Abstract
A novel gene of thermostable phytase, phyA, was isolated by polymerase chain reaction (PCR) techniques from Aspergillus aculeatus RCEF 4894. The full-length phyA gene comprises 1,404 bp and encodes 467 amino-acid residues, including a 19-residue putative N-terminal signal peptide. The phytase of A. aculeatus was a novel addition to the histidine-acid phosphatase family, as evidenced by both the conserved motifs RHGXRXP and HD in the amino-acid sequence, and 3D structure models. The recombinant phytase was overexpressed in Pichia pastoris, and its specific activity reached 3,000 U mL−1 at the optimum pH of 5.5. This recombinant, thermostable phytase was able to withstand temperatures of up to 90 °C for 10 min, with a loss of only 13.9% of initial enzymatic activity, and showed high activity with phytic-acid sodium salt at a pH range of 2.5–6.5. The broad pH optima and high thermostability of the phytase makes it a promising candidate for feed-pelleting applications.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Phytic acid is the primary storage form of phosphorus in cereals. Phytase (myo-inositol hexakisphosphate phosphohydrolase, EC 3.1.3.8 for 3-phytase and 3.1.8.26 for 6-phytase) catalyzes the hydrolysis of phytate into inorganic phosphates and lower forms of myo-inositol phosphates. Monogastric animals, such as pigs and poultry, are unable to utilize phytic-acid phosphorus, since they essentially lack enzymes for phytic-acid hydrolysis in the digestive tract. The supplementation of phytase in these animals’ diets enhances not only the nutritional quality of phytate-rich feed, but also the growth performance of the animals, thereby decreasing phosphorus pollution in the environment (Dvořáková 1998; Cho et al. 2005; Lei et al. 2007; Bohn et al. 2008; Selle and Ravindran 2008).
To date, the phytase produced by two fungi (A. niger or A. ficuum) that are currently used for commercial production are more specific and thermostable than phytases from other microorganisms (Hong et al. 2004). The phytase crystal structure of A. fumigatus and A. ficuum were also determined (Kostrewa et al. 1997; Liu et al. 2004). However, these phytases could not withstand high temperatures during the feed-pelleting process (Lei and Porres 2003). Many phytases from other microorganisms showed much higher thermostability, but were poorly active at 37 °C (Wyss et al. 1998; Vohra and Satyanarayana 2001), making them unsuitable as animal-feed additives. Therefore, it is necessary to utilize new phytase resources and obtain better phytases (Tseng et al. 2000; Miksch et al. 2002; Chadha et al. 2004; Cho et al. 2005; Guo et al. 2007; Nakashima et al. 2007; Huang et al. 2008; Pavlova et al. 2008; Shi et al. 2008; Zhang and Lei 2008).
In this paper, the gene phyA, encoding a thermostable phytase from A. aculeatus RCEF 4894, was cloned and heterologously expressed in Pichia pastoris, and the recombinant phytase was subjected to detailed assays of its thermostability and the pH dependence of its enzyme activity. Our study demonstrated that this phytase, from A. aculeatus RCEF 4894, is highly pH-stable at pH range of 2.5–6.5 and is thermostable with a loss of only 13.9% of the initial enzymatic activity after being heated at 90 °C for 10 min, providing significant advantages for the processing, transportation, storage, and application of this phytase.
Materials and methods
Strains and vectors
The A. aculeatus RCEF 4894 strain was isolated from a soil sample from Mt. Wuyishan, West Fujian Province, China. The Escherichia coli DH5α strain (Beyotime, China), used in all DNA manipulations, was grown in LB medium, when required, with 50 μg/mL of ampicillin. The P. pastoris GS115 (His4−) strain (Invitrogen, USA) was cultivated at 30 °C in yeast extract, peptone, and dextrose (YPD) medium. The plasmids pMD18-T (Takara, Dalian, China) and pPIC9 K (Invitrogen) were used as cloning and expression vectors, respectively.
Preparation of total RNA and genomic DNA
Aspergillus aculeatus RCEF 4894 was grown at 30 °C for three days on a shaker at 180 rpm in a culture medium containing (in g/L): corn starch 50, glucose 50, NaNO3 8.6, MgSO4·7H2O 0.5, KCl 0.5, FeSO4·7H2O 0.1, K2HPO4 0.1 g, at pH 5.0 (Gargova et al. 1997). Fungal mycelium was obtained by filtration and washed by DEPC-treated water, and the mycelium was ground into a fine powder in liquid nitrogen immediately. Isolation of genomic DNA and total RNA from A. aculeatus RCEF 4894 were prepared as described by Shirzadegan et al. (1991) and Pasamontes et al. (1997), respectively.
Cloning the phytase gene, phyA
The first cDNA strand was synthesized from total RNA by reverse transcription (RT)-PCR, using the M-MLV RTase cDNA Synthesis Kit (Takara) with oligo-dT primers. The obtained cDNA was PCR-amplified, using Taq DNA polymerase (Takara), and a pair of primers (PhyFor and PhyRev, Table 1) were designed based on the two conserved blocks from Aspergillus phytase (GenBank accession number: ATU60412, AY603416, AY745739, DQ192035, DQ198163, EF206311, XM-654197, Z16414). The PCR conditions employed were 94 °C, 4 min (once) + 94 °C, 1 min; 52 °C, 1 min; 72 °C, 1.5 min (35 cycles) + 72 °C, 10 min (once). The PCR amplicon was purified from agarose gel and subsequently cloned into a pMD18-T vector for sequencing. Based on the phyA fragment sequence, 5′-RACE primers (GSP1 and GSP2, Table 1) and 3′-RACE primers (NGSP1 and NGSP2, Table 1) were designed. The full-length cDNA of phyA was obtained using a SMART™ RACE cDNA amplification kit (Clontech, BD). All of the sequences were assembled to obtain a full-length gene of phyA, which was deposited in GenBank under the accession number GU120223.
Plasmid construction and transformation
To construct a recombinant expression vector, the phyA gene fragment encoding the mature enzyme and excluding the putative signal peptide was amplified with the Pfu DNA polymerase (Takara) by PCR, using primers PhyAFor and PhyARev (Table 1), with insertion of EcoRIand NotIsites at the 5′ and 3′ ends, respectively. PCR amplification was accomplished with conditions of 94 °C for 4 min, followed by 30 cycles of 94 °C for 1 min, 58 °C for 1 min, 72 °C for 1.5 min, and final extension at 72 °C for 10 min. The BglIIsite, 31 bp upstream of the TAG stop codon of the phyA gene, was removed by site-directed mutagenesis, replacing the T with a C and creating a samesense mutation of serine (TCT → TCC). The amplicons were digested with EcoRIand NotI and ligated into the pPIC9 K vector. The pPIC9K-phyA recombinant plasmid was transformed into Escherichia coli DH5α, and transformants were selected on an LB plate containing 50 μg/mL ampicillin. According to the Pichia expression-system manual, the pPIC9K-phyA plasmid was linearized by the restriction enzyme BglII and transformed into P. pastoris GS115 by electroporation, using an ECM 630 Electro Cell Manipulator (Genetronics, BTX Instrument Division, San Diego, CA, USA). These his+ transformants were plated onto MD medium (2% glucose, 1.34% YNB, 0.004% biotin, and 1.5% agar, pH 5.5) at 30 °C for two days. Subsequently, screening of multiple inserts was performed on YPD-Geneticin plates in vivo.
Expression of recombinant phytase
In preparation for fermentation, cultures of P. pastoris were grown on a shaker at 30 °C and 200 rpm for 30 h in BMGY media containing 1.34% yeast nitrogen base (YNB), 1% yeast extract, 2% Trypton, 100 mmol/L KH2PO4 (pH6.0), 0.4 mg/L biotin solution, and 1% (v/v) glycerol. Cells were harvested by centrifugation (3,000g, 5 min) and resuspended in 0.2 volumes of the same growth medium, but containing 0.5% methanol instead of glycerol, and supplied with methanol at 1% final concentration once per day to induce phytase expression. Completing a fermentation cycle required approximately 168 h.
Phytase assay
Phytase activity was estimated as described by Tseng et al. (2000). The fermentation supernatant was diluted with 250 mM sodium acetate buffer (pH 5.5) to bring phytase activity within the range of 0.1–0.7 FTU/mL solution and stored in melting ice until incubation. The incubation mixture (3 mL) contained 1 mL of the diluted enzyme solution and 5.0 mM sodium phytate (P3168, Sigma) as substrate. Incubation was carried out for one hour at 37 °C. The reaction was terminated by the addition of 2 mL of stop reagent containing 20 mM ammonium heptamolybdate and 5 mM ammonium vanadate. All sample tubes were centrifuged for 5 min at 3,000g. The absorbance was measured at 415 nm with a spectrophotometer, after zeroing the instrument with H2O. One unit of phytase activity was defined as 1 μmol of inorganic phosphorus produced from sodium phytate per min per mL of the diluted enzyme solution. Protein concentration was determined by Bradford assay, using bovine serum albumin as a standard (Sangon, China).
The profiles of pH versus activity were determined with the standard assay method at 37 °C in various pH buffers. The buffers used included glycine/HCl (pH 2–3.5), sodium acetate/acetic acid buffer (pH 3.5–6.5), and Tris/HCl (pH 7.0–7.5); thermal stability of the recombinant phytase was determined by incubation of the culture supernatant at 90 °C for 10, 20 and 30 min, respectively, then chilling the supernatant on melting ice for 30 min to assay the residual activity at 37 °C (Pasamontes et al. 1997; Wyss et al. 1998).
SDS–PAGE
Total soluble proteins from culture supernatants from the fermentation were run on 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) gels on the DYCZ-24DN Electrophoresis Cell (Beijing Liuyi Instrument Factory, China). Protein bands were detected by submerging gels in 0.1% (w/v) Coomassie brilliant blue R-250 in an aqueous methanol (40%) and acetic acid (10%) solution and destained in an aqueous methanol (40%) and acetic acid (10%) solution (Bae et al. 1999).
Real-time quantitative PCR assay
The number of phyA genes in the genome was identified by relative qRT-PCR, using the elongation factor 1 gene as reference gene. PCR reactions on samples containing equivalent copies of phyA and the elongation factor 1 gene on plasmid DNA templates were also performed as calibrator samples for each primer pair. The qRT-PCRs were carried out in 96-wells plates with the Applied Biosystems 7500 Real Time PCR System using SYBR® Premix Ex Taq™ (Takara, DRR081S) in a reaction volume of 20 μL (2 μL diluted DNAs, 10 μL of 2× SYBR® mix and primer pairs at 0.4 μM). Aliquots from the same genomic DNA solutions were used with all primer sets in each experiment, which were performed in quadruplicate. All PCR reactions were performed under the following conditions: 95 °C for 30 s, 45 cycles of 3 s at 95 °C, and 34 s at 60 °C. The experiments were analyzed with auto-baseline and manual thresholds chosen from the exponential phase of the PCR amplification. After the data analysis, the Ct number and DeltaRn were exported for statistical analyses. Data were analyzed using Sequence Detection Software (SDS 1.3) and 2−△△Ct method (De Preter et al. 2002). The real-time PCR reactions were carried out for both the phytase gene and the internal reference gene, with primers shown in Table 2.
Phylogenetic trees and homology modeling of phytases
Some phytase sequences closely related to Aspergillus spp. were obtained from GenBank for phylogenetic analysis, using MEGA 4.1 with the neighbor-joining method and bootstrap analysis. The phytase sequence from Penicillium oxalicum was selected as an outgroup. Sequences were aligned with the multiple sequence-alignment program ClustalW 1.81, and these alignments were adjusted manually. The structure of phosphomonoesterase (PDB code: 1ihpA) was used as a homology-modeling template. 3D models were generated by SWISS-MODEL (http://swissmodel.expasy.org/), using the program Alignment Mode (Arnold et al. 2006).
Results
Cloning of the phytase gene phyA and sequence analysis
The primers (PhyFor and PhyRev; Table 1) were designed to target two conserved motifs of phyA (5′-GGGGTATCAATGCTTCTC-3′ and 5′-GAGCTTTGCCAGATCTGG-3′) discovered among Aspergillus phytase sequences from the GenBank database, and the core region of the phytase gene phyA was amplified from the cDNA of A. aculeatus RCEF 4894 by PCR. The 5′ and 3′ end fragments of the mRNA of phyA was obtained by SMART RACE PCR. Assembly of the core region of 1240 bp, the 5′ end mRNA of 951 bp, and the 3′ end mRNA of 327 bp yielded a 1,642 bp fragment (GenBank no. GU120223) containing a 1,404 bp open reading frame encoding 467 amino acid residues with a typical signal peptide (residues 1–19) (http://www.cbs.dtu.dk/services/SignalP). Like all of the other fungal phytases, the phytase gene from A. aculeatus RCEF 4894 belongs to the family of histidine-acid phosphatases because it was found to have the active site septa-petide RHGXRXP and catalytically active dipeptide HD (Wang et al. 2007). In comparison with the sequences of phytases previously isolated by BLAST search, the deduced amino acid sequence of A. aculeatus RCEF 4894 phytase exhibited 69 and 66% identities with A. niger phytase (GenBank no. P34752) and A. fumigatus WY-2 phytase (GenBank no. AAU93517), respectively. Based on analysis of the deduced amino acid sequence, the A. aculeatus RCEF 4894 phytase had a theoretical pI value 6.70 that was much higher than the pI value of the A. niger phytase (4.72) and the A. fumigatus WY-2 phytase (6.51).
High-level expression of the recombinant phytase in P. pastoris
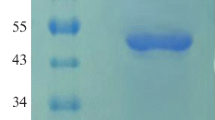
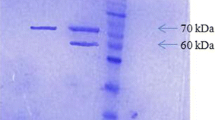
All of the transformants that grew normally on the MD medium were inoculated onto YPD-Geneticin medium plates containing each of the following concentrations of Geneticin: 0, 0.25, 0.5, 0.75, 1.0, 1.5, 1.75, 2.0, and 3.0 mg mL−1. Five transformants containing multiple copies of the phytase gene were selected from 3.0 mg mL−1 YPD-Geneticin medium plates for further analysis. Small-scale cultures of these transformants were screened for their ability to secrete the phytase of A. aculeatus RCEF 4894. Phytase was detected by SDS–PAGE (Fig. 1), and enzyme activity was measured. The results demonstrated that recombinants PA18 has the highest enzyme activity (2,260 U mL−1) of these transformants after 120 h of methanol induction. As the methanol induction time increased, phytase activity in the supernatant also increased, reaching 3,000 U mL−1 after 168 h of induction. Furthermore, crude protein concentration in the supernatant increased over time, eventually attaining a concentration of approximately 60.13 μg mL−1. Therefore, crude phytase activity in the supernatant could reach 52,700 U g−1. The molecular weights of the novel phytases, as determined by SDS–PAGE, were about 74.8 kDa (Fig. 1).
Thermostability and pH dependence of enzyme activity
The thermostability of the expressed phytase was tested, and results indicate that the enzyme retained 86.1% activity after heating at 90 °C for 10 min, and this phytase retained 74.6% activity after heat treatment for 30 min (Fig. 2).
The activity of the recombinant phytases were determined at various pH levels. The results indicated that they were active at pH 2.5–6.5, while over 50% activity remained at pH 3.0–6.0. The optimal pH for phytase activity tested at 37 °C was pH 5.5 (Fig. 3).
qRT-PCR analysis
As shown in Fig. 4, the results of the analysis clearly exhibit normalized reporter-dye fluorescence (Rn) as a function of cycle, and indicate that the phyA gene in the genome was a single copy, which was calculated according to Ct 19.524 in the genome, Ct 10.862 in the calibrator sample for elongation factor 1, Ct 32.947 in the genome, and Ct 20.288 in the calibrator sample for phytase gene.
Sequence analysis and homology modeling
The phytase phylogenetic tree of Aspergillus spp. was mainly composed of three clades with 100% boostrap support in the A. fumigatus clade, the A. oryzae clade, and the A. niger clade (Fig. 5). The phytase of A. aculeatus did not belong to one of the three clades. This result indicated that A. aculeatus has some distinct differences in the amino-acid sequence of phytase from those of A. fumigatus and A. niger.
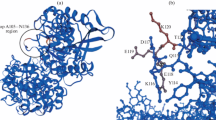
The three phytase sequences from Aspergillus spp. were aligned with the phosphomonoesterase (PDB code: 1ihpA) by ClustalW 1.81 (Fig. 6), which was submitted to SWISS-MODEL. These 3D models of phytase in A. fumigatus (GenBank no. AAU93517), A. aculeatus (GenBank no. GU120223) and A. niger (GenBank no. P34752) were conducted by homology modeling with SWISS-MODEL, using the 1ihpA structure as a template. The representative Beta-Alpha–Beta Units and Psi-Loops motifs are well conserved between phytases of A. fumigatus, A. aculeatus, and A. niger. However, their 3D-structure models have some distinct differences at amino-acid sequences 156 through 164 (Fig. 7A1, B1 and C1), 239 through 249 (Fig. 7A2, B2 and C2), and 353 through 370. Especially in the 353-through-370 sequence, there was an α-helix in A. fumigatus phytase at the 367-through-370 sequence (Fig. 7A3) and A. niger phytase at the 358-through-361 sequence (Fig. 7C3), but A. aculeatus phytase did not have an α-helix. All of these data demonstrate that the A. aculeatus phytase presented in this paper was a new addition to the histidine-acid phosphatase family.
Discussion
Cloning of the phyA gene from A. aculeatus RCEF 4894 was achieved by polymerase chain reaction (PCR) and Smart Race PCR. Gene sequencing suggested that this phytase was a histidine-acid phosphatase possessing the RHGXRXP and HD motifs. This phyA sequence was identical not only to A. niger phyA, with a similarity of 70%, but also to A. fumigatus phyA (a putative thermostable enzyme) with a similarity of 69%. In this study, P. pastoris was successfully used as a host organism to express phytase derived from A. aculeatus RCEF 4894. Moreover, the recombinant phytase exhibited both high enzyme activity and high thermostability, which the other phytases from A. fumigatus and synthetic thermostable constructs do not possess. The maximum phytase production level of P. pastoris was determined to be 3,000 U mL−1 in crude culture extracts and, in some cases, exceeded those from previously published studies.
As enzymes that are involved in animal feed should be able to withstand high temperatures during the feed-pelleting process, thermostable phytases are obvious candidates for feed supplements. The thermostability of fungal phytases has been extensively studied. Among all of the natural phytases of fungal origin, only that from A. fumigatus is able to withstand temperature up to 100 °C for 20 min, with a loss of only 10% of the initial activity (Pasamontes et al. 1997). Also, the phytase from P. anomla retained 85% activity when exposed to 80 °C for 15 min (Vohra and Satyanarayana 2001). However, other fungal phytases exhibited structural instability and a great loss in enzyme activity at high temperatures (Wyss et al. 1998). In contrast, the PhyA phytase studied here can retain 86.1, 77.7 and 74.6% of its activity after heat treatment at 90 °C for10, 20 and 30 min, respectively. Compared with a phytase derived from a quadruple mutant (A58E, P65S, Q191R, and T271R), which retained 20% greater (P < 0.05) activity after being heated at 90 °C for 10 min than that of the wild-type PhyA phytase (which demonstrated approximately 52% residual activity) (Zhang and Lei 2008), the PhyA phytase assayed here is more thermostable.
Deglycosylation has been shown to cause a substantial reduction in enzyme thermostability. While 62.6% of an enzyme’s activity remained after the nondeglycosylated enzyme was heated for 15 min at 80 °C, only 37.3% of the enzyme’s activity remained after the deglycosylated enzyme was treated under the same conditions (Han et al. 1999). The molecular weights of the recombinant phytases (~74.8 kDa) were much higher than the predicted values based on amino-acid sequences (49.6 kDa), indicating that the expressed phytase was hyperglycosylated. Moreover, the A. aculeatus RCEF 4894 phytase has 23 Asn amino-acid residues (potential glycosylation sites), which is more than the 19 Asn residues of the A. niger phytase. This difference may explain why the recombinant phytase is more thermostable than other phytases.
Phytase activity is limited to a relatively small region of the monogastric digestive tract, in which the pH ranges from 2 to 3 in the stomach and from 4 to 7 in the small intestine. The present recombinant phytases, active at pH values from 2.5 to 6.5, exhibited a broader pH optima, making them suitable as animal-feed additives. Therefore, the high thermostability and broad pH optima of this PhyA phytase makes it a promising candidate for feed-pelleting applications.
In order to study A. aculeatus phytase’s high enzymatic activity and high thermostability, the structures of the A. niger and A. fumigatus phytases were chosen as comparison models to elucidate the structural basis of the A. aculeatus phytase’s specific properties. Three regions, E29–S36, G157–R162, and R241–S246, were investigated as possible factors contributing to the heat resistance of the A. fumigatus phytase (Xiang et al. 2004). Comparison of the amino-acid sequences of the A. fumigatus, A. niger, and A. aculeatus phytases showed that the E29–S36 and R241–S246 sequences in the A. aculeatus phytase have the greatest homology to those in the A. fumigatus phytase, which may correlate with the high thermostability of the A. aculeatus phytase. Detailed inspection of the homology-modeled 3D structure provided a possible explanation for the A. aculeatus phytase’s high enzymatic activity. The α-helix in the K346–T370 region is close to the HD motif for substrate binding/product leaving. It may be inferred that the stability of the reaction product at the active site is related to its activity: the more stable the product, the less active the enzyme (Liu et al. 2004). The α-helix in the A. niger phytase at the 358-to-361 sequence (Fig. 7C3) has more similarity with the HD motif in the 3D structure than the α-helix in the A. fumigatus phytase at the 367-to-370 sequence (Fig. 7A3), and this effect may be attributed to the disruption of the interactions between the product and the residues, facilitating the release of the product PO4 3+ group. Therefore, given that the A. niger phytase showed more enzymatic activity than the A. fumigatus phytase because it lacked an α-helix in the K346–T370 region, it is not surprising that the A. aculeatus phytase’s enzymatic activity is similar to that of the A. niger phytase.
Abbreviations
- RCEF:
-
Research Center for Entomogenous Fungi of Anhui Agricultural University
References
Arnold K, Bordoli L, Kopp J et al (2006) The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 22:195–201. doi:10.1093/bioinformatics/bti770
Bae HD, Yanke LJ, Cheng KJ et al (1999) A novel staining method for detecting phytase activity. J Microbiol Methods 39:17–22. doi:10.1016/S0167-7012(99)00096-2
Bohn L, Meyer A, Rasmussen S (2008) Phytate: impact on environment and human nutrition. A challenge for molecular breeding. J Zhejiang Univ Sci B 9:165–191. doi:10.1631/jzus.B071064
Chadha BS, Harmeet G, Mandeep M et al (2004) Phytase production by the thermophilic fungus Rhizomucor pusillus. World J Microbiol Biotechnol 20:105–109. doi:10.1023/B:WIBI.0000013319.13348.0a
Cho J, Lee C, Kang S et al (2005) Molecular cloning of a phytase gene (phyM) from Pseudomonas syringae MOK1. Curr Microbiol 51:11–15. doi:10.1007/s00284-005-4482-0
De Preter K, Speleman F, Combaret V et al (2002) Quantification of MYCN, DDX1, and NAG gene copy number in neuroblastoma using a real-time quantitative PCR assay. Mod Pathol 15(2):159–166. doi:org/10.1038/modpathol.3880508
Dvořáková J (1998) Phytase: sources, preparation and exploitation. Folia Microbiol 43:323–338. doi:10.1007/BF02818571
Gargova S, Roshkova Z, Vancheva G (1997) Screening of fungi for phytase production. Biotechnol Tech 11:221–224. doi:10.1023/A:1018426119073
Guo MJ, Zhuang YP, Chu J et al (2007) Production and purification of a novel thermostable phytase by Pichia pastoris FPHY34. Process Biochem 42:1660–1665. doi:10.1016/j.procbio.2007.09.003
Han Y, Wilson DB, Lei Xg (1999) Expression of an Aspergillus niger phytase gene (phyA) in Saccharomyces cerevisiae. Appl Environ Microbiol 65:1915–1918
Hong CY, Cheng KJ, Tseng TH et al (2004) Production of two highly active bacterial phytases with broad pH optima in germinated transgenic rice seeds. Transgenic Res 13:29–39. doi:10.1023/B:TRAG.0000017158.96765.67
Huang H, Luo H, Wang Y et al (2008) A novel phytase from Yersinia rohdei with high phytate hydrolysis activity under low pH and strong pepsin conditions. Appl Microbiol Biotechnol 80:417–426. doi:10.1007/s00253-008-1556-5
Kostrewa D, Gruninger-Leitch F, Darcy A, Broger C, Mitchell D, van Loon APGM (1997) Crystal structure of phytase from Aspergillus ficuum at 2.5 a resolution. Nat Struct Mol Biol 4(3):185–190. doi:10.1038/nsb0397-185
Lei XG, Porres JM (2003) Phytase enzymology, applications, and biotechnology. Biotechnol Lett 25:1787–1794. doi:10.1023/A:1026224101580
Lei XG, Porres JM, Mullaney EJ et al (2007) Phytase: source, structure and application. In: Polaina J, MacCabe AP (eds) Industrial enzymes: structure, function and applications. Springer, Dordrecht, pp 505–529. doi:10.1007/1-4020-5377-0_29
Liu Q, Huang Q, Lei XG, Hao Q (2004) Crystallographic snapshots of Aspergillus fumigatus phytase, revealing its enzymatic dynamics. Structure 12(9):1575–1583. doi:10.1016/j.str.2004.06.015
Miksch G, Kleist S, Friehs K et al (2002) Overexpression of the phytase from Escherichia coli and its extracellular production in bioreactors. Appl Microbiol Biotechnol 59:685–694. doi:10.1007/s00253-002-1071-z
Nakashima B, McAllister T, Sharma R et al (2007) Diversity of phytases in the rumen. Microb Ecol 53:82–88. doi:10.1007/s00248-006-9147-4
Pasamontes L, Haiker M, Wyss M et al (1997) Gene cloning, purification, and characterization of a heat-stable phytase from the fungus Aspergillus fumigatus. Appl Environ Microbiol 63:1696–1700
Pavlova K, Gargova S, Hristozova T et al (2008) Phytase from antarctic yeast strain Cryptococcus laurentii AL27. Folia Microbiol 53:29–34. doi:10.1007/s12223-008-0004-3
Selle PH, Ravindran V (2008) Phytate-degrading enzymes in pig nutrition. Livestock Sci 113:99–122. doi:10.1016/j.livsci.2007.05.014
Shi P, Huang H, Wang Y et al (2008) A novel phytase gene appA from Buttiauxella sp. GC21 isolated from grass carp intestine. Aquaculture 275:70–75. doi:10.1016/j.aquaculture.2008.01.021
Shirzadegan M, Christie P, Seemann JR (1991) An efficient method for isolation of RNA from tissue cultured plant cells. Nucl Acids Res 19:6055
Tseng Y, Fang T, Tseng S (2000) Isolation and characterization of a novel phytase from Penicillium simplicissimum. Folia Microbiol 45:121–127. doi:10.1007/BF02817409
Vohra A, Satyanarayana T (2001) Phytase production by the yeast, Pichia anomala. Biotechnol Lett 23:551–554. doi:10.1023/A:1010314114053
Wang Y, Gao X, Su Q et al (2007) Cloning, expression, and enzyme characterization of an acid heat-stable phytase from Aspergillus fumigatus WY-2. Curr Microbiol 55:65–70. doi:10.1007/s00284-006-0613-5
Wyss M, Pasamontes L, Remy R et al (1998) Comparison of the thermostability properties of three acid phosphatases from molds: Aspergillus fumigatus phytase, A. niger phytase, and A. niger pH 2.5 acid phosphatase. Appl Environ Microbiol 64:4446–4451
Xiang T, Liu Q, Deacon AM, Koshy M, Kriksunov IA, Lei XG, Hao Q, Thiel DJ (2004) Crystal structure of a heat-resilient phytase from Aspergillus fumigatus, carrying a phosphorylated histidine. J Biol Chem 339(2):437–445. doi:10.1016/j.jmb.2004.03.057
Zhang WM, Lei XG (2008) Cumulative improvements of thermostability and pH-activity profile of Aspergillus niger PhyA phytase by site-directed mutagenesis. Appl Microbiol Biotechnol 77:1033–1040. doi:10.1007/s00253-007-1239-7
Acknowledgments
This research was supported by funds from the Department of Education of Anhui Province for Excellent Youth Teacher Program (2005JQ1199), the Anhui Provincial Key Project for Nature and Science (TD200708), and the Anhui Provincial Science Foundation for Excellent Youth Program for “NCET”(08040106902), China.
Conflict of interest statements
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ma, ZY., Pu, SC., Jiang, JJ. et al. A novel thermostable phytase from the fungus Aspergillus aculeatus RCEF 4894: gene cloning and expression in Pichia pastoris . World J Microbiol Biotechnol 27, 679–686 (2011). https://doi.org/10.1007/s11274-010-0506-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-010-0506-4