Abstract
Papaver bracteatum is an important medicinal plant valued for its high content of thebaine and an alternative to P. somniferum for benzylisoquinoline alkaloid production. Salutaridinol 7-o-acetyltransferase (SalAT) is a key gene in morphinan alkaloids biosynthesis pathway. Over expression of SalAT gene was used for metabolic engineering in P. bracteatum hairy root cultures. Transcript level of the salutaridinol 7-o-acetyltransferase gene in transgenic hairy root lines increased up to 154 and 128 % in comparison with hairy roots without SalAT over expression and wild type roots, respectively. High performance liquid chromatography analysis showed that the transgenic hairy roots relatively improved levels of thebaine (1.28 % dry weight), codeine (0.02 % dry weight) and morphine (0.03 % dry weight) compared to those hairy roots without SalAT over expression. This suggests that P. bracteatum hairy roots expressing the SalAT gene could be potentially used for the production of valuable morphinan alkaloids.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Papaver bracteatum (Iranian poppy) is naturally distributed in Iran and has some important medicinal alkaloids such as thebaine, codeine, morphine, noscapine and papaverine. However, thebaine is the major morphinan alkaloids in this plant (Tisserat and Berhow 2009). Thebaine is used as an intermediate for the semi-synthesis of other pentacyclic morphinan-based drugs such as oxycodone, naltrexone and buprenorphine, codeine is a cough suppressant, morphine is a narcotic drug, noscapine is an antitumor agent, sanguinarine and berberine are an antimicrobial drug and papaverine is used as a vasodilator for the treatment of vasospasms (Pienky et al. 2009).
In an investigation, Leonard et al. (2009) revealed a similar pathway for biosynthesis of thebaine in P. somniferum and P. bracteatum. During the 1970s, the United Nations requested studies into the utilization of Iranian poppy as a possible substitute for P. somniferum (United Nation Report 1976). This replacement would be difficult and uneconomical due to perennial nature of P. bracteatum. This has promoted researches into the production of a morphinan alkaloid using in vitro culture technology (Carolan et al. 2002). Many efforts have been done to optimize plant cell culture for improving the yield of several pharmaceutical alkaloids. However, because of the production of many alkaloids, biosynthetic pathways are tissue specific and dependent on the stage of development, rather than cell lines, therefore, plant tissue cultures are used as production platforms (Leonard et al. 2009).
Hairy root production mediated with Agrobacterium rhizogenes creates a rapid and simple means to integrate and express foreign genes in plant cells, which are capable of synthesizing specific secondary metabolites (Ono and Tain 2011).
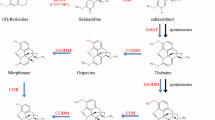
The acetyl coenzyme A-dependent enzyme salutaridinol 7-o-acetyltransferase (SalAT, EC 2.3.1.150) (Fig. 1) converted the salutaridinol to salutaridinol-7-0-acetate (Lenz and Zenk 1994). Grothe et al. (2001) reported molecular characterization of SalAT and indicated that the presence of SalAT transcript predicts the ability to accumulate morphinan alkaloids. Allen et al. (2008) demonstrated both over expression and suppression of SalAT in P. somniferum transgenic plants. Application of new biotechnological methods for improving of morphinane alkaloids compound has been reviewed by Dehghan et al. (2010).
Summary of the morphine biosynthesis pathway showing sites of action of salutaridinol 7-o-acetyltransferase (SalAT) enzyme. Double arrows indicate two or multiple enzyme step (adapted from Allen et al. 2008)
Agrobacterium rhizogenes can carry a binary vector containing an interest gene to be transferred into the plant genome. The bacterial T-DNA from the Ri plasmid and the T-DNA from binary vector integration (co-transformation) into a single explant cell induces a hairy root expressing the integrated gene (Cao et al. 2011). As A. rhizogenes is able to transfer T-DNA of binary vectors, it is enabling to produce transformed root including other foreign genes on a second plasmid. Transgenic hairy roots were produced in Corchorus eapsulqris with gusA reporter gene through A. rhizogenes mediated co-transformation (Chattopadhyay et al. 2011).
The present study focused on over expression of SalAT in transgenic hairy root by A. rhizogenes mediated co-transformation. The rapidly growing transformed hairy root cultures could serve as a simple system to study molecular regulation of SalAT gene encoding salutaridinol 7-o-acetyltransferase enzyme to evaluate its potential use in metabolic engineering of P. bracteatum.
Materials and methods
Plant material and vector
Seeds of P. bracteatum were surface-sterilized with 70 % (v/v) ethanol for 1 min and 2 % (v/v) sodium hypochlorite for 10 min then three times rinsed in sterilized water. The seeds were placed on agar solidified Murashige and Skoog (MS) medium containing 30 g sucrose l adjusted to pH 5.8 before autoclaving at 121 °C for 15 min. Seeds were incubated at 25 °C for 16 h photoperiod with 40 μmol m−2 s−1 until 4 weeks. A. rhizogenes strain LBA9402 harboring the binary vector pBI121SalAT was used for co-cultivation experiments. SalAT cDNA sequence was derived from P. somniferum. The pBI121SalAT vector was introduced into LBA9402 using freeze and thaw method of transformation. The T-DNA of this vector contains a neomycin phosphoteransferase gene and SalAT cDNA with CaMV35S promoter and nos terminator.
Bacterial culture
Agrobacterium rhizogenes strain LBA9402 harboring the binary vector pBI121SalAT was cultured to mid-log phase (A600 = 0.5) at 28 °C on a shaker at 160 rpm on LB medium containing selective antibiotics (50 mg kanamycin l−1 + 25 mg rifampicin l−1). The bacterial suspension was then centrifuged for 10 min of 3000 rpm and suspended at a final cell density of OD600 = 1.0 in liquid inoculation medium [MS salts — (KH2PO4, NH4NO3, KNO3, CaCl2), 50 g sucrose l−1, 100 μM acetocyringone and 10 mM MES] (Azadi et al. 2010; Sharafi et al. 2013b).
Transformation and production of transgenic hairy roots
Excised shoots (i.e. with the roots removed) from 3 weeks P. bracteatum seedlings were used as explants material for co-cultivation with A. rhizogenes. The excised shoots were randomly wounded using a sterile needle, dipped in A. rhizogenes inoculation medium for 10 min, blotted dry on sterile filter paper, and were cultured for 3 days on the co-cultivation medium, which was the same as inoculation medium but solidified with 7 g 1−1 agar at 25 °C in dark. After 2 days of co-cultivation, the explants were transferred to selection medium (hormone free MS medium containing 20 mg l−1 kanamaycin and 400 mg l−1 cefotaxime). Within 3 weeks numerous hairy root emerged from the wound sites. Two weeks later the hairy roots were excised on selection medium at 25 °C in the dark. Fast growing hairy root cultures were obtained after repeated transfer to fresh selection medium. For obtaining more hairy roots, about 1 g of hairy roots were separated from selection media and transferred to 50 ml MS liquid medium in 125 ml flasks. Also, wild type root culture was produced by inoculating liquid MS medium with excised roots from P. bracteatum seedling growth in vitro.
PCR analysis of transformation
For polymerase chain reaction (PCR) and southern blot analysis total DNA was extracted from roots (0.5 g) of the control and putative transgenic hairy root. Plant genomic DNA extraction kit iNtRON Biotechnology Co. was used according to the manufacturer’s instruction. Finally, the pellet of DNA resuspended in 50 μl of TE buffer. PCR was performed using rol B gene specific primer Forward rol B primer: 5′-gctcttgcagtgctagattt-3′ and Reverse rol B primer: 5′-gaaggtgcaagctacctctc-3′ (Sharafi et al. 2013b) for approving integration of T-DNA from Ri plasmid. Also, forward primer from the CaMV35S and reverse primer from the SalAT for approving the integration of T-DNA from binary vector as following set: 5′-agatacagtctcagaagacca-3′ (forward CaMV35S primer) and 5′-gagagctctcaaatctcaaggatttca-3′ (reverse SalAT primer) (Hosseini Khalifani 2007) were used.
Southern hybridization
For southern hybridization 25 μg of genomic DNA from putative hairy root lines was digested over night with Hind III, separated by electrophoresis on 0.8 % agarose gel and then blotted on to positively charged nylon membrane using capillary blotting. The following set of primers 5′-agatacagtctcagaagacca-3′ (CaMV35S forward) and 5′-gatagtgggattgtgcgtca-3′ (CaMV35S reverse) were used for probe synthesis. It was generated from plasmid DNA of pBI121SalAT by labeling with digoxigenin (DIG) using the PCR DIG probe synthesis kit (Roche). Hybridization (between digested DNA and labeled probe) washing and detection was done according to the manufactures instructions of DIG labeling and detection system.
Semi-quantitative reverse transcriptase (RT) PCR analysis
RNA was extracted from the hairy root without T-DNA of binary vector as control and two hairy root lines including the SalAT gene using Qiagen RNeasy plant mini kit. cDNA were synthesized from 1 μg of RNA from each samples using iNtRON Biotechnology Co. cDNA synthesis kit with oligo dT primers. Semi quantitative duplex reverse transcriptase PCR was performed by primers for amplification of SalAT transcript and primers for actin transcript. The PCR reaction was conducted as described above. The cycling conditions were as follows: 5 min at 95 °C, 1 min at 60 °C, 1 min at 72 °C and at the end of cycling, 10 min at 72 °C. The forward and reverse sequences utilized for the amplification of actin were: 5′-ggagaagatttggcatcacactttctacaatgag-3′ and 5′-cttcctgatatccacaatcacacttcatgatgg-3′. The forward and reverse sequences utilized for the amplification of SalAT were: 5′-cttcctgatatccacaatcacacttcatgatgg-3′ and 5′-gagagctctcaaatcaattcaaggatttca-3′. Levels of SalAT and actin transcript were determined by TotalLab software (v1.10).
HPLC analysis of morphinan alkaloids
Various plant tissues; wild type root (3 years old), hairy root and transgenic hairy roots line 1 (SalAT1) were ground and analyzed for thebaine, codeine and morphine contents. 1 g of each air-dried ground hairy root was extracted and analyzed as described by Park and Facchini (2000).
Results
Establishment of transgenic hairy root cultures
Excised shoots as explants showed induction of hairy root from a wound site after 3 weeks, while uninfected explants (control) had no root induction. Hairy roots were found to have a high rate of lateral branching (Fig. 2). The hairy roots were excised from explants and sub cultured on fresh selective medium.
Confirmation of genetically co-transformed hairy roots
The PCR was performed to determine the presence of T-DNA genes transferred from Ri plasmid (rolB) and the binary vector (SalAT). Among the 23 putative transgenic hairy root lines that were obtained, all of them were PCR analyzed and led to amplification of the rolB fragment (Fig. 3a). PCR using specific primers for amplification of A. rhizogenes VirD2 gene carried out in all hairy root genomic DNA to confirm hairy roots were not contaminated by A. rhizogenes, and amplification of virD2 was not observed (data not shown).
Molecular analysis in transgenic hairy roots of Papaver bracteatum. a PCR analysis for detection of the rolB gene. Lane M molecular size marker (100 bp ladder Fermentas), lane C an untransformed root, lane P positive control (pRiLBA9402), lanes 1–23 hairy roots. b PCR analysis for detection of the SalAT gene in transgenic hairy roots of Papaver bracteatum. Lane M molecular size marker (1 kb ladder iNtRON Biotechnology Co), lane C an untransformed root, lane P positive control (pBI121SalAT), lanes 1–23 transformed hairy roots
In four lines the integration of the SalAT gene into the hairy root genome was determined by PCR performed using forward primers specific for CaMV35S promoter and reverse primers specific for SalAT gene (Fig. 3b). This result demonstrates that co-transformation of T-DNA from both plasmids (Ri and pBI121SalAT) occurred in 4 putative hairy roots lines.
Southern blotting was performed with CaMV35S promoter probe and showed hybridization in two of the obtained transgenic hairy root lines (Fig. 4) while no hybridization was observed in the wild type sample.
Semi-quantitative reverse transcriptase PCR
Expression of SalAT gene was found at higher level compared to the both wild type roots (excised in flowering stage) and hairy root (induced by A. rhizogenes LBA9402) without SalAT gene over expression. Multiplex RT-PCR was performed by two pair primers (SalAT and actin primers) (Fig. 5).
Semi-quantitative reverse transcriptase PCR. Normal root (1); hairy root without SalAT gene over expression (2), SalATHR1&2 (3, 4), positive control of SalAT (pBI121 SalAT) (5), actin amplification (6) control negative (water) (c) and iNtRON Bio Co. 1 kb ladder (M). Actin gene was used as an internal control for RNA input
Results of analysis by TotalLab software (v1.10) revealed an increase from 131 to 154 % in terms of level of SalAT gene transcription for transgenic hairy root lines (co-transformed hairy root lines) in comparison with the hairy roots without SalAT over expression. Also, 107–128 % increase in level of SalAT gene transcription was recorded in transgenic hairy root lines compared to the wild type roots (Fig. 6).
Transcription level of SalAT gene to actin in two transgenic lines (SalAT1, 2), hairy root (HR), wild type roots (WTR) of Papaver bracteatum analyzed by TotalLab software. Expression of SalAT message in different samples was quantified relative to actin. Error bars represents standard error of the mean
HPLC analysis
Hairy roots without over expression of SalAT showed similar thebaine, codeine and morphine contents in comparison with wild type roots. Interestingly, in transgenic hairy root lines there was a higher level of thebaine and codeine (130 and 220 % increase respectively) and a novel peak of morphine was detected by HPLC (data not shown). In this study thebaine content showed a 30 % increase in transgenic hairy root line 1 (HRSalAT1) compared to wild type hairy roots. However, codeine and morphine contents were 0.02 and 0.03 DW in the transgenic HRSalAT1 line respectively. SalAT gene over expression was made and it obtained a transgenic hairy root line with more thebaine and morphine (Fig. 7).
Discussion
Agrobacterium rhizogenes mediated transformation is a rapid and convenient transformation system for functional characterization and manipulation of target genes to study plant secondary metabolism. Transgenic hairy root not only used as an efficient expression system in functional genomics but also as a source for developing transgenic plants (via direct or indirect plant regeneration) (Chattopadhyay et al. 2011). A. rhizogenes produce GALLS protein which is substitute for virE2 function in A. tumefaciens. It is an ATP-dependent strand transferase contained nuclear localization sequences to introduce T-DNA into the host plants nucleus (Geng et al. 2012). Therefore, in the present study A. rhizogenes was chosen as a tool to produce transgenic P. bracteatum over expressing SalAT gene.
Because of cytotoxicity of benzylisoquinoline alkaloid, accumulation of these compound is limited to specific cell types called laticifers which is adjacent to sieve elements. Therefore only differentiated organs such as roots can produced morphinan alkaloids (Le Flem-Bonhomme et al. 2004; Liscombe and Facchini 2008). Hairy roots originated from a single cell infected by A. rhizogenes are usually considered genetically stable in contrast with callus lines and, production of alkaloids is not repressed during the growth phase of the culture (Zhang et al. 2009). Induction of hairy root from excised shoot explants was observed after 3 weeks of co-cultivation in P. bracteatum. PCR analyzed were carried out for 5 putative transgenic hairy root lines and led to amplification of the rolB fragment. As it is possible that the Agrobacterium may still exist on explants or hairy roots even though it may be not alive after transfer to plates containing antibiotics (Ono et al. 2012), PCR was performed using specific primers for amplification of A. rhizogenes VirD2 gene in all hairy root genomic DNA to confirm absence of A. rhizogenes contamination in hairy roots. Integration of SalAT gene was determined by PCR and southern blot analysis demonstrating co-transformation of T-DNA from both plasmids (Ri and pBI121SalAT). Several studies indicated co-transformation of the foreign gene from the binary vector and T-DNA from Ri plasmid into the plant genome (Chattopadhyay et al. 2011).
Suppression of the gene encoding the morphinan pathway enzyme salutaridinol 7-o-acetyltransferase (SalAT) in opium poppy showed an average reduction of about 12 % of the control in SalAT transcript. The result revealed from RNA transcript analysis of 16 primary T0 transformants and their segregating T1 progeny (Allen et al. 2008). In our study, an increase in level of SalAT gene transcription for transgenic hairy root lines (co-transformed hairy root lines) in comparison with the hairy roots without SalAT over expression and wild type roots were observed.
Over expression of berberine bridge enzyme resulted in a fivefold to sixfold increase in total benzophenanthridines including a 33 fold increase in dihydrochelilutin in Eschscholiza californica hairy root culture. However, an up to tenfold decrease in total benzophenanthridine alkaloids occurred from down regulation of transgenic cell culture of E. californica with anti-sense berberine bridge enzyme (Park et al. 2002), confirming the role of enzymatic control of alkaloid contents.
Larkin et al. (2007) reported increased production of morphinan alkaloids by over-expression of another gene (codeinone reductase) involved in morphinan alkaloids pathway in transgenic P. somniferum plant. They reported an approximately tenfold greater level of CodR transcript in transgenic leaves compared to non-transgenic plants. Conversely, suppression of CodR with RNAi technology in opium poppy resulted in accumulation of (S)-reticuline (which is a precursor) in transgenic silenced plants at the expense of morphine, thebaine and codeine (Allen et al. 2004). In this study an increase rate (142 %) of SalAT gene transcription was observed in comparison with the control hairy roots.
In our previous study, amounts of codeine and morphine in transgenic hairy root lines of P. bracteatum increased by 160 and 86 % respectively, by over expression of codeinone reductase gene. This resulted in increasing codeine of up to 0.04 % DW and morphine of up to 0.28 % DW (Sharafi et al. 2013a).
Both over-expression and suppression of the gene encoding the morphinan pathway enzyme salutaridinol 7-o-acetyltransferase (SalAT) was reported by Allen et al. (2008) in transgenic opium poppy plants. Transgenic SalAT over expressing line with the highest alkaloid content showed on average a 40 % greater total alkaloids content compared to the control in three independent trials over 3 years. Novel accumulation of the alkaloid salutaridine at up to 23 % of total alkaloid was detected by DNA-encoded hairpin RNA-mediated suppression of SalAT, which was not detectable in the parental genotype. Salutaridine is not a substrate of SalAT but the substrate of the precursor enzyme in the pathway, salutaridine reductase. Detection of salotaridine, but not salutaridinol revealed the existence of a metabolic complex involving salutaridinol reductase (SalR) and SalAT in Papaver (Kempe et al. 2009). Correlation in transcript levels of SalR, SalAT and contents of salotaridine and thebaine supported the co-regulation of genes encoding enzymes operating in this part of the pathway (Desgagne-Penix et al. 2012). These results are in agreement with the cooperative analysis of ESTs from Papaver species with different alkaloid profiles (Ziegler et al. 2006).
Our results confirmed that over expression SalAT gene using A. rhizogenes mediated transformation could provide a fast, simple and reliable model system to investigate the molecular regulation of morphinan alkaloids biosynthesis in P. bracteatum. Moreover, similar strategy could be used to consider other candidate genes involving in benzylisoquinoline alkaloid pathway in Papaver species.
References
Allen RS, Millgate AG, Chitty JA, Thisleton J, Miller JA, Fist AJ, Gerlach WL, Larkin PJ (2004) RNAi-mediated replacement of morphine with the nonnarcotic alkaloid reticuline in opium poppy. Nat Biotechnol 22:1559–1566
Allen RS, Miller JAC, Chitty JA, Fist A, Gerlach WL, Larkin PJ (2008) Metabolite engineering of morphinan alkaloids by over expression and RNAi suppression of Salutaridinol 7-O-acetyltransferase in opium poppy. Plant Biotechnol J 6:22–30
Azadi P, Chin DP, Kuroda K, Khan RS, Mii M (2010) Macro elements in inoculation and co-cultivation medium strongly affect the efficiency of Agrobacterium-mediated transformation in Lilium. Plant Cell Tiss Organ Cult 101:201–209
Cao D, Hou W, Liu W, Yao W, Wu C, Liu X, Han T (2011) Overexpression of TaNHX2 enhances salt tolerance of composite and whole transgenic soybean plants. Plant Cell Tiss Organ Cult 107:541–552
Carolan JC, Hook ILI, Walsh JJ, Hodkinson TR (2002) Using AFLP markers for species differentiation and assessment of genetic variability of in vitro cultured Papaver bracteatum (section oxytona). In Vitro Cell Dev Biol Plant 39:300–307
Chattopadhyay T, Roy S, Mitra A, Maiti MK (2011) Development of a transgenic hairy root system in jute (Corchorus capsularis L.) with gusA reporter gene through Agrobacterium rhizogenes mediated co-transformation. Plant Cell Rep 30:485–493
Dehghan E, Hosseini B, Naghdi Badi H, Shariari Ahmadi F (2010) Application of conventional and new biotechnological approaches for improving of morphinane alkaloids production. J Med Plants 9:33–50
Desgagne-Penix I, Farrow SC, Cram D, Nowak J, Facchini PJ (2012) Integration of deep transcript and targeted metabolite profiles for eight cultivars of opium poppy. Plant Mol Biol 79:295–313
Geng L, Niu L, Gresshoff PM, Shu C, Song F, Huang D, Zhang J (2012) Efficient production of Agrobacterium rhizogenes-transformed roots and composite plants in peanut (Arachis hypogaea L.). Plant Cell Tiss Organ Cult 109:491–500
Grothe T, Lenz R, Kutchan TM (2001) Molecular characterization of the salutaridinol 7-O-acetyltransferase involved in morphine biosynthesis in opium poppy (Papaver somniferum). J Biol Chem 276:30717–30723
Hosseini Khalifani B (2007) Genetic manipulation of Papaver somniferum for increasing morphinan alkaloids. PhD Thesis, Ferdowsi University of Mashhad, Iran
Kempe K, Higashi Y, Frick S, Sabarna K, Kutchan TM (2009) RNAi suppression of the morphinan biosynthetic gene SalAT and evidence of association of pathway enzymes. Phytochemistry 70:579–589
Larkin PJ, Miller JA, Allen RS, Chitty JA, Gerlach WL, Frick S, Kutchan TM, Fist AJ (2007) Increasing morphinan alkaloid production by over-expressing codeinone reductase in transgenic Papaver somniferum. Plant Biotechnol J 5:26–37
Le Flem-Bonhomme V, Laurain-Matter D, Fliniavx MA (2004) Hairy root induction of Papaver somniferum var. album, a difficult-to-transform plant, by A. rhizogenes LBA 9402. Planta 218:890–893
Lenz R, Zenk MH (1994) Closure of the oxide bridge in morphine biosynthesis. Tetrahedron Lett 35:3897–3900
Leonard E, Runguphan W, Connor SO, Prather KJ (2009) Opportunity in metabolic engineering to facilitate scalable alkaloid production. Nat Chem Biol 5:292–300
Liscombe DK, Facchini PJ (2008) Evolutionary and cellular webs in benzylisoquinoline alkaloid biosynthesis. Curr Opin Biotech 19:173–180
Ono NN, Tain L (2011) The multiplicity of hairy root cultures: prolific possibilities. Plant Sci 180:439–446
Ono NN, Bandaranayake PCG, Tain L (2012) Establishment of pomegranate (Punica granatum) hairy root cultures for genetic integration of hydrolysable tannin biosynthetic pathway. Planta 236:931–941
Park SU, Facchini PJ (2000) Agrobacterium rhizogenes-mediated transformation of opium poppy, Papaver somniferum L., and California poppy, Eschscholzia californica cham., root cultures. J Exp Bot 51:1005–1016
Park SU, Yu M, Facchini PJ (2002) Antisense RNA-mediated suppression of benzophenanthridine alkaloid biosynthesis in transgenic cell cultures of California poppy. Plant Physiol 128:696–706
Pienky S, Brandt W, Schmidt J, Karmell R, Zigler J (2009) Functional characterization of involvement in papaverine biosynthesis in opium poppy (Papaver somniferum). Plant J 60:56–67
Sharafi A, Hashemi Sohi H, Mousavi A, Azadi P, Hosseini Khalifani B, Razavi K (2013a) Metabolic engineering of morphinan alkaloids by over expression of codeinone reductase in transgenic hairy root of Papaver bracteatum. Biotechnol Lett 35:445–453
Sharafi A, Hashemi Sohi H, Mousavi A, Azadi P, Razavi K, Ntui VO (2013b) A reliable and efficient protocol for inducing hairy roots in Papaver bracteatum. Plant Cell Tiss Organ Cult 113:1–9
Tisserat B, Berhow M (2009) Production of pharmaceutical from Papaver cultivars. In vitro Eng Life Sci 3:190–196
United Nations Document. Scientific Research on Papaver bracteatum. Report of the Fourth Working Group. ST/SOA/SER.J/23; 1976:6
Zhang HC, Liu JM, Lu HY, Gao SL (2009) Enhanced flavonoid production in hairy root cultures of Glycyrrhiza uralensis Fisch by combining the over-expression of chalcone isomerase gene with the elicitation treatment. Plant Cell Rep 28:1205–1213
Ziegler J, Voigtländer S, Schmidt J, Kramell R, Miersch O, Ammer C, Gesell A, Kutchan TM (2006) Comparative transcript and alkaloid profiling in Papaver species identifies a short chain dehydrogenase/reductase involved in morphine biosynthesis. Plant J 48:177–192
Acknowledgments
The authors thank to Dr. Houshang Ghasemi and Mr. Saeed Dini (TEMAD Co.) for providing HPLC services.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sharafi, A., Hashemi Sohi, H., Mousavi, A. et al. Enhanced morphinan alkaloid production in hairy root cultures of Papaver bracteatum by over-expression of salutaridinol 7-o-acetyltransferase gene via Agrobacterium rhizogenes mediated transformation. World J Microbiol Biotechnol 29, 2125–2131 (2013). https://doi.org/10.1007/s11274-013-1377-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-013-1377-2