farm specific. Both Staphylococcus and Corynebacterium were common among BM and BMC of Aanthmile and Pathgaon farm respectively. Few species of Staphylococcus can withstand pasteurisation by the production of heat-stable enterotoxins (Balaban and Rasooly 2000). However, both these bacterial genera are also involved in aroma, flavour and colour development in cheese (Verdier-Metz et al. 2009). The success of such fermentation depends on the particular blend of microorganisms present in the previous batch along with the surrounding (Parente and Cogan 2004). Therefore, the primeval milk microbiota together with those from the environment directly impact microbiome of curd.
As conventional culture dependent methods cannot detect uncultivable bacteria, NGS analysis with the metagenomics DNA was performed (Jany and Barbier 2008). Previous reports suggest that Firmicutes, Proteobacteria, Bacteroidetes and Actinobacteria were the most prevalent phyla in naturally fermented dairy products (Zhong et al. 2016). Interestingly, in our study, the curd prepared in either way had prevalence of Firmicutes and Proteobacteria in comparison to milk samples where Actinobacteria, Bacteroidetes, Firmicutes and Proteobacteria were prevalent. Firmicutes and Proteobacteria can survive high temperature and humidity respectively, while Actinobacteria and Bacteroidetes cannot (Li et al. 2018). Additionaly, in few important gut bacterial genera such as Prevotella, Oscillospira, Phascolarctobacterium and Akkermansia were also identified in BM. Prevotella is a commensal genus of rumen as well as human and is involved in metabolizing protein and carbohydrates (Quigley et al. 2013a). Akkermansia is regarded as a next generation beneficial microbe and is inversely associated with certain metabolic disorders (Cani and de Vos 2017; Everard et al. 2013). Oscillospira belongs to Ruminococcaceae family and is abundant in human faecal microbes and was reported to be positively associated with leanness (Gophna et al. 2017). Phascolarctobacterium is also a gut bacteria which produces short chain fatty acids (Wu et al. 2017).
The dominance of Lactobacillus, a common LAB involved in milk fermentation (Van de Casteele et al. 2006) was observed in the RMC and BMC samples of Aanthmile. However, dominance of Lactococcus over Lactobacillus was observed in RMC and BMC of Jagiroad. This might be due to the fact that few LAB are negatively correlated to each other which were also reflected in the correlation studies. Lactobacillus were found to be negatively correlated with other LAB such as Enterococcus, Streptococcus and Leuconostoc along with few other environmental microbes present in these curd samples which suggested that there was a competition within LAB for the ecological niches. The RMC samples of Pathgaon had co-occurrence of Lactococcus, Leuconostoc, Streptococcus and Enterococcus. In dairy industries, Streptococcus thermophillus is widely used as a starter culture (Hols et al. 2005) and are also bacteriocins producers which protect the dairy products from microbial spoilage (Kabuki et al. 2009). Leuconostoc strains are reported to be in synergestic functional relationship with acid producing Lactococcus (Hemme and Foucaud-Scheunemann 2004). Leuconostoc initiates citrate metabolism and aroma production in acidic conditions (Hemme and Foucaud-Scheunemann 2004). Enterococcus has proteolytic and hydrolytic activities that contribute to milk fermentation along with aroma production as reported earlier (Franz et al. 1999). Bacillus detected in the BM and BMC samples of Pathgaon might have originated from the farms including teat surfaces, dust, hay air and milking parlours (Vacheyrou et al. 2011).
Zhong et al. (2016) reported Lactobacillus, Streptococcus and Lactococcus as core bacteria in NFM products of the Mongolian diet. Interestingly, RMC prepared by natural fermentation in Assam had prevalence of Lactobacillus, Lactococcus, Leuconostoc, Chryseobacterium, Enterococcus, Streptococcus, Klebsiella, Acinetobacter, Enhydrobacter and Pseudomonas as core bacterial genera. Studies on the traditionally fermented milk products of Inner Mongolia in China have reported that Enterococcus sp., Lactococcus lactis, Leuconostoc sp. and Saccharomyces cerevisae predominated in the fermented cow’s milk (Naer and Kitamoto 1995; Watabe 1998). A variety of genera such as Lactobacillus, Lactococcus, Streptococcus and Leuconostoc were reported to be dominant in naturally fermented cow’s milk (NFCM) of Russia which is also a traditional Mongolian fermented dairy product (Liu et al. 2015). Previous studies reported that Lactobacillus species was dominant in curd prepared with back sloping technique which is widely preferred in daily diet in the southern part of India (Balamurugan et al. 2014). Interestingly, in our study the bacterial genera in the curd prepared from boiled milk using back sloping technique which is generally preferred by few other communities of Assam were Lactobacillus, Leuconostoc, Lactococcus and Acetobacter. The culture independent technique also depicted that common bacterial genera found in both RMC and BMC were Lactobacillus, Leuconostoc and Lactococcus. In oligotyping analysis previously unexplored diversity of the genera Lactobacillus, Lactococcus and Leuconostoc in the curd samples were revealed. Diverse strains of Lactobacillus brevis and L. paracasei were found in the RMC of Jagiroad and Pathgaon respectively. However, Aanthmile farm had prevalence of L.delbrueckii and L. fermentum in RMC as well as BMC. It was earlier reported that few strains of L. brevis is a nonstarter LAB species which are responsible for flavour and texture development in cheese (Smit et al. 2005) and also has probiotic properties that improves human bowel function (Nobuta et al. 2009). Previous reports suggests that Lactobacillus conferred health benefits such as increase in immune response, lactose tolerance and colon cancer (Maragkoudakis et al. 2006).
During fermentation, microorganisms increases nutritional value of the dairy products by improving the organoleptic attributes (Von Mollendorff et al. 2006). The results of metabolomics study indicated that RMC and BMC had significantly different metabolites which were farm specific, as in the case of microbes. Presence of ascorbic acid in the RMC samples of Aanthmile might be responsible for enhancing flavour in the curd sample as microbial, enzymatic or chemical transformation of ascorbic acid found in milk produce flavour active compounds such as acetaldehyde, ethyl butyrate etc. (Boelrijk et al. 2003; McGorrin 2001; Tamime and Robinson 1999). Hexadecanoic acid and octadecanoic acid was found in the RMC samples of Jagiroad and Pathgaon, respectively. Hexadecanoic acid is involved in bile acid biosynthesis pathway and are a highly variable lipid component of human breast milk (Jensen et al. 1978). Unlike octadecanoic acid, hexadecanoic acid increases serum cholesterol (Serafeimidou et al. 2013). The Streptococcus and Lactobacillus bind to the free bile acids in the intestine for its removal from the human body (Pigeon et al. 2002). It is reported that milk fermented with Lactobacilli may have hypocholesteromic effect in human (Mann and Spoerry 1974).
Conclusion
To our knowledge, this is the first report of microbial and metabolite profiles of curd prepared from raw milk (RMC) and boiled milk (BMC) using culture dependent, independent and metabolomic approaches. This study reveals that LAB was prevalent in raw milk (RM) and hence in respective RMC. Few strains of Lactobacillus were found to be farm specific. Additionally, metabolites detected in curd samples were also farm specific. These suggest that indigenous and environmental microbes present in dairy products might play an important role in metabolite production to increase its organoleptic and nutritional properties. Few important uncultivable bacterial genera were detected in boiled milk (BM) that might have some beneficial role in human gut health. Therefore the microbial and metabolites compositions of the dairy products should be a focus of research to know their probable implications on health.
References
Balaban N, Rasooly A (2000) Staphylococcal enterotoxins. Int J Food Microbiol 61:1–10
Balamurugan R, Chandragunasekaran AS, Chellappan G, Rajaram K, Ramamoorthi G, Ramakrishna BS (2014) Probiotic potential of lactic acid bacteria present in home made curd in southern India. Indian J Med Res 140:345
Boelrijk AEM, Jong C, Smit G (2003) Flavour generation in dairy products. In: Smith G (ed) Dairy processing and improving quality. CRC Press and Wood Head Publishing Limited, Cambridge, pp 130–148
Bose M (1989) Social history of Assam: being a study of the origins of ethnic identity and social tension during the british period, 1905–1947. Concept Publishing Company, New Delhi
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinform 10:421
Cani PD, de Vos WM (2017) Next-generation beneficial microbes: the case of Akkermansia muciniphila. Front Microbiol 8:1765
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335
Dehingia M, Talukdar NC, Talukdar R, Reddy N, Mande SS, Deka M, Khan MR (2015) Gut bacterial diversity of the tribes of India and comparison with the worldwide data. Sci Rep 5:18563
Dehingia M, Sen S, Bhaskar B, Joishy TK, Deka M, Talukdar NC, Khan MR (2017) Ethnicity influences gut metabolites and microbiota of the tribes of Assam. India. Metabolomics 13:69
Delgado S, Rachid CT, Fernández E, Rychlik T, Alegría Á, Peixoto RS, Mayo B (2013) Diversity of thermophilic bacteria in raw, pasteurized and selectively-cultured milk, as assessed by culturing, PCR-DGGE and pyrosequencing. Food Microbiol 36:103–111
Dewan S, Tamang JP (2007) Dominant lactic acid bacteria and their technological properties isolated from the Himalayan ethnic fermented milk products. Anton Leeuw Int J G 92:343–352
Dutta M, Joshi M, Srivastava S, Lodh I, Chakravarty B, Chaudhury K (2012) A metabonomics approach as a means for identification of potential biomarkers for early diagnosis of endometriosis. Mol BioSyst 8:3281–3287
Eren AM, Maignien L, Sul WJ, Murphy LG, Grim SL, Morrison HG, Sogin ML (2013) Oligotyping: Differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol 4:1111–1119
Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, De Vos WM (2013) Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA 110:9066–9071
Franz CM, Holzapfel WH, Stiles ME (1999) Enterococci at the crossroads of food safety? Int J Food Microbiol 47:1–24
Giannino ML, Marzotto M, Dellaglio F, Feligini M (2009) Study of microbial diversity in raw milk and fresh curd used for Fontina cheese production by culture-independent methods. Int J Food Microbiol 130:188–195
Gophna U, Konikoff T, Nielsen HB (2017) Oscillospira and related bacteria–From metagenomic species to metabolic features. Environ Microbiol 19:835–841
Hemme D, Foucaud-Scheunemann C (2004) Leuconostoc, characteristics, use in dairy technology and prospects in functional foods. Int Dairy J 14:467–494
Hols P, Hancy F, Fontaine L, Grossiord B, Prozzi D, Leblond-Bourget N, Decaris B, Bolotin A, Delorme C, Dusko Ehrlich S, Guédon E (2005) New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiol Rev 29:435–463
Jany J-L, Barbier G (2008) Culture-independent methods for identifying microbial communities in cheese. Food Microbiol 25:839–848
Jensen RG, Hagerty MM, McMahon KE (1978) Lipids of human milk and infant formulas: a review. Am J Clin Nutr 31:990–1016
Kabak B, Dobson AD (2011) An introduction to the traditional fermented foods and beverages of Turkey. Crit Rev Food Sci Nutr 51:248–260
Kabuki T, Uenishi H, Seto Y, Yoshioka T, Nakajima H (2009) A unique lantibiotic, thermophilin 1277, containing a disulfide bridge and two thioether bridges. J Appl Microbiol 106:853–862
Li N, Wang Y, You C, Ren J, Chen W, Zheng H, Liu Z (2018) Variation in raw milk microbiota throughout 12 months and the impact of weather conditions. Sci Rep 8:2371
Liu W, Zheng Y, Kwok LY, Sun Z, Zhang J, Guo Z, Hou Q, Menhe B, Zhang H (2015) High-throughput sequencing for the detection of the bacterial and fungal diversity in Mongolian naturally fermented cow’s milk in Russia. BMC Microbiol 15:45
Mann GV, Spoerry A (1974) Studies of a surfactant and cholesteremia in the Maasai. Am J Clin Nutr 27:464–469
Maragkoudakis PA, Zoumpopoulou G, Miaris C, Kalantzopoulos G, Pot B, Tsakalidou E (2006) Probiotic potential of Lactobacillus strains isolated from dairy products. Int Dairy J 16:189–199
McGorrin RJ (2001) Advances in dairy flavor chemistry. RSC Adv 274:67–84
Motato KE, Milani C, Ventura M, Valencia FE, Ruas-Madiedo P, Delgado S (2017) Bacterial diversity of the Colombian fermented milk “Suero Costeño” assessed by culturing and high-throughput sequencing and DGGE analysis of 16S rRNA gene amplicons. Food Microbiol 68:129–136
Naer S, Kitamoto Y (1995) Microbial flora of “Edosensuu”, a traditional fermented milk of Inner-Mongolia in China. Anim Sci Technol 66:555–563 (in Japanese with English abstract)
Nobuta Y, Inoue T, Suzuki S, Arakawa C, Yakabe T, Ogawa M, Yajima N (2009) The efficacy and the safety of Lactobacillus brevis KB290 as a human probiotics. Int J Probiotics Prebiotics 4:263–270
Parente E, Cogan T (2004) Starter cultures: general aspects. In: Fox PF, McSweeney PLH, Cogan TM, Guinee TP (eds) Cheese: chemistry, physics and ***microbiology. Elsevier Academic Press, London, pp 123–148
Pigeon R, Cuesta E, Gilliland S (2002) Binding of free bile acids by cells of yogurt starter culture bacteria. J Dairy Sci 85:2705–2710
Quigley L, O'sullivan O, Stanton C, Beresford TP, Ross RP, Fitzgerald GF, Cotter PD (2013) The complex microbiota of raw milk. FEMS Microbiol Rev 37:664–698
Quigley L, McCarthy R, O'Sullivan O, Beresford TP, Fitzgerald GF, Ross RP, Stanton C, Cotter PD (2013) The microbial content of raw and pasteurized cow milk as determined by molecular approaches. J Dairy Sci 96:4928–4937
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold spring Harbor Laboratory Press, Plainview
Serafeimidou A, Zlatanos S, Kritikos G, Tourianis A (2013) Change of fatty acid profile, including conjugated linoleic acid (CLA) content, during refrigerated storage of yogurt made of cow and sheep milk. J Food Compos Anal 31:24–30
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504
Smit G, Smit BA, Engels WJ (2005) Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol Rev 29:591–610
Tamang JP, Dewan S, Thapa S, Olasupo NA, Schillinger U, Wijaya A, Holzapfel WH (2000) Identification and enzymatic profiles of the predominant lactic acid bacteria isolated from soft-variety chhurpi, a traditional cheese typical of the Sikkim Himalayas. Food Biotechnol 14:99–112
Tamime AY, Robinson RK (1999) Yoghurt: science and technology. Woodhead Publishing, Cambridge
Vacheyrou M, Normand AC, Guyot P, Cassagne C, Piarroux R, Bouton Y (2011) Cultivable microbial communities in raw cow milk and potential transfers from stables of sixteen French farms. Int J Food Microbiol 146:253–262
Van de Casteele S, Vanheuverzwijn T, Ruyssen T, Van Assche P, Swings J, Huys G (2006) Evaluation of culture media for selective enumeration of probiotic strains of lactobacilli and bifidobacteria in combination with yoghurt or cheese starters. Int Dairy J 16:1470–1476
Verdier-Metz I, Michel V, Delbes C, Montel M-C (2009) Do milking practices influence the bacterial diversity of raw milk? Food Microbiol 26:305–310
Von Mollendorff J, Todorov S, Dicks L (2006) Comparison of bacteriocins produced by lactic-acid bacteria isolated from boza, a cereal-based fermented beverage from the Balkan Peninsula. Curr Microbiol 53:209–216
Watabe J (1998) Comparison of microbiological and chemical characteristics among types of traditionally fermented milk in Inner Mongolia in China and Calpis sour milk (Sannyuu). Milk Sci 47:1–8
Wolfe BE, Button JE, Santarelli M, Dutton RJ (2014) Cheese rind communities provide tractable systems for in situ and in vitro studies of microbial diversity. Cell 158:422–433
Wu F, Guo X, Zhang J, Zhang M, Ou Z, Peng Y (2017) Phascolarctobacteriumáfaecium abundant colonization in human gastrointestinal tract. Exp Ther Med. 14:3122–3126
Xia J, Sinelnikov IV, Han B, Wishart DS (2015) MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic Acids Res 43:W251–W257
Zhang J, Wang X, Huo D, Li W, Hu Q, Xu C, Liu S, Li C (2016) Metagenomic approach reveals microbial diversity and predictive microbial metabolic pathways in Yucha, a traditional Li fermented food. Sci Rep 6:32524
Zhong Z, Hou Q, Kwok L, Yu Z, Zheng Y, Sun Z, Menghe B, Zhang H (2016) Bacterial microbiota compositions of naturally fermented milk are shaped by both geographic origin and sample type. J Dairy Sci 99:7832–7841
Acknowledgements
We thank the Institutional level Biotech Hub (DBT, Govt. of India) and Central instrumentation facility of IASST for providing the facilities. The author T.K.J. (IF150782) is thankful to Department of Science and Technology, Ministry of Science and Technology, Govt. of India, for supporting with DST-INSPIRE fellowship. This work was funded by Unit of Excellence project (BT/550/NE/U-Excel/2014) (DBT, Govt. of India). We thank all the households for providing the dairy samples.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Joishy, T.K., Dehingia, M. & Khan, M.R. Bacterial diversity and metabolite profiles of curd prepared by natural fermentation of raw milk and back sloping of boiled milk. World J Microbiol Biotechnol 35, 102 (2019). https://doi.org/10.1007/s11274-019-2677-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-019-2677-y