Abstract
This study reports the isolation of 63 endophytic fungal isolates from two traditional medicinal plants, Ocimum sanctum and Sapindus detergens from different locations of Amritsar, India. The functional characterization of the fungi for their ability to produce anti bacterial and anti cancer agent was carried out. Sixteen strains were characterized at molecular level by sequencing the amplified ITSI-5.8-ITSII region of rDNA. The phylogenetic tree resolved the endophytic fungi into different clades. The fungal endophytes belonging to order Pleosporales (Alternaria sp., Phoma sojicola and Exserohilum sp.) were functionally versatile as they produced diverse biomolecules including antibacterial agent active against Mycobacterium smegmatis, as well as cytotoxic activity against different human cancer cell lines of lung, ovary, breast, prostrate, neuroblastoma and colon.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The increasing emergence of resistant pathogenic strains to the existing drugs and new diseases has necessitated the need for searching novel molecules with better anticancer or antimicrobial properties than the existing ones. Fungi have always been a source of pharmaceutically important compounds. Although soil fungi have provided a broad spectrum of secondary metabolites with diverse chemical structure, the most recent exciting discoveries have come from exploration of fungi living in unusual ecological niches such as endophytic fungi (Strobel 2000; Schulz et al. 2002; Tulp and Bohlin 2004). It appears that all higher plants are hosts to one or more endophytic microbes. These fungi reside in the tissues between and among living plant cells. The relationship that they establish with the plant varies from symbiotic to pathogenic (Schulz and Boyle 2005). The importance of endophytes as potential sources of novel bioactive compounds can be assessed from the fact that of the 300,000 plant species that exist on the earth, each individual plant is host to one or more endophytes. Plants produce a diverse range of structurally novel bioactive molecules, making them a rich source of different types of medicines. It is likely that some of the relatively rare bioorganic molecules made by specific higher plants can be produced by certain endophytes as well. This is exemplified with the case of anticancer compound taxol from yews and also taxol being produced by a series of endophytes from yews as well as other plants (Stierle and Strobel 1995; Gangadevi and Muthumary 2007). Besides taxol other compounds having anticancer activity have also been reported to be produced by endophytic fungi (Radu and Kqueen 2002). Antimicrobial activities have been demonstrated in a variety of metabolites biosynthesized by the plant endophytes (Li et al. 2001; Strobel et al. 2001, 2002; Harper et al. 2003). It is evident that fungal endophytes have now been recognized as a repository of secondary metabolites including novel antibiotics, anti cancer and immunosuppressant compounds (Strobel and Daisy 2003). Despite this anticipated diversity, relatively few of these organisms have been characterized. Endophytic fungi are still relatively unstudied and potential sources of novel natural products for exploitation in medicine, agriculture, and industry.
The use of endophytic fungi for biotechnological exploitations has necessitated the isolation, cultivation and identification of these organisms. Extensive research on endophytes of diverse plants has identified distinct communities for each plant species usually with a small number of dominant fungi which may be evident as saprobes following death of the host tissues (Petrini 1986; Huang et al. 2008). Endophytic assemblages are influenced by a number of factors such as geographic location, age and specificity of the colonized tissue (Collado et al. 1999; Ganley and Newcombe 2006; Arnold 2007). Endophytes are often studied at the morphological level, but, many of the endophytes either fail to sporulate or they are rare and difficult to identify. Therefore, molecular analysis based on DNA sequences is recognized as the most reliable method to reveal genetic relationship between the strains which could be unambiguously used to identify and evaluate the isolates at any taxonomic rank (Burns et al. 1990).
In view of their immense potential as sources of natural products including new and innovative biologically active compounds, the present study was taken up to investigate the anticancer and antimicrobial activities of the endophytic fungi isolated from traditional medicinal plants Ocimum sanctum L. (Holy Basil) and Sapindus detergens (Soapnut). O. sanctum has been used for thousands of years in ayurveda (Indian traditional medicine system) for its diverse healing properties in curing common colds, headaches, stomach disorders, inflammation, heart disease, various forms of poisoning, and malaria. The O. sanctum has also been suggested to possess antifertility, anticancer, antidiabetic, antifungal, antimicrobial, hepatoprotective, cardio protective, antispasmodic and analgesic actions (Karthikeyan et al. 1999; Prakash and Gupta 2005; Tiwari et al. 2010). In view of this, O. sanctum was selected for isolation of endophytes and their functional characterization. S. detergens is rich in saponins which are reported to possess biological activities (Park et al. 2005), but there is no report of isolation of endophytic fungi from S. detergens. The present study reports the functional diversity of the endophytes from these plants with an aim to obtain novel pharmaceutically active biomolecules.
Materials and methods
Isolation of endophytic fungi
Different samples (stems, leaves) from healthy traditional medicinal plants (O. sanctum and S. detergens) were collected from Guru Nanak Dev University campus of Amritsar (India), for the isolation of endophytic fungi. The material was thoroughly washed using distilled water followed by treatment with 70% ethanol for 2 min and 5% sodium hypochlorite for 5 min to accomplish surface sterilization. It was again rinsed in sterile distilled water prior to plating. The water obtained after the last wash was plated on PDA to ensure complete surface sterilization. The samples were cut into 5–6 pieces (2–6 mm size) and were placed onto water agar plates (distilled water, 1.5% agar–agar) supplemented with ampicillin (100 μl/ml) and incubated at 30°C for 3–4 days to few weeks till the growth initiated (Hijri et al. 2002). The hyphal tips, that emerged from the plant parts were picked, purified and maintained on PDA plates for further studies.
Molecular characterization of endophytic fungi
Isolation of DNA
For DNA extraction, the fungi were grown on potato dextrose broth (PDB) for 72 h at 30°C under shaking conditions (120 rpm) and the resultant mycelium was harvested by vacuum filtration and stored at −70°C. The chilled mycelia (200 mg) was ground in 550 μl of extraction buffer (50 mmol l−1 Tris HCl, pH 8.0; 700 mmol l−1 NaCl; 10 mmol l−1 EDTA, 1% (v/v) β-mercaptoethanol and 10% (w/v) SDS and then 300 μl of equilibrated phenol and extraction buffer was added. The contents were homogenized and incubated for 15 min at 65°C. The DNA in the aqueous phase was purified with repeated extractions using equal volumes of saturated phenol, chloroform, iso-amyl alcohol (PCI) mixture (25:24:1). The DNA was precipitated with 9 parts of ice cold isopropyl alcohol and 1 part of sodium acetate (3 mol l−1; pH 8.0) and kept at −20°C for 2 h, followed by centrifugation for 15 min at 8,000g. The DNA pellet was rinsed with 70% (v/v) ethanol, air dried, suspended in 50 μl of sterilized double distilled water and stored at 4°C (Sharma et al. 2008).
PCR amplification
DNA coding for internal transcribed spacers (ITS I & ITS II) and the intervening 5.8S rDNA region was amplified using universal primers, ITS1 (5′ TCCGTAGGTGAACCTGCGG 3′) and ITS4 (5′ TCCTCCGCTTATTGATATGC 3′) (White et al. 1990). The PCR amplification was carried out in 0.2 ml PCR tubes, using Master cycler personal (Eppendorf). The PCR reaction mixture (50 μl) contained 25 μl of PCR master mix (Genei, Bangalore, India), 2.5 μl of DMSO, 1 pmol l−1 of each primer and 100 ng of template DNA. Thermal cycling conditions were as follows: initial denaturation (4 min at 95°C), followed by 30 cycles of denaturation (94°C for 50 s), annealing (51°C for 1 min), and primer extension (72°C for 1 min), followed by final extension step for 10 min at 72°C. Amplification products were electrophoretically resolved on 1.4% (w/v) agarose gel containing ethidium bromide, using 1× TAE buffer at 70 V.
Internal transcribed spacer sequence analysis
The purified amplified internal transcribed spacer (ITS) region was sequenced by single primer analysis (SPA) services (Genei, Bangalore, India). The ITS sequences of different fungi were aligned to each other as well as the sequences retrieved from NCBI databases, using multiple sequence alignment software (CLUSTAL X). Dendrogram was generated using neighbour joining (NJ) plot and the boot strapping was carried out using 100 replications. The ITS sequences were deposited with NCBI gene bank (Table 1). The cultures used in the study have been deposited to Microbial Type culture collection (MTCC), Chandigarh and National Bureau of Agriculturally Important Microorganisms (NBAIM), Mau Nath Bhanjan, Uttar Pradesh.
Culture conditions and preparation of extracts
Erlenmeyer flasks (250 ml) containing 50 ml of liquid production medium that contained (per litre): 50 g, lactose; 5.41 g, defatted soybean meal (containing 8% w/w nitrogen content); 0.8 g, KH2PO4; 0.4 g, NaCl; 0.52 g, MgSO4·7H2O; 1 mg, ZnSO4·H2O; 0.04 mg, biotin and 1 ml of trace element solution of following composition (per litre): 100 mg, Na2B4O7·10H2O; 50 mg, MnCl2·4H2O; 50 mg, Na2MoO4·2H2O and 250 mg, CuSO4·5H2O, were inoculated with three pieces (6 mm diameter) of agar plugs taken from the periphery of actively growing cultures of endophytic isolates grown on PDA plates. The flasks were incubated at 30°C and 250 rpm on a rotary shaker for 7 days. Thereafter, 50 ml of ethyl acetate was added to each of the flask and extraction was carried at 45°C for 2 h at 150 rpm. The upper organic phase thus obtained was separated and concentrated on rotary evaporator (BUCHI). The concentrated extracts were then re-suspended in ethyl acetate.
Anti-bacterial activity
The agar plate diffusion assay was used to evaluate the antimicrobial activity. The EtOAc (Ethyl acetate) extracts from isolated endophytic fungi were screened for anti-bacterial activity against Pseudomonas paucimobilis, Escherichia coli, Enterobacter faecalis, Salmonella typhii and Mycobacterium smegmatis. Fifty μl of each extract was added in the wells of nutrient agar plates seeded with 100 μl of activated test cultures (P. paucimobilis, E. coli, E. faecalis, S. typhii and M. smegmatis). The plates were incubated at 30°C for 24–48 h and the clear zones of inhibition around the fungal extracts were measured and compared with negative control.
In vitro cytotoxicity against human cancer cell lines
The human cancer cell lines [lung (A-549 & HOP-62), ovary (IGR-OV-1) breast (MCF-7) prostrate (PC-3 & DU-145) neuroblastoma (IMR-32) and colon (colo-205 & HCT-15)] procured from National Cancer Institute, Frederick, USA was used in the present study (Sharma et al. 2010). Cells were grown in tissue culture flasks in complete growth medium (RPMI-1640 medium with 2 mM glutamine, pH 7.4 supplemented with 10% fetal bovine serum, 100 μg/ml streptomycin and 100 units/ml penicillin) in a CO2 incubator (5% CO2; 90% RH) at 37oC. The cells at sub-confluent stage were harvested from the flask by treatment with trypsin (0.05% w/v in PBS (pH 7.4) containing 0.02% EDTA). Cells with viability of more than 98%, as determined by trypan blue exclusion, were used for determination of cytotoxicity. In vitro cytotoxicity against various human cancer cell lines was determined using 96-well cell culture plates. Cell suspension (100 μl) was added to each well and the cells were grown in CO2 incubator (37oC, 5% CO2, 90% RH) for 24 h. Suitable concentrations of test materials (30/100 μl) were added thereafter to each well containing cell suspension. Cells were also incubated with 10 μM mitomycin-c, 1 μM doxorubicin and 20 and 100 μM 5-FU which were used as positive controls. Suitable control with equivalent concentration of CV1 (normal monkey kidney cell line) was also included. The plates were further incubated for 48 h (37°C in an atmosphere of 5% CO2 and 90% RH). The cell growth was stopped by gently layering trichloroacetic acid (50% TCA, 50 μl) on top of the medium in all the wells. The plates were incubated at 4°C for 1 h to fix the cells attached to the bottom of the wells and the cell growth was measured using sulforhodamine B dye (Skehan et al. 1990; Sharma et al. 2010).
Results
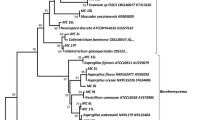
Sixty three endophytic fungal strains were isolated from two traditional medicinal plants O. sanctum and S. detergens. The number of endophytic fungal strains isolated from O. sanctum and S. detergens were 31 and 32, respectively. All the isolates were screened for their antimicrobial potential. Of these sixteen endophytic fungal isolates that showed antimicrobial activity were further taken up for assessment of anticancer activity and molecular characterization. The amplified ITS rDNA region of the selected isolates was sequenced and compared with the ITS sequences of organisms represented in the NCBI database gene bank using BLAST search to generate a phylogenetic tree (Fig. 1). The sequences that showed E = 0.0 and highest % similarity with the amplified sequences were included for alignment and bootstrapping using CLUSTAL X. The generated dendrogram showed that the isolates belonging to diverse fungal group are distributed within Ascomycota and were dominated by three classes (Eurotiomycetes, Sordariomycetes and Dothideomycetes).
One of the major clade placed at the top of the tree was represented by members of Alternaria that belongs to family Pleosporaceae and order Pleosporales (Figs. 1, 2). The isolates Pj-5 (Alternaria sp.), RL-8 (A. mali), RL-14 (A. compacta) and RL-15 (Alternaria sp.) originated from S. detergens whereas isolates Gul-5 (A. tenuissima) and TL-6 (Alternaria sp.) originated from O. sanctum. Isolates RL-2 and RL-3, identified as Phoma sojicola, Exserohilum sp. respectively, on the basis of morphological features (Table 1) and closest match in database were clustered close to Alternaria clade as they also belong to same family Pleosporaceae.
Two closely related isolates (TL-1 and Gul-16) showing 99% sequence similarity with Cladosporium cladosporioides, belonging to the order Capnodiales were nested together. The isolate RL-9 from plant S. detergens showing 96% sequence similarity with Guignardia vaccinii, is a member of family Botrysphaeriales. Isolate RL-17 from S. detergens identified as Pseudocercospora sp. which also belongs to the same order, however, showed only 87% sequence similarity to nearest match in database. The isolate Gul-17 from O. sanctum was identified as Aspergillus fumigatus that showed high sequence similarity (above 95%) to the strains in the database. Whereas, isolates RL-5 and RL-18 showed sequence similarity with genus Colletotrichum sp. belongs to the order hypocreales.
Anti microbial activity of endophytes
The results in Table 2 show that endophytic fungi included in this study exhibited antibacterial activity against variety of test organisms. The isolates RL-5, RL-14 and Pj-5 from S. detergens showed broad spectrum anti-bacterial activity. Isolate RL-5, however, showed no activity against M. smegmatis whereas, isolates RL-14 and Pj-5 showed appreciable activity against M. smegmatis but did not show activity against S. typhii. On the other hand the extracts from RL-13 and RL-3 were highly specific and showed antibacterial activities only against M. smegmatis and P. paucimobilis, respectively. Extracts of the isolates RL-8 and RL-18 exhibited activity against M. smegmatis and also against E. coli and P. paucimobilis, respectively.
Endophytic isolates Gul-17, Gul-5, TL-1 and TL-6 from O. sanctum were also found to possess antibacterial activity. Isolate Gul-17 produced antibacterial agent active against M. smegmatis, E. coli and S. typhii, whereas isolates Gul-5, TL-1 and TL-6 exhibited similar spectrum of anti-bacterial activity, being active against E. faecalis and M. smegmatis. On the other hand extract from isolate Gul-16 inhibited growth of S. typhii only (Fig. 3).
Anticancer activity of endophytes
Out of the sixteen isolates selected on the basis of antibacterial activity, only eight exhibited anti-cancerous activity. These isolates were represented by two orders Pleosporales (RL-2, RL-3, RL-13, RL-14, RL-15 and Pj-5) and Capnodiales (TL-1 and RL-17). All these isolates, except for TL-1were isolated from S. detergens and produced bioactive molecules, which were active against different cancerous cell lines (Table 3). The extract from TL-1 (C. cladosporioides) at a concentration of 30 μg/ml was active (more than 50% growth inhibition) against all the tested cell lines, except for HOP-62 lung cancer cell line. TL-1 showed 74, 72, 67, 64 and 58% inhibition of (A-549) lung, COLO-205 (colon), IMR-32 (neuroblastoma), MCF-7 and DU-145 (prostate) cell lines, respectively. The extract of RL-3 (Exserohilum sp.) showed 93% cytotoxicity against PC-3 (prostrate) followed by 83% against A-549 (lung) and MCF-7 (breast) cancer cell lines, whereas, the extracts from RL-13 (Alternaria sp.) and RL-14 (Alternaria compacta) showed selective inhibition of A-549 lung cancer cell line corresponding to 66 and 56% at (30 μg/ml) concentration (Table 3). On the other hand, isolate RL-15 (Alternaria sp.) produced anti cancer agent active against different cell lines viz. lungs, ovary, breast and prostrate where it exhibited 87, 74 and 69% against A-549 (lung), MCF 7 (breast) and PC-3 (prostrate) cell lines at 30 μg/ml. Similarly, isolate Pj-5 (Alternaria sp.) was active only against two cell lines, breast (85%) and ovary (71%). These extracts however, showed insignificant cytotoxicity against normal monkey kidney cell line (CV1), even when tested at a higher concentration of (100 μg/ml).
Discussion
Endophytic fungi represent an important genetic resource in search for novel biomolecules (Guimaraes et al. 2008). The endophytic fungi isolated from traditional medicinal plants (O. sanctum and S. detergens) from Guru Nanak Dev University Amritsar, a region located in Indo Gangetic plains of North West part of Indian subcontinent were included in this study. The plants included in this study have been used as traditional medicants in curing cough, cold, respiratory disorder, malaria, lack of appetite, constipation, acidity and gum pain (Arulmozhi et al. 2005; Sharma et al. 2010). The plants were found to harbour a variety of endophytic fungi including previously reported strains of various genera i.e. Alternaria, Colletotrichum, Guignardia, Phoma, Aspergillus and Cladosporium (Firakova et al. 2007; Gunatilaka 2006) and few others like Exserohilum sp. being reported for the first time. The endophytes that exhibited characteristic colony morphology and microscopic identification features (hyphal and spore arrangement) were putatively identified at genus level (Larone 2002). The endophytes which could not be clearly identified on morphological basis were subjected to molecular characterization based on sequencing the ITSI-5.8S-ITSII region as well as link these strains to their closely related isolates using NJ plot. The amplified ITSI-5.8S-ITSII region sequences have also been previously used for detecting intra-specific variability between Candida and Penicillium species (Skouboe et al. 1999; Korabecna et al. 2003; Morakotkarn et al. 2007).
A phylogenetic tree was constructed on the basis of these sequences along with those aligned from databases (Fig. 1). The dendrogram showed that the isolates belonging to diverse fungal group are distributed within Ascomycetes. They were dominated by three classes (Eurotiomycetes, Sordariomycetes and Dothideomycetes). The clade represented by class Dothideomycetes included 14 isolates related to order Pleosporales and Capnodiales. Of these Alternaria sp. was the dominating group of fungi represented by A. compacta, A. tenussima and A. mali in addition to few more isolates that were identified up to genus level (Fig. 2). Most of these isolates were very potent and produced antibacterial and anti cancer agents. Differences in the functional characteristics were observed among the isolates from the same species, with respect to their ability to produce metabolites (Pelaez et al. 1998). Even, Alternaria from different parts of the same plant showed variation in their metabolites production capabilities. The observed differences in these strains to produce the metabolites can be ascribed to the high variability among the strains as the branch nodes of the dendrogram were supported with low bootstrap values, indicating horizontal gene transfer (Hampl et al. 2001) and genetic variability as possible mechanisms for adapting to the host system. Our studies suggest that the members of Dothideomycetes (order Pleosporales) showed higher functional versatility. The members of this group produced a wide variety of biomolecules including anti-bacterial agent active against M. smegmatis, in addition to activity against a variety of cancer cell lines. The anti-bacterial activity of endophytic fungi (Pleosporales) against M. smegmatis is being reported for the first time, although, unidentified endophytic fungi with activity against M. tuberculosis have been previously reported by Wiyakrutta et al. (2003). The other members of this clade i.e., P. sojicola and Exserohilum sp. are also being reported for the first time for their ability to produce anticancer molecules. In the present study more isolates from S. detergens possessed anticancer activity as compared to those from O. sanctum. Previous studies have also reported the production of taxol, an anticancer compound, from fungal endophytes isolated from a related species Ocimum basilicum (Gangadevi and Muthumary 2007). It is evident from the results obtained that the isolated cultures have the potential to be exploited as sources of novel cytotoxic molecules as some of them showed significantly high anticancer activity even at low concentration. Further research is now being focused on understanding the chemical nature of biomolecules being produced by members of Pleosporales for their anticancer and anti-bacterial activities.
References
Arnold AE (2007) Understanding the diversity of foliar endophytic fungi: progress, challenges and frontiers. Fungal Biol Rev 21:51–66
Arulmozhi DK, Veeranjaneyulu A, Bodhankar SL, Arora SK (2005) Pharmacological studies of the aqueous extract of Sapindus trifoliatus on central nervous system: possible anti migraine mechanisms. J Ethnopharmacol 97:491–496
Burns TD, Vilgalys R, Barns SM (1990) Evolutionary relationships within the fungi: analyses of nuclear small subunit rRNA sequences. Mol Phylogenet Evol 1:231–241
Collado J, Platas G, González I, Peláez F (1999) Geographical and seasonal influences on the distribution of fungal endophytes in Quercus ilex. New Phytol 144:525–532
Firakova S, Sturdikova M, Muckova M (2007) Bioactive secondary metabolites produced by microorganisms associated with plants. Biologia Bratislava 62:251–257
Gangadevi V, Muthumary J (2007) Endophytic fungal diversity from young, mature and senescent leaves of Ocimum basilicum L. with special reference to Taxol production. Indian J Sci Technol 1:1–15
Ganley RJ, Newcombe G (2006) Fungal endophytes in seeds and needles of Pinus monticola. Mycol Res 110:318–327
Guimaraes DO, Borges WS, Kawano CY, Ribeiro PH, Goldman GH, Nomizo A, Theimann OH, Oliva G, Lopes NP, Pupo MT (2008) Biological activities from extracts of endophytic fungi isolated from Viguiera arenaria and Tithonia diversifolia. FEMS Immunol Med Microbiol 52:134–144
Gunatilaka AAL (2006) Natural products from plant associated microorganisms: distribution, structural diversity, bioactivity and implications of their occurrence. J Nat Prod 69:509–526
Hampl V, Pavlick A, Flegr J (2001) Construction and bootstrap analyses of DNA fingerprinting based phylogenetic trees with freeware program free tree: application to trichomonad parasites. Int J Syst Evol Microbiol 51:731–735
Harper JK, Ford EJ, Strobel GA, Arif A, Grant DM, Porco J, Tomer DP, Oneill K (2003) Pestacin: a 1, 3-dihydro isobenzofuran from Pestalotiopsis microspora possessing antioxidant and antimycotic activities. Tetrahedron 59:2471–2476
Hijri M, Redecker D, Jean A, Petetot MC, Voigt K, Wöstemeyer J, Sanders IR (2002) Identification and isolation of two ascomycete fungi from spores of the arbuscular mycorrhizal fungus Scutellospora castanea. Appl Environ Microbiol 68:4567–4573
Huang WY, Cai YZ et al (2008) Biodiversity of endophytic fungi associated with 29 traditional Chinese medicinal plants. Fungal divers 33:61–75
Karthikeyan K, Gunasekaran P, Ramamurthy N, Govindasamy S (1999) Anticancer activity in Ocimum sanctum. Pharm Biol 37:285–290
Korabecna M, Liska V, Fajfrlik K (2003) Primers ITS1, ITS2 and ITS4 detect the intraspecies variability in the internal transcribed spacers and 5.8 rRNA gene regions in clinical isolates of fungi. Folia Microbiol 48:233–238
Larone DH (2002) Medically important fungi: a guide to identification, 4th edn. ASM Press, Washington
Li JY, Harper JK, Grant M, Tombe BO, Bashyal B, Hess WM, Strobel GA (2001) Ambuic acid, a highly functionalized cyclohexenone with antifungal activity from Pestalotiopsis sp. and Monochaetia sp. Phytochemistry 56:463–468
Morakotkarn D, Kawasaki H, Saki T (2007) Molecular diversity of bamboo associated fungi isolated from Japan. FEMS Microbiol Lett 266:10–19
Park J, Rhee D, Lee Y (2005) Biological activities and chemistry of saponins from Panax ginseng CA Meyer. Phytochem Rev 17:19–20
Pelaez F, Callado J, Arenal F, Basilio A, Cabello A, Matas MTD, Garcia JB, Del Val AG, Gonzalez V, Gorrochategui J, Hernandez P, Martin I, Platas G, Vicente F (1998) Endophytic fungi from plants living on gypsum soils as a source of secondary metabolites with antimicrobial activity. Mycol Res 102:755–761
Petrini O (1986) Taxonomy of endophytic fungi of aerial plant tissues. In: Fokkema NJ, Heuvel JVD (eds) Microbiology of the Phyllosphere. Cambridge University Press, New York, pp 175–187
Prakash P, Gupta N (2005) Therapeutic use of Ocimum sanctum Linn. (Tulsi) with a note on eugenol and its pharmacological actions: a short review. Indian J Pharmacol 49:125–131
Radu S, Kqueen CY (2002) Preliminary screening of endophytic fungi from medicinal plants in Malaysia for antimicrobial and antitumor activity. Malays J Medical Sci 9:23–33
Schulz B, Boyle C (2005) The endophytic continuum. Mycol Res 109:661–686
Schulz B, Boyle C, Dreager S, Rommert AK, Krohn K (2002) Endophytic fungi: a source of novel biologically active secondary metabolites. Mycol 106:996–1004
Sharma M, Chadha BS, Kaur M, Ghatora SK, Saini HS (2008) Molecular characterization of multiple xylanase producing thermophilic/thermotolerant fungi isolated from composting materials. Lett Appl Microbiol 46:526–535
Sharma M, Agarwal SK, Sharma PR, Chadha BS, Khosla MK, Saxena AK (2010) Cytotoxic and apoptotic activity of essential oil from Ocimum viridae towards COLO 205 cells. Food Chem Toxicol 48:336–394
Skehan P, Storeng R, Scudiero D, Monks A, McMohan J, Vistica D, Warren JT, Bokesch H, Kenney S, Boyd MR (1990) New colorimetric cytotoxic assay for anticancer-drug screening. J Nat Cancer Inst 82:1107–1112
Skouboe P, Frisvad JC, Taylor JW, Lauritsen D, Boysen M, Rossen L (1999) Phylogenetic analysis of nucleotide sequences from the ITS region of terverticillate Penicillium species. Mycol Res 103:873–881
Stierle A, Strobel G (1995) The search for a taxol producing microorganisms among the endophytic fungi of the Pacific Yew, Taxus brevifolia. J Nat Prod 58:1315–1324
Strobel GA (2000) Microbial gift from rain forests. Can J Plant Pathol 24:14–20
Strobel G, Daisy B (2003) Bioprospecting for microbial endophytes and their natural products. Microbiol Mol Biol Rev 67:491–502
Strobel GA, Dirksie E, Sears J, Markworth C (2001) Volatile antimicrobials from a novel endophytic fungus. Microbiol 147:2943–2950
Strobel GA, Ford E, Worapong J, Harper JK, Arif AM, Grant DM, Fung PCW, Chan K (2002) Isopestacin, an isobenzofuranone from Pestalotiopsis microspora, possessing antifungal and antioxidant activities. Phytochemistry 60:179–183
Tiwari R, Kalra A, Darokar MP, Chandra M, Aggarwal N, Singh AK, Khanuja SPS (2010) Endophytic bacteria from Ocimum sanctum and their yield enhancing capabilities. Curr Microbiol 60:167–171
Tulp M, Bohlin L (2004) Unconventional natural resources for future drug discoveries. Drug Discov Today 9:450–458
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols. A guide to methods and applications. Academic Press, San Diego
Wiyakrutta S, Sriubolmas N, Panphut W, Thongon N, Danwiset K, Ruangrungsi N, Meevootisom V (2003) Endophytic fungi with anti-microbial, anti-cancer and anti-malarial activities isolated from Thai medicinal plants. World J Microbiol Biotechnol 20:265–272
Acknowledgments
The author financial support in form of project fellowship awarded by Guru Nanak Dev University to Ms. Jyoti Bhagat is duly acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bhagat, J., Kaur, A., Sharma, M. et al. Molecular and functional characterization of endophytic fungi from traditional medicinal plants. World J Microbiol Biotechnol 28, 963–971 (2012). https://doi.org/10.1007/s11274-011-0894-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-011-0894-0