Abstract
A two-phase integrated sludge thickening and digestion (TISTD) reactor composed of an inner and an outer reactor was developed. Acidification of natural organic material was the primary process in the outer reactor, whilst methane production was the dominant bioreaction occurring in the inner one. The special structure of TISTD thus enables the effective separation of the acid production phase and methane production phase during sludge processing. Molecular biological technology, including 16S rRNA gene and PCR-TGGE, was utilized to investigate the overall microbial community structure and diversity, as well as the processes of dynamic change. Analysis was also conducted on succinate dehydrogenase and coenzyme F420 change trends at each dosing ratio. The microbial community structure of the system exhibited disorder gradually and led to collapse when the dosing ratio increased above 30 %.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The sludge processing industry has experienced rapid development recently in China, with the growth in sludge processing capabilities and processing rate. The processed sludge in China by 2012 had reached 42.28 × 109 m3, with dewatered sludge annual production volume approximately 22 × 106 tons. Environmental issues caused by sludge from sewage works has been highlighted. It has been revealed that 80 % of the sludge produced by China’s sewage treatment works (STWs) is not appropriately disposed of and this has led to secondary contamination attracting severe environmental health concerns.
China currently has 20,000 small cities and towns, 70 % of which are located in the Western mountainous regions. The production capacity of the STWs in these small cities and towns is low, characterized by tight facility footprint, and a dearth of general operations technology, management, and funding. In addition, as a result of the overly complex processes of traditional sludge processing, and the issue of the high cost of construction and operations, sewage plants in these small cities and towns produce lower sludge processing rates, severely influencing the ecological environment of rural regions in China.
The TISTD reactor was developed to deal with sludge processing by STWs of small cities and towns in the Three Gorges Reservoir Regions on Yangtze River. The sludge is handled by this reactor through the integration of a gravitational sludge thickener, a rapid digester, and a rapid anaerobic reactor IC, UASB, and EGSB, among other technologies. It not only allows for a simplified sludge handling process, but also solves high cost and technical issues involved in sludge processing, thus achieving sludge reduction and stability. Laboratory-scale researches reveal that the TISTD reactor sludge thickening digestion is more effective than regular thickeners and digesters (He et al. 2012). Although the optimum design and operational parameters of the reactors were obtained, the operational microbial mechanisms also needed to be known in order to further optimize reactor design and application.
The TISTD reactor moves the anaerobic digestion process production of acid and methane into separate inner and outer chambers of the reactor, respectively. The objective is to permit optimal development conditions for bacterial cultures that produce acid and methane, respectively, and to allow freedom of activity to each and, thus, improve sludge anaerobic digestion efficiency (Pahl et al. 2008). The bacterial cultures for acid and methane production are hardly separated according to the three-phase theory of anaerobic digestion, and a dynamic equilibrium state can be obtained through mutual constraint, mutual influence and coordinative effects. This is the microbial foundation of the TISTD reactor’s stable operation (Wen-Tso et al. 2002; Ji et al. 2009). From the perspective of microbial ecology, even though the acid-producing stage primarily consists of acid-producing fermentative bacteria, and the methane-producing stage consists of methane-producing bacteria, the overall system nonetheless includes various other community (Yoshida et al. 2009; Song et al. 2010). The reactor’s operational stability and efficiency depends to a large degree on the microbial diversity and activity of dominant types. Different conditions of the reactor’s operational stability and efficiency are necessarily related to system community structure succession (Liu et al. 2010; Shin et al. 2010). Traditional microbial isolation and cultivation techniques can isolate a portion of functional bacterial strains; however, it is unable to fully reflect environmental microbial diversity in terms of abundance and scope. Fortunately, modern molecular biology technology offers new routes through research into uncultivatable microbes (Gomez-Villalba et al. 2006; Ivnitsky et al. 2007). Temperature Gradient Gel Electrophoresis(TGGE) is used to investigate microbial communities through detecting the information of nucleic acid, and can allow the observation of uncultivatable bacteria and their functional genes (Muyzer et al. 1998; Riesner et al. 1989). TGGE is widely utilized in active sludge, biological membranes, biological reactors, and other environmental sample microbe diversity testing and species succession research (Calderón et al. 2012; Molina-Muñoz et al. 2009; Wan et al. 2011).
This research, through the use of molecular biology techniques, including 16S rRNA gene sequencing and PCR-TGGE, investigated complete TISTD reactor cycles from initiation to failure in terms of microbial diversity and successful microbial community structure dynamic processes. The objective of the research is to promote reactor applications, as well as to further optimize reactor system structure and operational parameters, thereby improving the sludge-processing efficiency and the theoretical basis for microbiology.
Materials and methods
TISTD reactor structure and principles
The TISTD reactor is composed of five components: an outer reactor, an inner reactor, a precipitate liquid outlet area, a sludge compression area, and a gas collection chamber (see Fig. 1 for structure and operation principles). The design of the reactor aims to utilize the outer reactor area to conduct sludge thickening and hydrolysis-acidification once thickened. The goal of oxygen exhausting within the sludge is also achieved, together with the hydrolysis-acidification in the outer chamber. The sludge drops from the outer reactor portion through holes and into the inner reactor after hydrolysis-acidification. Anaerobic digestion occurs in the inner reactor, with third-stage separation of gas, water, and mud completed in the third step. Digested sludge is expelled from the reactor’s base sludge-thickening area. The gas is separated by a gas–liquid separator, from whose top portion the gas is exhausted after passing through gas-washing bottles and a wet-type gas flow meter. Based on the principle that sludge acid production rates are higher than methane production rates, the ratio of volume between outer and inner reactors is 1:3.
Inoculation of sludge was brought to the preliminary sedimentation tank, with a sludge inoculation amount of 50 l. On the same day, the remaining 20 l of sludge was added to initiate inoculated sludge domestication. The temperature of the inner reactor was gradually raised to approximately 35° and then maintained at 35° ± 2°. The sludge were taken from the same wastewater treatment plant and the parameters of the entry sludge and the excreted sludge such as pH, VFA, water content were detected everyday during each dosing ratios so the range of values of each parameters were obtained. The sludge was stirred for 30 min each time and 4 times per day at the steady temperature. Stable operating status was achieved at dosing ratios of 20, 30, 40, and 50 %. Stable gas production, expelled sludge water content, and relatively balanced VS/TS are the references of stable operation. One month after each stage was stabilized in effect, the dosing ratio was raised to the next stage conduct culturing. The construction parameters of the reactor are shown in Table 1.
Strain separation and identification
At the same time and under the same conditions, we consulted Hungate et al. (1980) for culture medium preparation. Microbe calculation utilized the MPN method (Lu Fudi et al. 1996). Anaerobic bacterial strain identification used the 16S rDNA series analysis method. The expanded PCR reaction primer employed in 16S rDNA was a general primer. Forward primer 8F: AGAGTTTGATCCTG GCTCAG; and reverse primer 1495R: CTACGGCTACCT TGTTACGA. The PCR method was utilized as the reference described (Molina et al. 2014). PCR products were tested by agarose electrophoresis, and underwent extraction of DNA from agarose gel, PCR amplification, and TGGE identification prior to sequencing.
Sludge total DNA extraction and purification
The phenol–chloroform protocol was employed in sludge total DNA extraction. In a 10-ml sterilized centrifuge tube, 8 ml of sludge liquid mixture was added, centrifuged at 8000 rev min−1 at 4 °C for 10 min, and the supernatant removed. 8 ml of sterile double-distilled water was added into the centrifuge tube, oscillating the suspended sludge, and finally centrifuged for another 10 min with 8000 rev min−1 speed and at 4 °C, removing the supernatant. These steps were repeated 3 times. After the sludge was cleaned, 2 ml DNA extraction buffer liquid was added to the centrifuge tube (STE buffer liquid: 20 ml 5 M NaCl; 10 ml 1 M Tris–HCl pH = 8.0; 2 ml 0.5 M EDTA pH = 8.0), as well as 40 μl of protease (10 mg ml−1). The sludge was uniformly mixed with the solution and, at 37 °C, placed it on a horizontal oscillator to oscillate for 30 min at a speed of 225 rev min−1. Once completed, 2 ml 10 % SDS was added, mixed until uniform, and the mixture placed in a 65 °C bath for 2 h. During this time, it was lightly shaken once every 10 min. Finally 10 min centrifugation of the sample at a speed of 8000 rev min−1 was followed by removal of the supernatant and it was put on ice for future use.
An equal amount of phenol–chloroform was added to the DNA serum (1:1), with light mixing to make the solution become a milky-white color. We centrifuged the milky-white liquid at room temperature at a speed of 8000 rev min−1 for 5 min, transferring the supernatant to another sterile centrifuge tube. This step was repeated 3 times. Finally, an equal amount of chloroform was added to the water, and mixed evenly until it became a milky-opaque color. It was then centrifuged on the at room temperature at a speed of 8000 rev min−1 for 5 min, transferring the supernatant to another sterile centrifugal tube. 40 μl of 2 M ammonium acetate and 4 ml of anhydrous ethanol precipitated DNA were utilized, and allowed to stand for 30 min at −20 °C; and then centrifuged for 20 min at a speed of 10,000 rev·min−1. 2 ml of 70 % ethanol cleaning precipitate was employed, and centrifuged for 10 min at a speed of 10,000 rev min−1. We carefully removed the supernatant, and all of the remaining liquid was allowed to evaporate at room temperature until dry. The DNA precipitate was then dissolved into 100 μl of sterile double-distilled water. The DNA solution was stored at −20 °C.
The rough DNA utilized a DNA-purifying test kit (Shanghai Sangon Biotech) for purification, and DNA samples were stored at −20 °C.
16S rDNA V3 area amplification
The primers which were produced by Invitrogen Biotechnology Co., Ltd (Shanghai) were utilized in this paper. Primer P1 sequence was: 5′-ATTACCGCGGCTGCTGG-3′; and primer P2 sequence was: 5′-CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGGCCTACGGGAGGCAGCAG-3′. The PCR reactor system composition and reactor conditions were as follows: total reaction volume of 25 μl, of which 16 μl of double-distilled water was included, 2.5 μl buffer (including Mg2+), primer was diluted to 10 pmol μl−1, P1 and P2 each of 1 μl, DNA template 2 μl, and 2U Taq polymerase (Dingguo Biotechnology Co., Ltd, Shanghai). The TE buffer solution (100 mM Tris–HCl (pH = 8.0) and 10 mM EDTA (pH = 8.0) were mixed together and then autoclaved) was chosen to substitute template DNA to act as a blank control. The temperature setting: initial denaturation for 5 min at 94 °C;denaturation for 45 s at 94°; annealing for 1 min at 55°;extension for 45 s at 72°. There were a total of 30 cycles; finally, completing the reaction with extension for 10 min at 72 °C.
PCR product analysis: the 1.5 % agarose gel was used to conduct electrophoresis with PCR product (25 μl). The electrophoresis liquid was TBE buffer liquid(0.M Tris–HCl,0.1 M boric acid, 0.002 M EDTA,pH = 8.0). The voltage was 4 V cm−1, the electrophoresis time was 1 h, and 1 Kb gradient DNA was utilized as standard for molecular weight. The results of electrophoresis were observed in the GEL imaging system.
TGGE analysis
PCR amplification product used 1 % agarose to conduct gel electrophoresis qualitative analysis. A 42–60 °C temperature gradient for the 16S rDNA fragment was used to conduct analysis, with a voltage of 155 V, and a time of 18 h. Silver staining was employed to conduct staining of the TGGE gel, including using 400 ml of stabilizing liquid (30 % EtOH 10 % glacial acid), stabilized for 30 min, 400 mL 30 % ethanol enhancer was utilized for 30 min, and this process was then repeated 2 times. The 400 ml staining liquid (0.1 % AgNO3, 0.1 % formaldehyde) staining was completed for 30 min and developing solution (2.5 % Na2CO3, 0.1 % formaldehyde, 0.02 % Na2CO3) was used to reveal color. Once the required bands appeared, 400 ml 10 % glacial acetic acid was utilized to cease development.
Gel bands and system phylogeny analysis
Return and analysis of cut gel bands was conducted on sludge sample TGGE patterns. The returned cut gel bands were transferred to 1.5 ml centrifugal tubes, and an appropriate amount of TE buffer liquid was added, crushed, and left overnight at 4 °C. PCR amplification was again conducted after the gel DNA precipitate obtained 1 μl as a template. The TGGE technology was used to inspect the purity of the returned band and condition of separation. The above steps were repeated until there were no longer any nonspecific amplifications.
The final PCR product was brought to a company for gene sequencing. Organization of the sequences was completed using DNAMAN software. Non-chimera sequence data from GenBank (http://www.ncbi.nlm.nih.gov/BLAST/) was obtained for homologue-related classic bacterial strain 16S rDNA sequences. Relevant characteristic sequences were chosen from GenBank, with multiple sequence comparisons conducted according to the largest homologue principle together with sequencing results in this paper using Clustal W (www.ebi.ac.uk/clustalw/). Comparison results were transmitted into the MEGA 4 (Molecular Evolutionary Genetics Analysis, MEGA) software package, and a phylogenetic tree was built using the Neighbour-Joining method. The bootstrap appraisal value was set to 1,000.
Enzymatic activity testing
SDH activity testing method
A 50 ml amount of anaerobic sludge was obtained from the reactor and placed into a 250-ml Erlenmeyer flask. Rigorous shaking with glass beads was applied to break apart and shatter the sludge; once centrifugal force was applied, the supernatant was removed. A saline solution (0.9 % concentration NaCl) was then employed to shake and clean, and this was repeated 3 times. Finally, we used the saline solution to dilute the original volume, and agitated until the sludge floated; it was then placed to one side. The standard arc of the SDH was drawn out according to standard methods, with active sludge liquid of 2 ml placed in a capped tube. 2 ml of Tris–HCl buffer liquid, 0.1 M glucose liquid, and 0.5 % TTC (2,3,5-triphenyltetrazolium chloride), respectively, were then added. This was left at 37 ± 1 °C in an incubator to grow for 4 h. Upon extraction, 2 drops of concentrated sulfuric acid were added to stop the reaction, and 5 ml of methylbenzene was added. The mixture was centrifuged at 4,000 rev min−1 for 5 min. The organic solution color layers were compared. Under the above-described conditions, 1 h of produced 1 µg (Triphenylformazan, TF) per unit volume is one unit of enzyme activity.
Coenzyme F420 activity confirmation method
10 ml of reaction liquid was taken and added to the saline solution, and mixed and diluted. The liquor was then centrifuged for 15 min under the conditions of 6,000 rev min−1. The precipitate was extracted and, after using the saline solution for dilution, it was heated at 95 °C for 30 min. Once cooled, ethanol solution was added [V (ethanol):V (substrate) = 2.5:1, and centrifuged at 6,000 rev min−1 for 15 min], and the top layer of serum was collected. 4 M NaOH solution was utilized to adjust the top serum, so that pH = 13.5. After 15 min centrifugation at 8,000 rev min−1, the supernatant was extracted; this was then separated evenly into two halves. 6 M HCl was utilized to maintain a pH of just below 3 for one half, and HCl usage amount was recorded. Distilled water in a similar volume to that of the HCl was added to the second half to be used as a reference sample. The two above-mentioned solutions underwent colorimetric estimation at wavelength 420 nm, with coenzyme F420 activity calculated. The coenzyme F420 calculation formula is C = Af/sL. Where, C is the coenzyme F420 concentration, μmol l−1; A represents the value which use pH < 3 sample solution as reference, pH = 13.5 sample solution at 420 nm extinction value, g/(cm-mol); f is the dilution factor; L is the cuvette thickness, cm; and s is the millimolar extinction coefficient of F420 at pH = 13.6, s = 54.3.
Results and discussion
TISTD reactor operation results
The pH value and VFA are important influencing factors for the process of anaerobic digestion. In addition, VFA was an important intermediate product during the process of anaerobic digestion. Based on the three-phase theory of methane fermentation, 72 % of methane came from acetic acid splitting, and only 28 % came from H2 oxidation and CO2 restoration (Wen-Tso et al. 2002; Ivnitsky et al. 2007). The majority of the acetic acid, moreover, was produced from acetogenic bacteria oxidizing propionic acid, butyric acid, and valeric acid, among other VFA. The anaerobic digestion reactor in good operation had a VFA concentration <500 mg/l, with the ideal being 300 mg/l (Medina-Pons et al. 2008).
In order to observe the reactor’s ideal processing capabilities, sludge dosing ratios of 20, 30, 40, and 50 % were selected for operating loads, with sludge and water added and removed in batches. The results of the experiments are displayed in Table 2. The reactor’s sludge entry VS/TS was between 0.53 and 0.77, with expelled sludge VS/TS between 0.21 and 0.28 when the dosing ratio is ≤30 %. Gas production was between 18.72 and 48.38 l day−1. This demonstrates that sludge organic matter in the reactor received relatively effective anaerobic digestion. The organic matter decomposition ratio reached between 60 and 65 %. When the dosing ratio was 40 %, the reactor’s sludge entry VS/TS was between 0.58 and 0.78, with the expelled sludge VS/TS between 0.38 and 0.44. Experimental results indicated that the 30 % dosing ratio was the critical value for sludge digestion efficiency and the reactor’s optimal operating load thus was set as 30 %.
30 % dosing rate microbial diversity and distribution characteristics
Operating results revealed that the TISTD reactor was able to achieve sludge thickening and digestion simultaneously within a single reactor at a fair efficacy. From a microbiological perspective, the acid production stage occurs primarily through the existence of acid fermentative bacteria, and the methane stage occurs primarily through methanogenic bacteria, the entire system thus includes various bacterial species. The reactor’s operational stability and efficiency largely depend on the microbial diversity and activity of dominant types. Observation was conducted of the microbial diversity of the reactor’s inner and outer compartments and distributions at a dosing ratio of 30 % in order to observe the microbial conditions under various dosing ratios and to better understand reactor operating performance. An S3400 scanning electron microscope was firstly applied to observe the reactor’s inner and outer reactor microbes. Results are presented in Fig. 2 (a is the inner reactor top section; b is the inner reactor mid-section; and c is the outer reactor).
Some bacteria found in the inner reactor upper portion were similar to methanogenic species. They were numerous (Fig. 2a), including Methanosaeta, Methanobacteriales, and Methanococcus. Of these, those similar in structure to Methanosaeta strains were relatively numerous. Methanosaeta existed in large numbers within the anaerobic digestion reactor, and its growth environment requires a relatively low concentration of methane. The inner reactor mid-portion scanning electron microscopic image is as seen in Image b.
Some strains were similar to Methanosaeta and Methanosarcina were relatively numerous within the inner reactor, with also many cells resembling zoogloea, with relatively large shaped volumes, and dominant colonies were relatively marked. Methanosarcina is a bacterium that converts acetic acid to methane, and its growth environment requires relatively high concentrations of acetic acid. The outer reactor’s lower portion scanning electron microscope image is as seen in Fig. 2c. The outer reactor lower portion has numerous types of bacteria, and the microbes have various types which were similar to cocci, long and short bacilli, filamentous bacteria, and Acetobacter. These bacterial colonies existed in groups, or were distributed sporadically. The outer reactor lower portion thickening sludge content was especially complex. A diversity of bacterial strains is beneficial to the acidic decomposition of the sludge.
Through separated, purified and cultured using the method as showed above, 10 strains of bacteria were selected from the system’s inner and outer reactor sludge by separation and purification. Their respective labels were inner reactor: N1–N10; and outer reactor: W1–W10. Through colony counts, there were 6 strains with higher count: N4, N5, N6, W3, W4, and W9. Results were compiled as shown in Table 3. Of these, N4, W4, and W9 strain cell shapes were as seen in Fig. 3a. From the image, the shape of the separated culture anaerobic strains also showed diversified distribution, and the strain shapes obtained were relatively uniform, with good purification results. This indicates that the anaerobic work station provided good conditions for the anaerobic microbial culturing, and it was possible to conduct anaerobic bacteria calculation analysis. The reactor at its optimal dosing ratio (30 %) stage was able to separate and culture the highest number of dominant strains, with the possibility of relatively numerous non-cultured bacteria strains still existing to maintain the reactor operating functionality. The TISTD reactor microbes that could be cultured were primarily anaerobic and facultative anaerobic functional types, offering primary support to its good processing performance.
Six strains of dominant bacteria were subjected to extraction of DNA from agarose gel, PCR amplification, TGGE identification, and gene sequencing and finally three of these strains’ 163 rDNA sequences (W4, W9, N4) were obtained. Using the MEGA 4 software package, system growth analysis was conducted on the obtained 3 dominant strain sequences to establish a phylogenetic tree (Fig. 3b). The results show that the W4 and W9 strains, and bacillus homologies are relatively good, and are above 98 %. The N4 strain and Methanospirillum homologies were relatively good, at more than 97 %. It was possible to preliminarily identify W4 and W9 as bacillus; N4 was Methanospirillum hungatei. Of these, the W4 and W9 strains were obtained from the outer reactor. Based on the results presented in Table 2, these two strains were the most prevalent two strains in the outer reactor. Their quantities were 1.3 × 105 and 1.2 × 105, respectively. This shows that W4 and W9 were the primary movers in terms of functional strains in the outer reactor. Summarizing Figs. 2 and 3 analyses, it is clear that the outer reactor is the primary location for hydrolysis acidification. In the same manner, the N4 strain was obtained from the inner reactor, proving that the inner reactor facilitates the growth of methanogenic bacteria and was the primary location for production of gas and anaerobic digestion. Therefore, the reactor can culture anaerobic methane-producing bacteria and effectively achieve sludge anaerobic digestion biological stage separation processes.
Reactor microbial community structure succession process
Remarkable drops in operating efficiency were observed when the dosing rate was increased from recommended 30 to 40 % or even 50 %, and this may lead to system failure. In order to explain this, research was conducted on TISTD reactor processes in terms of microbial community structure characteristics and microorganism activity succession rules. TGGE technology has its advantages in researching the inheritance diversity and type difference of the microorganisms. However, this technology is unable to offer information regarding microbiological colony metabolic activity, microorganism quantity, and gene expression (Ginige et al. 2004). Biological and enzymological analysis can provide colony metabolic activity characteristics (Yun et al. 2010). The energy necessary for microorganisms in anaerobic digestion sludge to maintain activity comes from cell component material oxidized decomposition, or endogenous metabolism. Succinate dehydrogenase is a microbial cell internal enzyme catalysing dehydrogenation, and electrons are transmitted to the final electron acceptor oxidoreductase. To a great extent, this reflects active microorganism quantity and its endogenous metabolism, in regards to organic matter. SDH activity is often ascertained through adding artificial hydrogen acceptors, or TTC. Coenzyme F420 itself is a kind of coenzyme that produces methane, and is utilized as a vehicle for low potential electricity; it plays an important role in the formation of methane. Therefore, this paper explains microorganism characteristic change rules in the TISTD reactor at various dosing ratios through results analysis from SDH and coenzyme F420 in summary together with TGGE illustrations.
Under various dosing ratios, the tested reactor inner chamber coenzyme F420 change trends and SDH of the inner and outer reactors change trends are seen in Fig. 4.
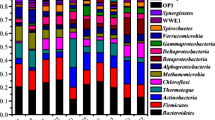
Quantity one software was utilized to conduct band analysis and cluster analysis of inner and outer reactor TGGE illustrations at various dosing ratios; the results are presented in Table 4 and Fig. 5.
From the Table 4 we can see the similarity between every two dosing, for example, the similarity between 20 % dosing and 30 % dosing in the inner reactor was 64.3 %, it got the relative higher similarity than others. When the dosing raised from 30 to 40 %, the similarity was dropped to 36.5 % in inner reactor and 31.0 % when raised from 30 to 50 %. This is probably because when the dosing ratio was raised to 50 % the inner reactor lost its function of anaerobic digestion, as its microbial construction was changed significantly. From the results, it can be predicated that the construction of the microbial population was changed when the dosing ratio was above 30 %.
Results from Fig. 5 show that the system’s microorganism strain similarity was highest under conditions of 20 and 30 % dosing ratio in the inner reactor, whilst the type structure similarity differences were marginal in the outer reactor. At the dosing ratio 10, 20, and 30 %, similarity was high. In the inner reactor, each of the bands 2, 3, 8, 9, 10, 12, 15, 21, 24, 28, and 33 were markedly dominant. Their proportional changes were not significant in relation to reactor load changes. Of these, bands 9, 21, and 33 lacked or had weak strains at dosing ratios of 10 and 50 %, which were likely to be methanogenic bacteria. The outer reactor bands 3, 6, 7, 8, 11, 15, 16, 17, 26, 34, and 35 were dominant. Of these, bands 6, 8, and 34 became weak or even lost at dosing ratios of 10 and 50 %, and these were likely characteristic spectral band strains maintaining outer reactor hydrolysis acidification, key strains that support reactor sludge digestion stabile operation. In addition, the outer reactor’s 2, 7, and 34 bands did not appear in the inner reactor, and became weak at 50 % dosing rate, which showed that this strain can only have an effect in the hydrolysis acidification stage, and its ability to resist load pressure is not strong. The inner reactor 26, and strains between the outer reactor 8 and 10, 23 and 25, and 28–29 all had system failure periods. For this period’s special strains, they were likely primarily dominant bacteria that led to the system failure. Inner reactor band 9 strain basically vanished during the system failure period. This also shows that this bacterium cannot suit itself to growth conditions during the failure period; rather, it is a supporting strain for normal operations.
According to Table 1, the reactor’s processing efficiency was further improved and reached an optimal processing performance, when the dosing ratio was raised from 20 to 30 %. Also from Table 3 and Fig. 5, the inner reactor strain quantity basically remained unchanged, with strain structure similarity relatively high at 64.3 %. The outer reactor strain quantity also remained basically unchanged, with strain structure similarity of 60.9 %, slightly lower than the inner reactor. This shows that the overall reactor system microorganism strain structure and quantities did not change significantly and were relatively stable at this stage with the increase of dosing ratio. This shows the efficacy of the reactor in dealing with load pressure. However, it is difficult to completely understand the reactor’s marked improvements in performance at this stage by simply looking at microbial community structure information. It is also necessary to analyse a summation of enzyme activity changes. According to the results from Fig. 4, the SDH activity and F420 were both increasing with the increasing of dosing ratio during this period, showing that the reactor’s system microorganism strain metabolic activity was rising and that the reactor’s processing performance was rising accordingly.
When the dosing ratio was raised from 30 to 40 %, the inner reactor band quantity was reduced from 48 to 35, strain structure similarity dropped to 36.5 % and, observed from Table 3, the inner reactor SDH and coenzyme F420 contents both dropped. Of these, SDH dropped from 80.1 to 79.3 µg l−1 h−1, while coenzyme F420 dropped from 0.184 to 0.171 µmol g−1. The outer reactor band quantity rose from 36 to 43, and the SDH rose from 97.7 to 101.3 µg l−1 h−1. The above results reveal that as the dosing ratio is raised, bacteria involved in hydrolysis acidification in the outer reactor increase, leading to an increase in organic acid entering the inner reactor. This agrees with the VFA determinations in Table 2. With the excessive organic acid entering the inner reactor, a portion of Methanosaeta is subjected to suppression, which influences the final digestion rate.
When the dosing ratio was raised from 40 to 50 %, the outer reactor band quantity dropped from 43 to 42, basically remaining unchanged. SDH also dropped slightly, to 95.5 µg l−1 h−1, which reveals that an increase in dosing ratio did not result in changes to outer reactor dominant strain quantity changes, and that strain structure was relatively stable. The inner reactor band quantity rose steeply from 35 to 44, but SDH and coenzyme F420 contents both dropped. Of these, coenzyme F420 content lowered from 0.171 to 0.129 µmol g−1, marking a drop of 24.6 %. Based on reactor-processing performance, it is possible to determine that when the dosing ratio reaches 50 %, a portion of sludge directly enters the inner reactor from the outer reactor, with a large buildup of volatile organic acid. Methane-producing bacteria activity is arrested; whereas, at the same time, entry sludge microorganisms also form a certain amount of competitive restriction on the inner reactor microorganism strains, resulting in a disordered inner reactor microorganism metabolism. The balance of microorganism strain structure is lost, sludge anaerobic digestion trends toward stagnation, and the reactor’s processing performance drops up to and including collapse.
Conclusions
Based on principles of sludge anaerobic digestion, and with the goal of effectively achieving the separation of acid and gas production during anaerobic digestion processes, the TISTD was developed targeting China’s small cities and towns experiencing underdeveloped economies and technical shortcomings. Operational results reveal that this reactor operates at an optimal performance at a 30 % dosing ratio, with expelled sludge average water content reduced to 92.09 %, and with expelled sludge VS/TS lowered from entry sludge 0.68 to 0.24. PCR, TGGE, enzyme activity testing, and other molecular biology technology were utilized for analysis of reactor microorganism physical principles. Results indicate that the reactor can effectively achieve the separation of sludge hydrolysis acidification and methane production. The microorganism strain structure is stable, diverse, based on testing of the conditions inside the reactor’s inner and outer reactors under each dosing ratio. Microorganism and enzyme content testing results support that 30 % is the optimal dosing ratio for TISTD reactor.
References
Calderón K, González-Martínez A, Montero-Puente C, Reboleiro-Rivas P, Poyatos JM, Juárez-Jiménez B, Martínez-Toledo MV, Rodelas B (2012) Bacterial community structure and enzyme activities in a membrane bioreactor (MBR) using pure oxygen as an aeration source. Bioresour Technol 103:87–94
Ginige MP, Hugenholtz P, Daims H (2004) Use of stable-isotope probing, full-cycle rRNA analysis, and fluorescence in situ hybridization-microautoradiography to study a methanol- fed denitrifying microbial community. Appl Environ Microbial 70:588–596
Gomez-Villalba B, Calvo C, Vilchez R (2006) TGGE analysis of the diversity of ammonia oxidizing and denitrifying bacteria in submerged filter biofilms for the treatment of urban wastewater. Appl Microbiol Biotechnol 72:393–400
He Q, Li J, Liu H (2012) Efficiency of a pilot-scale integrated sludge thickening and digestion reactor in treating low-organic excess sludge. Environ Technol 33:1403–1408
Ivnitsky H, Katz I, Minz D, Volvovic G, Shimoni E, Kesselman E, Semiat R, Dosoretz CG (2007) Bacterial community composition and structure of biofilms developing on nanofiltration membranes applied to wastewater treatment. Water Res 41:3924–3935
Ji GD, Sun TH, Ni JR (2009) Anaerobic baffled reactor(ABR) for treating heavy oil produced water with high concentrations of salt and poor nutrient. Bioresour Technol 100:1108–1114
Liu S, Song F, Zhu N (2010) Chemical and microbial changes during autothermal thermophilic aerobic digestion (ATAD) of sewage sludge. Bioresour Technol 101:9438–9444
Medina-Pons FJT, Errados J, Rossello-Mora R (2008) Application of temperature gradient gel electrophoresis technique to monitor changes in the structure of the eukaryotic leaf-epiphytic community of posidonia oceanica. Mar Biol 155:451–460
Molina M, Hunter S, Cyterski M (2014) Factors affecting the presence of human-associated and fecal indicator real-time quantitative PCR genetic markers in urban-impacted recreational beaches. Water Res 64:196–208
Molina-Muñoz M, Poyatos JM, Sánchez-Peinado M (2009) Microbial community structure and dynamics in a pilot-scale submerged membrane bioreactor aerobically treating domestic wastewater under real operation conditions. Sci Total Environ 407:3994–4003
Muyzer G, Smalla K et al (1998) Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Van Leeuwenhoek 73:127–141
Pahl O, Firth A, MacLeod I (2008) Anaerobic co-digestion of mechanically biologically treated municipal waste with primary sewage sludge-A feasibility study. Bioresour Technol 99:3354–3364
Riesner D, Steger G, Zimmat R (1989) Temperature-gradient gel electrophoresis of nucleic acids: analysis of conformational transitions, sequence variations, and protein-nucleic acid interactions. Electrophoresis 10:377–389
Shin SG, Lee S, Lee C (2010) Qualitative and quantitative assessment of microbial community in batch anaerobic digestion of secondary sludge. Bioresour Technol 101:9461–9470
Song M, Shin SG, Hwang S (2010) Methanogenic population dynamics assessed by real time quantitative PCR in sludge granule in upflow anaerobic sludge blanket treating swine wastewater. Bioresour Technol 101:23–28
Wan C-Y, De Wever H, Diels L (2011) Biodiversity and population dynamics of microorganisms in a full-scale membrane bioreactor for municipal wastewater treatment. Water Res 45:1129–1138
Wen-Tso L, On-Chim C, Fang HHP (2002) Microbial community dynamics during start-up of acidogenic anaerobic reactors. Water Res 36:3203–3210
Yoshida H, Tokumoto H, Ishi K (2009) Efficient, high speed methane fermentation for sewage sludge using subcritical water hydrolysis as pretreatment. Bioresour Technol 100:2933–2939
Yun JL, Ma AZ, Li YM, Zhuang GQ, Wang YF, Zhang HX (2010) Diversity of methanotrophs in Zoige wetland soils under both anaerobic and aerobic conditions. J Environ Sci 22:1232–1238
Acknowledgments
The financial support of the Natural Science Foundation, China (NSF 51108480) is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Qiang, H., Xingfu, S., Li, G. et al. Two-phase integrated sludge thickening and digestion (TISTD) reactor microbial diversity and community structure succession rules. World J Microbiol Biotechnol 30, 3137–3147 (2014). https://doi.org/10.1007/s11274-014-1741-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-014-1741-x