Abstract
In the present study 14 bacteriocinogenic strains of Enterococcus faecium isolated from dairy foods and faecal sample were evaluated for the presence of virulence determinants, production of biogenic amines and their susceptibility to various antibiotics. Genetic diversity among them was evaluated by RAPD-PCR method. Further, they were evaluated for their probiotic potential under in vitro trials. The efaAfm was the only virulence trait detected in all E. faecium and tyramine was the only biogenic amine produced by 9 tested strains. No strain was resistant to all antibiotics and for some strains, multiple resistances were observed. E. faecium FH 99 showed highest good ability to tolerate acid and bile, while good bile salt hydrolase activity and were able to assimilate cholesterol from growth media. These results suggest that the tested E. faecium are generally free from virulence traits and having good probiotic potential and may be exploit in dairy industry and probiotic preparations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The enterococci are lactic acid bacteria (LAB) and form an important part of environmental, food and clinical microbiology. They are normal inhabitants of the gastrointestinal tracts of both humans and animals (Bhardwaj et al. 2008). Among several enterococcal species Enterococcus faecium and Enterococcus faecalis are the two predominant species in the human intestine (Giraffa 2003). Enterococci also occur in or are deliberately added to fermented foods, in which they contribute to the organoleptic properties (Giraffa 2002, 2003). Moreover, several strains of genus Enterococcus have also been used as probiotics, which may improve the microbial balance of the intestine or can be used in the treatment of gastroenteritis in humans and animals (Giraffa 2003; Foulquie Moreno et al. 2006; Bhardwaj et al. 2008). In contrast to most other genera of the lactic acid bacteria, not all enterococcal species have “generally recognized as safe” status (Ogier and Serror 2008). Indeed, some of the enterococcal species especially E. faecalis, are typical opportunistic pathogens that cause disease mostly in the nosocomial settings (Kayser 2003; Franz et al. 2003; Eaton and Gasson 2001; Ogier and Serror 2008), which may in part be linked to the presence of virulence determinants and antibiotic resistance. Several putative virulence factors have been identified in enterococci, such as aggregation substance (encoded by agg) (Galli et al. 1990), cytolysin (encoded by cylA) (Gilmore et al. 1994), gelatinase (encoded by gel E) (Su et al. 1991), hyaluronidase (encoded by the hyl gene) (Rosan and Williams 1964), sex pheromone (encoded by cpd, ccf, cob) (Eaton and Gasson 2001), adhesin of collagen (encoded by ace) (Kayaoglu and Orstavik 2004) and enterococcal surface protein (encoded by esp) (Shankar et al. 1999). FAO–WHO has recommended that antimicrobial resistance patterns and opportunistic virulence properties should be tested to document the safety of probiotic strains (FAO/WHO 2002). In our earlier work we isolated and characterized 60 strains of enterococcal species as potent bacteriocin producer (Gupta and Malik 2007). Recently, safety of some strains among them was evaluated by testing their susceptibility to killing under in vitro opsonophagocytic assay (Bhardwaj et al. 2009a). Now the aim of the present study was to investigate the genetic diversity, presence of virulence determinants and probiotic potential of selected bacteriocinogenic strains of Enterococcus faecium.
Materials and methods
Bacterial strains and growth conditions
All the selected 14 E. faecium strains and others reference bacterial species and strains used in this study are listed in Table 1.
DNA isolation
Genomic DNA of all tested strains was extracted by method described by phenol–chloroform extraction as previously described Pospiech and Neumann (1995).
RAPD-PCR
RAPD-PCR was performed using the 2 primers viz; M13, 5′-GAGGGTGGCGGTTCT-3′ (Andrighetto et al. 2001) and AP4, 5′-TCACGCTGCA-3′ M13, 5′-GAGGGTGGCGGTTCT-3′ and AP4, 5′-TCACGCTGCA-3′ (Omar et al. 2004) in separate reaction. E. faecium NCDC 124 and E. faecium BFE 900 were used as reference strains. DNA was amplified with M13 primer at 95°C for 5 min [94°C, 60 s; 37.3°C, 45 s; and 72°C, 60 s], 40 cycles and then 72°C, 4 min while for AP4 it is: 94°C, 120 s [94°C, 60 s; 34.5°C, 60 s; and 72°C, 60 s], 40 cycles; 72°C, 4 min. PCR amplified products obtained were electrophoresed on the agarose gels (1.5%). The images were normalized and subsequently analyses of the patterns were carried out with the NTSYSpc 2.02 software package (Applied Biostatistics Inc., NY, USA). Grouping of the RAPD-PCR profiles was performed by calculating the similarity coefficients for pairs of tracks by using Jaccard coefficient and strains were grouped by Sequential agglomerative hierarchal nested cluster analysis (SAHN) using the unweighted pair group method with arithmetic averages (UPGMA) clustering algorithm. The RAPD-PCR fingerprints obtained with both the primers were analyzed together as a single data set by calculating the average matrix from the two separate similarity matrices of the different primers to obtain a single dendrogram.
Safety assessment
PCR screening for virulence determinants
The detection of different virulence genes involved in the expression of enterococcal surface protein (esp), cytolysin (cylA, cylB, cylM), gelatinease (gelE), aggregation substance (agg), endocarditis antigen (efaAfm, efaAfs), sex pheromone (cpd, ccf, cob), adhesin of collagen (ace) and hyaluronidase (hyl) was done by PCR using PCR conditions as per reference given in Table 2. The primers used and their amplification products are reported in (Table 2). PCR products were detected after electrophoresis on 1.5% agarose gels.
Detection of aggregation substance by clumping assay
Detection of the aggregation substance (AS) of the enterococci was performed by clumping assay. E. faecalis JH2-SS An 18-h incubated culture of E. faecalis JH2-SS in Todd-Hewitt broth (THB) was centrifuged at 10,000 rpm (15 min, 4°C) and the supernatant was recovered and filter sterilized. A 0.5 ml volume of the supernatant was mixed with equal volumes of fresh Todd-Hewitt broth and 18-h grown tested culture. The mixture was then incubated at 37°C and visually examined for cell clumping after 2, 4, 8, and 24 h. In order to account for constitutive clumping in the absence of AS, culture were also tested in THB without sex pheromone.
Molecular detection of tyrosine and histidine decarboxylase gene
Multiplex PCR method was used for the simultaneous detection of histidine decarboxylase (hdc) and tyrosine decarbolylase (tdc) gene as described by Coton and Coton (2005). Primer sets HDC3 (5′-GATGGTATTGTTTCKTATGA-3′)/HDC4 (5′-CAAACACCAGCATCTTC-3′) and TD2 (5′-ACATAGTCAACCATRTTGAA-3′)/TD5 (5′-CAAATGGAAGAAGAAGTAGG-3′) (Coton et al. 2004) were used for the detection of hdc and tdc gene, respectively. Whereas, 16S rRNA universal primers BSF8 (5′-AGAGTTTGATCCTGGCTCAG-3′) and BSR1541 (5′-AAGGAGGTGATCCAGCCGCA-3′) (Wilmotte et al. 1993) were used as the PCR internal control. The PCR reaction mixture consisted of 1 ng of total DNA, Primer concentrations were 20 pmol for HDC3, HDC4, TD2 and TD5 and 5 pmol for BSF8 and BSR1541 (synthesised by Immperialls life science), 1 U of Taq DNA polymerase, 5 μl of 10× Taq buffer, and 1 μl dNTPs (50 pmol) in a final volume of 50 μl. The amplification was performed as follows: 95°C for 5 min, then 35 cycles at 95°C for 45 s, 48°C for 45 s, and 72°C for 1 min, and a final extension at 72°C for 10 min in thermo cycler, EP Gardient (Eppendorf Master Cycler, Hamburg, Germany). The integrity of PCR products was assayed by the development of single bands following electrophoresis in 1.0% (w/v) agarose gels.
Antibiotic susceptibility
Antibiotic susceptibility profile of the all tested strains was evaluated against Bacitracin (10 units), Polymyxin B (300 units), Nitrofurantoin (200 mcg), Rifampicin (30 mcg), Teicoplanin (30 mcg), Neomycin (30 mcg), Ciprofloxacin (30 mcg) and Vancomycin (30 mcg) were determined by a disc diffusion method on Mueller–Hinton Agar No. 2 using antibiotic discs (HiMedia, Mumbai, India). The results were recorded according to the interpretative criteria of the Clinical and Laboratory Standards Institute (CLSI 2007).
In vitro probiotic attributes
Gastro-intestinal stress tolerance
The E. faecium strains were grown in MRS broth at 37°C and growth was monitored at 550 nm. Cells were harvested by centrifugation and resuspended in an equal volume of MRS broth, adjusted at pH 6.5, 3.0, 2.0 and 1.0 using 1N HCl and containing 1.0, 2.0 and 3.0% of dehydrated fresh bile (Oxgall, HiMedia Laboratories, Ltd., Mumbai) for acid and bile tolerance, respectively. Viable cells counts were determined after 0, 30, 60, and 120 min in case of acid and after 0, 1, 3 and 12 h in case of bile tolerance. The data presented for all tests are mean values ± standard deviation of assays conducted in triplicate.
Cell surface hydrophobicity
The cell-surface hydrophobicity of all E. faecium towards three hydrocarbons viz. n-hexadecane, octane or xylene was measured as described by Rosenberg et al. (1980). Cells were harvested in their early log growth phase, washed twice and resuspended in PUM buffer and their absorbance was determined at 450 nm (recorded as ODinitial). Then variable volumes (30–300 μl) of hydrocarbons were added to resuspended cells in PUM buffer. The cells were pre-incubated at 30°C for 10 min and then mixture was agitated using vortex mixer. The hydrocarbon layer was allowed to rise completely. After about 15 min the aqueous phase was taken carefully and absorbance was determined at 450 nm (recorded as ODfinal). Percent hydrophobicity was calculated according to the formula: {ODinitial − ODfinal/ODinitial} × 100.
Bile salt hydrolase assay
Cultures (10 μl) were spotted onto bile salt hydrolase (BSH) screening medium, which consisted of MRS agar plates supplemented with 0.5% (w/v) two different bile salts of taurocholic acid (TC), taurodeoxycholic acid (TDC) and 0.37 g/l of CaCl2. Plates were incubated in anaerobic jars under CO2 atmosphere at 37°C for 72 h after which the BSH activity was detected by the presence of precipitation zones. BSH activity of grown culture was quantified by a two-step procedure as described by Nguyen et al. (2007) by determination of the amount of the liberated amino acids from the bile salts.
Analysis of cholesterol assimilation from growth media
The efficiency of all 14 E. faecium strains to remove cholesterol from growth media was evaluated in triplicate by the method adopted by Gilliland and Walker (1990). Freshly prepared MRS-Thio (MRS broth with 0.2% sodium thioglycollate) broth was supplemented with 0.2% sodium taurocholate. A filter-sterilized cholesterol solution (10 mg/ml in ethanol) was added to the broth to a final concentration of 70 μg/ml. The broth was inoculated with 1% of culture and incubated anaerobically by using GasPak Anaerobic System (Becton-Dickinson, Cockeysville, MD) for 24 h at 37°C. After incubation, cells were removed by centrifugation for 7 min at 5,400×g and 4°C. The method described by Rudel and Morris (1973) was used to determine the amount of cholesterol in the spent broth and in uninoculated broth. Percent cholesterol removal from growth media was calculated according to the formula: {(Chouninoculated − Choinoculated)/Chouninoculated} × 100.
Results and discussion
Genetic diversity among E. faecium
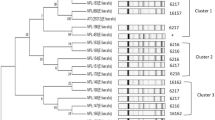
All 14 bacteriocinogenic E. faecium isolates, were subjected to RAPD-PCR, to study the interspecies diversity among them. The single two-dimensional dendrogram presented in Fig. 1 reflects the relationships within the strains. The isolates were grouped into two well separated clusters (cluster A and B) at a similarity level of 62.5 and 59%, respectively. Three E. faecium isolates (KH 24, DH 28 and RH 33) were grouped into cluster A. Whereas, 11 E. faecium isolates of cluster B were divide into two subgroups B1 and B2 at a similarity level of 60.4 and 64.1%, respectively. B1 group consisted of the type strain BFE 900 which has a 68% similarity with RH 31 of its subgroup. While, second type strains, i.e. E. faecium NCDC 124 was placed in B2 cluster with similarity level of 76%. Thus, results showed a high degree of genetic diversity among the profiles of B cluster as compared to cluster A. Out of all the strains studied, the strains RH 38 and FH 102 were the most similar with a similarity level of approximately 93%. E. faecium isolates from milk, dahi, cream and faecal samples did not form separate clusters.
RAPD-PCR technique is a rapid method used for taxonomic and genotypic analyses of the natural bacterial isolates at the genus, species and strain. Several workers have successfully used RAPD-PCR for the discrimination between Enterococcus strains from different geographical, food faecal and clinical origin (Vancanneyt et al. 2004; Yousif et al. 2005; Serio et al. 2007; Templer and Baumgartner 2007). Thus, comparison of RAPD-PCR data obtained with a combination of several primers designed for either conservative or variable regions of bacterial genome, makes it possible to get an idea on interspecies diversity of E. faecium.
Safety assessment
Virulence determinants
All the 14 E. faecium strains were found to be positive for efaAfm (human endocarditis antigen) and only one strain, i.e. E. faecium RH 38 gave a positive result for ccf gene which is responsible for sex pheromone (Table 3). In the present study a very low prevalence of virulence traits among E. faecium strains isolated from food and faeces is in agreement with the previous reports (Franz et al. 2001a; Eaton and Gasson 2001; Omar et al. 2004; Perez-Pulido et al. 2006; Templer and Baumgartner 2007; Serio et al. 2007; Abriouel et al. 2008; Billstroma et al. 2008; Valenzuela et al. 2009). High prevalence of the gene for endocarditic antigen (efaAfm) in E. faecium strains was also reported by other workers (Sanchez et al. 2007; Cariolato et al. 2008). The role of efaAfm has not yet been clearly demonstrated. The high prevalence of efaAfm gene among E. faecium strains indicates that this gene may have some important role in the persistence of such species in environments other than human tissues.
Clumping assay
Sex pheromone-inducible conjugation is an important mechanism for horizontal transfer genes such as genes antibiotic resistance and virulence traits in enterococci (Dunny et al. 1995). In the present study clumping assay was used to investigate sex pheromone-inducible conjugation in tested E. faecium strains. No visible macroscopic cellular aggregate was observed even after 12 h incubation in presence of supernatant of E. faecalis JH2-SS by any of the isolates tested, except E. faecium FH 115. Further to confirm this, FH 115 was grown in THB without sex pheromone and found that strain clumped constitutively not the result of the presence of aggregation substance. Similar kind of results were also observed by Franz et al. (2001a) and Omar et al. (2004).
Molecular detection of tyrosine and histidine decarboxylase gene
Results of the multiplex PRC used for the detection of tyrosine and histidine decarboxylase gene (tdc and hdc, respectively) are presented in Fig. 2. Nine out of all fourteen strains (i.e. 65%) of E. faecium found to be positive for the tyrosine decarboxylase gene, i.e. gave two amplicons of 1,100 and 1560 bp (for tdc gene and internal control, respectively) similar to the positive control. Whereas, no strain was found to be positive for hdc gene, i.e. no 440 bp amplicon could be observed. Finally, as expected the negative control only showed amplification of the internal control while the positive control showed amplification of the 3 targets.
Detection of tyrosine decarboxylase (tdc) gene 1,100 bp and histidin decarboxylase gene (hdc) 440 bp by multiplex PCR. 1 = Marker 100 bp, 2 = Negative Control (Water), 3 = KH 24, 4 = DH 28, 5 = RH 31, 6 = RH 33, 7 = RH 38, 8 = DH 56, 9 = KH 79, 10 = DH 59, 11 = FH 99, 12 = FH 102, 13 = KH 106, 14 = DH 110, 15 = KH 115, 16 = FH 133, 17 = E. faecalis NCDC 114 (positive control): 18 = L. lactis subspp. cremoris NCDC 61 (negative control), 19 = Marker 500 bp
Previously, we have also been reported that there is a 100% correlation between the biochemical method, molecular method (PCR) and HPLC methods (Bhardwaj et al. 2009b). Thus, in the present study a multiplex PCR method was used for the detection of tdc and hdc gene. In the present study tyramine was the only biogenic amine produced by the E. faecium strains. Similar, kind of results were also observed by several workers (Yousif et al. 2005; Perez-Pulido et al. 2006; Psoni et al. 2006; Serio et al. 2007). With respect to the number of tested enterococcal strains found to be positive for tdc gene, our results (i.e. 65% strains were positive for tdc) were found to be quite similar to the result observed by Psoni et al. (2006) who reported that tyrosine decarboxylase activity was exhibited by 75% of the strains after 72 h of incubation and 88.9% strains after 7 days.
Antibiotic susceptibility profile
The results of antibiogram of the all 14 E. faecium is shown in Table 4. Among all the selected strains not even a single strain was found to be sensitive or resistant to all the tested antibiotics. Mostly all the tested strains of E. faecium were found to be sensitive to Bacitracin, Ciprofloxacin, Tetracyclin, Ampicilin, Nitrofurantoin and Teicoplanin except a few strains which showed moderate sensitivity towards one or more antibiotics. Only two strains (E. faecium KH 106), was found to be resistant to amplicillin. Not a single tested strain showed resistance towards vancomycin. Besides this, most of the tested strains showed resistance towards Polymyxin B, Neomycin and Rifampicin.
In vitro probiotic attributes
Acid tolerance
The viability of 14 bacteriocinogenic E. faecium strains under different pH conditions is presented in Table 5. Out of 14 isolates, only FH 99 showed viability of about 2.4 ± 0.2 at pH 2.0 even after 120 min. At pH 2.5, most of the strains showed a progressive reduction in viable counts, while at pH 3.0, 12 out of 14 strains showed a large variation from 2 to 4 log cycles. Thus, among all E. faecium FH 99, however, was found to be relatively better survivor to acidic conditions as compare to others in the selected pH range.
The pH of gastric juice (2.0–3.0) in stomach generally causes destruction of most of the ingested microorganisms (Charteris et al. 1998) and hence is one of the first major physiological challenges faced by probiotic cultures upon oral administration. In this sense, tolerance to acidic conditions is an important selection criterion for probiotic organisms. Reports related to the in vitro acid tolerance, are scarce in case of enterococci. Lewenstein et al. (1979) studied the probiotic attributes of a commercially exploited E. faecium SF 68, and showed only 66% survival at pH 3.0 and 40% survival in pH 2.5 in KH2PO4–HCl solution after 60 min of incubation. Gardiner et al. (1999) had shown that a probiotic strain of E. faecium Fargo 688® could survive the porcine gastric juice at pH 2.0 only for 8 min. Similarly, Strompfova et al. (2004) observed that 57% E. faecium strains showed appreciable survival at pH 2.0 and pH 3.0 in the range between 76 and 87% after 3 h. Recently, Ruiz-Moyano et al. (2008) reported that strains of E. faecium showed had good survival rate after exposure at pH 2.5.
Bile tolerance
The bile salt tolerance pattern of the 14 isolates is presented in Table 6. There was almost no variation in behavior of different strains for varying concentration of bile. Almost all strains were able to tolerate 1, 2 and 3% bile concentrations showing even less than 2 log cycle reduction in their cell counts (Table 6). Hence, it may be concluded that enterococci use bile as a source of nutrients for their survival.
Once bacteria reach the small intestinal tract, their ability to survive depends on their resistance to bile acids (Gilliland et al. 1984). Bile acids synthesized in liver from cholesterol (500–700 ml/d) entering the duodenal section of the small intestine have been reported to reduce the survival of bacteria (Jin et al. 1998). Thus, the success of a probiotic organism also depends on its bile-tolerance characteristics. Enterococci are well known to be commensals of the gastrointestinal tract of human and animals, and in this ecological niche, these bacteria come in contact and interact with bile salts (Franz et al. 2001b). Thus, it is not surprising to find Enterococcus spp. resistant to bile acid. Several reports were also state that E. faecium strains are strongly resistant to bile conditions (Wijaya 2002; Strompfova et al. 2004; Harun-ur-Rashid et al. 2007; Ruiz-Moyano et al. 2008) indicating the potentiality of E. faecium group to overcome the gastrointestinal environment.
Cell surface hydrophobicity
The 14 strains of E. faecium under study were evaluated for their cell surface hydrophobicity (CSH) towards the three different hydrocarbons, i.e. n-hexadecane, n-octane and xylene (Table 7). E. faecium KH 24, DH 59 and RH 106 showed relatively more affinity for n-hexadecane than the rest of the hydrocarbons as the maximum percent hydrophobicity values observed for the three strains were in range of 9.5–8.5%. In case of xylene, maximum hydrophobicity was observed with RH 38 (91.0%) than the rest. Similarly, for n-octane, a higher affinity was depicted by strains RH 38, DH 56 and FH 99 (Table 7).
It may be observed from these results that the strains showed maximum adherence towards xylene while lowest towards n-hexadecane. The variation in hydrophobicity to solvents has been reported in other probiotic bacteria also and has been explained by the fact that adhesion depends upon the origin of strains as well as surface properties (Morata De Ambrosini et al. 1998). The hydrophobic nature of the outermost surface of microorganism has been implicated in the attachment of bacteria to host tissue (Ljungh and Wadstom 1982; Kiely and Olson 2000) and hence is an essential feature in order to impart beneficial effects to the host. The information regarding the hydrophobic interactions as well as adherence ability of the enterococcal isolates is very sparse. A study conducted by Wijaya (2002) has indicated that enterococci in general did not have a high hydrophobicity, as 92.9% of E. faecium and 79.2% of the tested E. faecalis strains showed weak hydrophobicity (0–30%) values for n-hexadecane depending on their source. In our study also, three best strains (DH 56, DH 59 and FH 99) exhibited hydrophobicity values in the range of (10–87%) for all the three organic solvents viz., n-hexadecane, xylene and n-octane.
Bile salt hydrolase activity
All the 14 selected E. faecium strains grow well on medium supplemented with 0.5% conjugated bile salts. None of them showed deconjugation of primary salt, i.e. taurocholate (TC). However, among them, 12 strains (except RH 31 and KH 79) showed positive results against secondary bile salt namely taurodeoxycholate (TDC), with zone of precipitation differing in size (Table 8). The positive E. faecium strains were further selected for their quantitative estimation of BSH activity against TDC. BSH activity was expressed in terms of AU/ml. All the 12 tested cultures showed efficient BSH activity, in a variable range between 2.5 and 32.2 AU/ml (Table 8). Wijaya (2002) investigated the incidence of BSH activity among enterococci isolated and reported that 67.4% of E. faecalis, 58.1% of E. faecium and 50% E. durans strains exhibited BSH activity against TCD. Interestingly, none of our E. faecium strains showed detectable deconjugation of TC, whereas they clearly deconjugated DC. Previously, Vinderola and Reinheimer (2003) and Noriega et al. (2006) also observed similar results in case of Bifidobacterium spp.
The precise function(s) of microbial BSH is currently unknown, although several hypotheses have been proposed in this regards such as bile detoxification, gastrointestinal persistence, nutritional role, cholesterol lowering and activation of carcinogens (Begley et al. 2007). Deconjugation of bile salts has been included by World Health Organization (WHO) experts as one of the main activities of intestinal microbiota for them to be considered as probiotic microorganisms (FAO/WHO 2002).
Cholesterol assimilation from growth media
Cholesterol assimilation ability from growth media of all the 14 selected bacteriocinogenic strains of E. faecium is presented in Table 8. The assimilated cholesterol in the range from 1.42% (for KH 79) to 61.36% (for DH 59). Interestingly, strains RH 38 and FH 102 which did not assimilate as much cholesterol as the other cultures, were among the most active in deconjugating sodium taurodeoxycholate.
There are very few reports on the in vitro cholesterol removal by Enterococcus spp. as compared to other LAB. Rossi et al. (1999) also reported that E. faecium used as a single or mixed starter was able to reduce cholesterol in vitro. It has been found that E. faecalis assimilated 1.5 times more cholesterol than the other LAB strains (Pereira and Gibson 2002). A high level of cholesterol in blood is generally considered to be a risk factor for coronary heart disease. From these results, it may be suggested that these strains have an excellent hypocholesterolemic effect and thus may be use as probiotics to prevent hypercholesterolemia in human health. However, the mechanisms of regulating serum cholesterol and the effect on the serum cholesterol level in vivo animal experiment needs further extensive investigations.
Conclusion
Results of the present investigation also support that enterococci isolated from dairy food and faecal sample are generally free from potential virulence traits and sensitive to clinically relevant antibiotics. Among all tested bacteriocinogenic E. faecium strains, E. faecium FH 99 strain isolated from human faecal that was found to be the safe with outstanding probiotic attributes and positive health effect as it showed a relatively high tolerance to gasterointestinal stress and exhibited significant BSH and cholesterol assimilation activity. Hence, E. faecium FH 99 may offer exciting opportunities in food and dairy sector for its use either as a starter/adjunct culture to produce fermented products. Although further in vivo studies are necessary in order to evaluate its role as a probiotic which may help in strengthening the immune response and lowering the blood cholesterol level.
References
Abriouel H, Omar NB, Molinos AC, Lopez RL, Grande MJ, Martinez-Viedma P, Ortega E, Canamero MM, Galvez A (2008) Comparative analysis of genetic diversity and incidence of virulence factors and antibiotic resistance among enterococcal populations from raw fruit and vegetable foods, water and soil, and clinical samples. Int J Food Microbiol 123:38–49
Andrighetto C, Knijff E, Lombardi A, Torriani S, Vancanneyt M, Kersters K, Swings J, Dellaglio F (2001) Phenotypic and genetic diversity of enterococci isolated from Italian cheeses. J Dairy Res 68(2):303–316
Begley M, Hill C, Gahan CGM (2007) Bile salt hydrolase activity in probiotics. Appl Environ Microbiol 72(3):1729–1738
Bhardwaj A, Malik RK, Chauhan P (2008) Functional and safety aspects of enterococci in dairy foods. Indian J Microbiol 48:317–325
Bhardwaj A, Kapila S, Mani J, Malik RK (2009a) Comparison of susceptibility to opsonic killing by in vitro human immune response of Enterococcus strains isolated from dairy products, clinical samples and probiotic preparation. Int J Food Microbiol 128:513–515
Bhardwaj A, Gupta H, Iyer R, Kumar N, Malik RK (2009b) Tyramine-producing enterococci are equally detected on tyramine production medium, by quantification of tyramine by HPLC, or by tdc gene-targeted PCR. Dairy Sci Technol 89:601–611
Billstroma H, Lunda B, Sullivana A, Norda CE (2008) Virulence and antimicrobial resistance in clinical Enterococcus faecium. Int J Anti Agents 32:374–377
Cariolato D, Andrighetto C, Lombardi A (2008) Occurrence of virulence factors and antibiotic resistances in Enterococcus faecalis and Enterococcus faecium collected from dairy and human samples in North Italy. Food Control 19:886–892
Charteris WP, Kelly PM, Morelli L, Collins JK (1998) Antibiotic susceptibility of potentially probiotic Lactobacillus species. J Food Prot 61:1636–1643
Clinical and Laboratory Standards Institute (CLSI) (2007) Performance standards 636 for antimicrobial disk susceptibility tests, 27(1)
Coton E, Coton M (2005) Multiplex PCR for colony direct detection of Gram-positive histamine- and tyramine-producing bacteria. J Microbiol Methods 63:296–304
Coton M, Coton E, Lucas P, Lonvaud A (2004) Identification of the gene encoding a putative tyrosine decarboxylase of Carnobacterium divergens 508. Development of molecular tools for the detection of tyramine-producing bacteria. Food Microbiol 21:125–130
Dunny GM, Leonard BAB, Hedberg PJ (1995) Pheromone-inducible conjugation in Enterococcus faecalis: interbacterial and host-parasite chemical communication. J Bacteriol 177(4):871–876
Dupre I, Zanetti S, Schito AM, Fadda G, Sechi LA (2003) Incidence of virulence determinants in clinical Enterococcus faecium and Enterococcus faecalis isolates collected in Sardinia (Italy). J Med Microbiol 52:491–498
Eaton TJ, Gasson MJ (2001) Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl Environ Microbiol 67:1628–1635
FAO/WHO (2002) Joint FAO/WHO working group report on drafting guidelines for the evaluation of probiotics in food, London, 30 April and 1 May 2002
Foulquie Moreno MR, Sarantinopoulos P, Tsakalidou E, De Vuyst L (2006) The role and application of enterococci in food and health. Int J Food Microbiol 106:1–24
Franz CMAP, Muscholl-Silberhorn AB, Yousif NMK, Vancanneyt M, Swings J, Holzapfel WH (2001a) Incidence of virulence factors and antibiotic resistance among enterococci isolated from food. Appl Environ Microbiol 67:4385–4389
Franz CMAP, Specht I, Haberer P, Holzapfel WH (2001b) Bile salt activity of enterococci isolated from food: screening and quantitative determination. J Food Prot 64:725–729
Franz CMAP, Stiles ME, Schleifer KH, Holzapfel WH (2003) Enterococci in foods—a conundrum for food safety. Int J Food Microbiol 88:105–122
Galli D, Lottspeich F, Wirth R (1990) Sequence analysis of Enterococcus faecalis aggregation substance encoded by the sex pheromone plasmid pAD1. Mol Microbiol 4:895–904
Gardiner GE, Ross RP, Wallace JM, Scanlan FP, Jagers P, Fitzgerald GF, Collins JK, Stanton C (1999) Influence of a probiotic adjunct culture of Enterococcus faecium on the quality of cheddar cheese. J Agric Food Chem 47(12):4907–4916
Gilliland SE, Walker DK (1990) Factors to consider when selecting a culture of Lactobacillus acidophilus as a dietary adjunct to produce a hypochlosterolemic effect in humans. J Dairy Sci 73:905–911
Gilliland SE, Staley TE, Bush LJ (1984) Importance of bile tolerance of Lactobacillus acidophilus used as dietary adjunct. J Dairy Sci 67:3045–3055
Gilmore MS, Segarra RA, Booth MC, Bogie CP, Hall LR, Clewell DB (1994) Genetic structure of the Enterococcus faecalis plasmid pAD1-encoded cytolytic toxin system and its relationship to antibiotic determinants. J Bacteriol 176:7335–7344
Giraffa G (2002) Enterococci from foods. FEMS Microbiol Rev 26:163–171
Giraffa G (2003) Functionality of enterococci in dairy products. Int J Food Microbiol 88:215–222
Gupta H, Malik RK (2007) Incidence of virulence in bacteriocin-producing enterococcal isolates. Lait 87:587–601
Harun-ur-Rashid M, Togo K, Ueda M, Miyamoto T (2007) Probiotic characteristics of lactic acid bacteria isolated from traditional fermented milk ‘dahi’ in Bangladesh. Pak J Nutr 6(6):647–652
Jin LZ, Ho YW, Abdullah N, Jalaludin S (1998) Acid and bile tolerance of Lactobacillus isolated from chicken intestine. Lett Appl Microbiol 27:183–185
Kayaoglu G, Orstavik D (2004) Virulence factors of Enterococcus faecalis: relationship to endodontic disease. Crit Rev Oral Biol Med 15(5):308–320
Kayser FH (2003) Safety aspects of enterococci from the medical point of view. Int J Food Microbiol 88:255–262
Kiely LJ, Olson NF (2000) The physicochemical surface characteristics of Lactobacillus casei. Food Microbiol 17:277–291
Lewenstein A, Frigerio G, Moroni M (1979) Biological properties of SF 68, a new approach for the treatment of diarrhoeal diseases. Curr Ther Res 26:967–981
Ljungh A, Wadstom T (1982) Salt aggregation test for measuring cell surface hydrophobicity of urinary Escherichia coli. Eur J Clin Microbiol 1:383–393
Morata De Ambrosini V, Gonzalez S, Pesce De Ruiz Holgando A, Oliver G (1998) Study of morphology of cell of some strains of lactic acid bacteria and related species. J Food Prot 61:557–562
Nguyen TDT, Kang JH, Lee MS (2007) Characterization of Lactobacillus plantarum PH04, a potential probiotic bacterium with cholesterol-lowering effects. Int J Food Microbiol 113:358–361
Noriega L, Cuevas I, Margolles A, de los Reyes-Gavila CG (2006) Deconjugation and bile salts hydrolase activity by Bifidobacterium strains with acquired resistance to bile. Int Dairy J 16:850–855
Ogier JC, Serror P (2008) Safety assessment of dairy microorganisms: the Enterococcus genus. Int J Food Microbiol 126:291–301
Omar BN, Castro A, Lucas R, Abriouel H, Yousif NMK, Franz CAMP, Holzapfel WH, Perez-Pulido R, Martinez-Canamero M, Galvez A (2004) Functional and safety aspects of enterococci isolated from different Spanish foods. Syst Appl Microbiol 27:118–130
Pereira DIA, Gibson GR (2002) Effects of consumption of probiotics and prebiotics on serum lipid levels in humans. Crit Rev Biochem Mol Biol 37:259–281
Perez-Pulido R, Abriouel H, Omer BN, Lucas R, Martinez-Canamero M, Galvez A (2006) Safety and potential risks of enterococci isolated from traditional fermented capers. Food Chem Toxicol 44:2070–2077
Pospiech A, Neumann B (1995) A versatile quick-prep of genomic DNA from Gram-positive bacteria. Trends Genet 11:217–218
Psoni L, Kotzamanides C, Andrighetto C, Lombardi A, Tzanetakis N, Litopoulou-Tzanetaki E (2006) Genotypic and phenotypic heterogeneity in Enterococcus isolates from Batzos, a raw goat milk cheese. Int J Food Microbiol 109:109–120
Rosan B, Williams NB (1964) Hyaluronidase production by oral enterococci. Arch Oral Biol 9:291–298
Rosenberg M, Gutnick D, Rosenberg E (1980) Adherence of bacteria to hydrocarbons: a simple method for measuring cell-surface hydrophobicity. FEMS Microbiol Lett 9:29–33
Rossi EA, Vendramini RC, Carlos IZ, Pei YC, de Valdez GF (1999) Development of a novel fermented soymilk product with potential probiotic properties. Eur Food Res Technol 209:305–307
Rudel LL, Morris MD (1973) Determination of cholesterol using O-phthaladehyde. J Lipid Res 14:364–366
Ruiz-Moyano S, Martin A, Benito MJ, Nevado FP, de Guia Cordoba M (2008) Screening of lactic acid bacteria and bifidobacteria for potential probiotic use in Iberian dry fermented sausages. Meat Sci 80:715–721
Sanchez J, Basanta A, Gomez-Sala B, Herranz C, Cintas LM, Hernandez PE (2007) Antimicrobial and safety aspects and biotechnological potential of bacteriocinogenic enterococci isolated from mallard ducks (Anas platyrhynchos). Int J Food Microbiol 117:295–305
Serio A, Paparalla A, Chaves-Lopez C, Corsetti A, Suzzi G (2007) Enterococcus populations in Pecorino Abruzzese cheese: biodiversity and safety aspects. J Food Prot 70:1561–1568
Shankar V, Baghdayan AS, Huycke MM, Lindahl G, Gilmore MS (1999) Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect Immun 67:193–200
Strompfova V, Laukova A, Ouweh AC (2004) Selection of enterococci for potential canine probiotic additives. Vet Microbiol 100:107–114
Su YA, Sulavik MC, He P, Makinen KK, Makinen PL, Fiedler S, Wirth R, Clewell DB (1991) Nucleotide sequence of the gelatinase gene (gelE) from Enterococcus faecalis subsp. liquefaciens. Infect Immun 59:415–420
Templer SP, Baumgartner A (2007) Enterococci from appenzeller and schabziger raw milk cheese: antibiotic resistance, virulence factors, and persistence of particular strains in the products. J Food Prot 70(2):450–455
Valenzuela AS, Omar NB, Abriouel H, Lopez RL, Veljovic K, Cañamero MM, Topisirovic MKL, Galvez A (2009) Virulence factors, antibiotic resistance, and bacteriocins in enterococci from artisan foods of animal origin. Food Control 20:381–385
Vancanneyt M, Zamfir M, Devriese LA, Lefebvre K, Engelbeen K, Vandemeulebroecke K, Amar M, De Vuyst L, Haesebrouck F, Swings J (2004) Enterococcus saccharominimus sp. nov., from dairy products. Int J Syst Evol Microbiol 54:2175–2179
Vankerckhoven V, Autgaerden TV, Vael C, Lammens C, Chapelle S, Rossi R, Jabes D, Goossens H (2004) Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. J Clin Microbiol 42(10):4473–4479
Vinderola CG, Reinheimer JA (2003) Lactic acid starters and probiotic bacteria: a comparative “in vitro” study of probiotic characteristics and biological barrier resistance. Food Res Int 36:895–904
Wijaya A (2002) Investigation into the influence of a bacteriocin-producing Enterococcus strain on the intestinal microflora. PhD thesis, An der Fakultät für Chemie und Biowissenschaften der Universität, Karlsruhe
Wilmotte A, Van der Auwera G, De Wachter R (1993) Structure of the 16S ribosomal RNA of the thermophilic cynobacterium chlorogloeopsis HTF (‘Mastigocladus laminosus HTF’) strain PCC7518, and phylogenetic analysis. FEBS Lett 317:96–100
Yousif NMK, Dawyndt P, Abriouel H, Wijaya A, Schillinger U, Vancanneyt M, Swings J (2005) Molecular characterization, techonological properties and safety aspects of enterococci from “Hussuwa”, an African fermented sorghum products. J Appl Microbiol 98:216–228
Acknowledgments
The authors are grateful to Prof. Annalisa Serio (Universita degli Studi di Teramo, Stazione TE, Italy), Prof. Haruyoshi Tomita (Gunma University, Gunma, Japan) and Prof. Vanessa Vankerckhoven (University of Antwerp, Antwerp, Belgium) for gifting E. faecalis ATCC 29212 & E. faecalis Lab 47/2, E. faecalis MMH594 and E. faecalis JH2-SS, respectively.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bhardwaj, A., Kaur, G., Gupta, H. et al. Interspecies diversity, safety and probiotic potential of bacteriocinogenic Enterococcus faecium isolated from dairy food and human faeces. World J Microbiol Biotechnol 27, 591–602 (2011). https://doi.org/10.1007/s11274-010-0494-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-010-0494-4