Abstract
Eleven sourdoughs from Molise region (Southern-Italy) were subjected to microbiological analyses in order to select predominant lactobacilli species to be utilised as starter culture for bread production. A multiple approach was used, consisting of the growth in different culture media, DGGE analysis, 16S rRNA gene sequencing and RAPD-PCR typing. Forty-three lactobacilli were identified and four different species, facultatively or obligately heterofermentative lactobacilli, were found. Lactobacillus plantarum and Lactobacillus brevis represented the prevailing lactobacilli, while Lactobacillus casei and Lactobacillus paracasei ssp. paracasei were detected only in few samples. The use of different media demonstrated that there is no efficient medium for the study of sourdoughs and the cultivation in different substrates remains the best tool to obtain a picture of lactic acid bacteria population. DGGE and 16S rRNA gene sequencing allowed to obtain a reliable identification of strains, while RAPD-PCR resulted a suitable method for typing lactobacilli at strain level.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Many sourdough breads exist in the world and they differ in the type of flour, the added ingredients, the applied technology and the environmental conditions for bread production. These aspects cause the selection of a characteristic microflora that generally contains a complex mixture of lactic acid bacteria (LAB), in association with yeasts. Microbiological studies revealed that more than 50 LAB species have been isolated from this ecosystem, mainly belonging to the genus Lactobacillus (about 30 species) and more than 20 yeast species belonging to the genera Saccharomyces and Candida (Iacumin et al. 2009; De Vuyst and Vancanneyt 2007; Reale et al. 2005; Succi et al. 2003; De Vuyst et al. 2002). However, several works report also the presence of non-Lactobacillus species, such as Enterococcus, Lactococcus, Leuconostoc, Pediococcus, Streptococcus and Weissella (Corsetti and Settanni 2007; Valmorri et al. 2006).
During sourdough fermentation, a selection of microbial population with specific nutrient requirements and growth conditions occurs; lactobacilli, among all the bacteria inhabitant of sourdoughs, are highly adapted to the environmental conditions (temperature, pH, acidity, antimicrobial products, etc.) of sourdough (Vera et al. 2009). For this reason, lactobacilli represent the dominant microbial group and commonly they occur with the highest concentration, especially in mature sourdoughs.
In many cases, subdominant LAB species, such as Enterococcus faecium and Pediococcus pentosaceus, are stronger acidifiers than other species at the beginning of sourdough production. These bacteria, inhibiting indigenous microorganisms by lowering the pH, might prepare the environment for the establishment of the typical microbiota of mature sourdough (Corsetti et al. 2007). When dominant species colonise the matrix, the subdominant species have a rapid decrement and the selective pressures that arise during the sourdough fermentation provoke the predominance of a species respect to other ones (Corsetti et al. 2001, 2007). Numerous works (Galli et al. 1988; Rocha and Malacata 1999; Corsetti et al. 2001; De Vuyst et al. 2002; Valmorri et al. 2006) reported the importance of dominant and subdominant microorganisms occurring in sourdough fermentation but in mature sourdoughs the role of lactobacilli appears to have major relevance for the characterisation of the final baked products. Differences other than acetic acid production in the overall aroma profile of final bread are due to the type of dominating lactobacilli (Corsetti and Settanni 2007). Also, lactobacilli play an important role in the development of the dough structure through the production of exopolysaccharides (EPS) that positively influence the viscosity of sourdough (Vogel et al. 2002). Furthermore, many lactobacilli improve the nutritional quality and healthy aspects of bread and produce antimicrobial substances (Corsetti and Settanni 2007; Reale et al. 2007).
However, the description of the whole diversity of a sourdough LAB consortium remains difficult, often unattainable, and the study of the microbial diversity, in terms of species richness and species abundance, could be underestimated using both culture-dependent and culture-independent methods (Robert et al. 2009). In fact, cultivation-dependent methods have limitations such as antagonism, growth requirements between different organisms and selectivity of the agar media. As reported in literature (De Vuyst and Vancanneyt 2007), lactobacilli cultivation on laboratory media is difficult to achieve and different type of media should be used in order to study the predominant lactobacilli population from a quantitative and qualitative point of view (Corsetti et al. 2001; Catzeddu et al. 2006; Valmorri et al. 2006).
Cultivation-independent methods allow to visualise only predominant species of a microbial community and could be biased by genome size, by the efficiency of DNA extraction or by preferred amplification of certain templates in PCR (Wintzingerode et al. 1997; Meroth et al. 2003).
Moreover, as reported by other Authors (Ercolini 2004; Temmerman et al. 2003; Muyzer and Smalla 1998) the detection limit depends on the species and perhaps even the strain considered. The number and the concentration of the members of the microbial community, along with the nature of the food matrix, all represent variables influencing the detection limit of DGGE by affecting both the efficiency of DNA extraction and the PCR amplification due to the possible competition among templates.
On these bases, the present work was planned with the aim to isolate and identify predominant lactobacilli naturally occurring in sourdoughs of Molise region. Several culture media were used to detect cultivable species, subsequently identified by DGGE and 16S rRNA gene sequencing. Finally, RAPD-PCR allowed to visualise the presence of different biotypes. The objective was to collect strains to be subsequently characterised and used as starter cultures for bread production.
Materials and methods
Samples
Eleven samples of sourdoughs were collected from 11 different artisanal bakeries located in various areas of Molise region. All the samples were type I sourdoughs, according to the description of Corsetti and Settanni (2007). Sourdoughs were produced after regular propagation by backslopping at 20–28°C to keep the microorganisms in an active state and were collected before the last refreshment, according to the artisanal producers.
Microbiological analysis and pH measurement
Ten grams of each sourdough was diluted in 90 ml of physiological sterile solution (9 g/L NaCl) and homogenised in a Stomacker 400 Lab Blender (PBI International, Milan, Italy) (1 min agitation, 1 min pause, 1 min agitation). As suggested by other Authors (De Vuyst and Vancanneyt 2007), several culture media were used for LAB enumeration and isolation. Briefly, LAB were enumerated and isolated by plating serial decimal dilutions on MRS agar medium (Oxoid, Milan, Italy) adding 40 mg/L actidione, on modified MRS containing 1% maltose, 5% fresh yeast extract, pH 5.5 (MRS-M), on SDB agar medium (Kline and Sugihara 1971) and on Rogosa agar (Oxoid, Milan, Italy). Plates were incubated at 28°C for 72 h under anaerobic conditions using an anaerobic system (Anaerogen, Oxoid, Milan, Italy). Three colonies randomly picked from each assayed medium, were obtained from plates with the highest dilution having positive growth, following the procedure described by Valmorri et al. (2006). Isolates were than purified by streaking on the fresh medium and incubated as above. After morphological examination, presumptive lactobacilli were maintained frozen at −80°C in MRS medium with 15% glycerol. Yeasts were counted in YPD Agar plates (10 g/L yeast extract, 20 g/L bacteriological peptone, 20 g/L glucose, 20 g/L agar) after incubation at 28°C for 72 h.
For each sourdough, pH was measured by a spin electrode pH-meter (Crison model 2001).
Identification of lactobacilli
Gram staining, catalase testing and microscope observation were used to screen the isolates and to presumptively identify those belonging to the Lactobacillus genus. Lactobacilli were then identified by PCR-DGGE and 16S rRNA gene sequencing and biotyped by RAPD-PCR.
DNA extraction and purification from pure culture
Two milliliters of each overnight culture was centrifuged at 14,000g for 10 min at 4°C to pellet the cells and the pellet was subjected to DNA extraction according to Querol et al. (1992), with the addition of lysozyme (25 mg/ml, Sigma) and mutanolysin (10 U/ml, Sigma) for bacterial cell-wall digestion. Quantity and purity of the DNA were assessed by optical reading at 260 and 280 nm, as described by Sambrook et al. (1989).
DGGE analysis
The DNA from each strain was prepared for DGGE by amplifying the V1 region of 16S rRNA using the following primers: P1V1 (5′-GCG GCG TGC CTA ATA CAT GC-3′) (Cocolin et al. 2001) and P2V1 (5′-TTC CCC ACG CGT TAC TCA CC-3′) (Rantsiou et al. 2005). A GC clamp (5′- CGC CCG CCG CGC CCC GCG CCC GTC CCG CCG CCC CCG CCC G-3′) (Sheffield et al. 1989) was attached to the 5′ end of the P1V1 primer. PCR was performed in a Mastercycler gradient (Eppendorf, Hamburg, Germany). The reaction mixture (50 μl) consisted of 10 mmol/L Tris–HCl (pH 8.3), 50 mmol/L KCl, 200 μmol/L of each dATP, dGTP, dCTP and dTTP, 1.5 mmol/L MgCl2, 0.2 μmol/L of each primer, 200 ng DNA and 1.25 U Taq-DNA polymerase (Finnzymes, Finland). The amplification program consisted of a 1 min denaturation step at 95°C, a 1 min annealing step at 45°C and a 1 min extension step at 72°C. The first cycle was preceded by an initial step at 95°C for 5 min. After 35 cycles, there was a final 7 min extension step at 72°C. Negative controls without DNA template were included in parallel. PCR products were separated in 1.5% (w/v) agarose gel (Sigma) by electrophoresis for 45 min at 120 V in TBE 0.5× (Sigma) and were subsequently visualised by UV illumination after ethidium bromide (50 μg/ml) staining (Sigma).
PCR products obtained from amplification of V1 region of 16S rRNA were subjected to DGGE analysis, using a DCode Universal Mutation Detection System (BioRad, Hercules, CA, USA). Electrophoresis was performed in a 0.8-mm polyacrylamide gel (8% [w/v] acrylamide-bisacrylamide [37.5:1]) by using two different ranges of denaturant to optimise separation of the products. Two denaturant gradients, from 40 to 60% (100% denaturant was 7 M urea plus 40% [w/v] formamide) increasing in the direction of electrophoresis run, were used. The gels were subjected to a constant voltage of 120 V for 5 h at 60°C, and after electrophoresis they were stained for 20 min in 1.25× TAE containing 50 μg/ml ethidium bromide and visualised under UV illumination.
DGGE gels were digitally captured by GEL DOC XR System (Bio-Rad, Hercules, CA, USA) using the software Quantity One Analysis (Bio-Rad) and analysed with the pattern analysis software package, Gel Compare II Version 2.0 (Applied Maths, Kortrijk, Belgium). Calculation of similarities in the profiles of bands was based on Pearson product-moment correlation coefficient. Dendrograms were obtained by mean of the Unweighted Pair Group Method using Arithmetic Average (UPGMA) clustering algorithm (Vauterin and Vauterin 1992).
Sequence analysis
One to four representative strains of each cluster obtained by DGGE analysis were amplified with primers P1 and P4, as described by Klijn et al. (1991), targeting 700 bp of the V1–V3 region of the 16S rRNA gene. After purification, (QIAquick PCR purification kit, QIAGEN GmbH, Hilden), products were sent to a commercial facility for sequencing (Eurofins MWG Biotech Company, Ebersberg, Germany). Sequences were aligned with those in GeneBank with the Blast program (Altschul et al. 1997) to determine the closest known relatives, based on the partial 16S rRNA gene homology.
RAPD-PCR
Amplification reactions were performed in a 25 μl reaction volume containing 10 mmol/L Tris–HCl (pH 8.3), 50 mmol/L KCl, 200 μmol/L of each dATP, dGTP, dCTP and dTTP, 1.5 mmol/L MgCl2, 1 μmol/L primer, 80 ng DNA and 1.25 U Taq-DNA polymerase (Finnzymes, Finland). Amplifications were performed in a Mastercycler gradient (Eppendorf, Hamburg, Germany) using the following primers and amplification conditions: (a) M13: 5′GAGGGTGGCGGTTCT 3′ (Huey and Hall 1989); the amplification was carried out for 35 cycles of 94°C for 1 min, 40°C for 20 s, ramp to 72°C at 0.5°C/s, 72°C for 2 min; (b) D8635: 5′GAGCGGCCAAAGGGAGCAGAC 3′ (Akopyanz et al. 1992); after an initial step of 94°C for 2 min the amplification was performed for 35 cycles of 94°C for 1 min, 42°C for 1 min, 72°C for 1 min and 30 s, and a final step at 72°C for 10 min.
The amplification products were separated by electrophoresis on 1.5% (w/v) agarose gel (Sigma-Aldrich, Steinheim, Germany) in 0.5× TBE buffer and then subjected to ethidium bromide staining.
RAPD-PCR gels were digitally captured and analysed as previously described for DGGE analysis.
Results and discussion
The main characteristics of the samples and the number of isolates recovered from the different sourdoughs are reported in Table 1. The sourdoughs were characterised by pH ranging from 3.65 (sample B) to 4.14 (sample M). These values closely respected those of typical ripe sourdoughs traditionally produced in Molise (Reale et al. 2005; Succi et al. 2003; Iorizzo et al. 1995). Yeast/LAB ratio was 1:100 or 1:10 in almost all the samples, reflecting the typical proportion existing in mature sourdoughs (Corsetti and Settanni 2007; Corsetti et al. 2001; Gobbetti et al. 1994).
Counts registered on the four media used for LAB cultivation highlighted very different results in microbial load. For instance, LAB counts were higher in MRS medium for samples B, Q, R and T, whereas samples A, D, L and N showed the highest counts in SDB medium. MRS plus maltose (MRS-M) allowed to register the best counts for samples M and W. The sole sample R showed comparable counts in all the media used. These results confirm the importance to use different media in combination. As reported by a recent study on type I sourdough (Vera et al. 2009), microbial population of sourdough can be evaluated through cultivation on agar medium, but there is no universal medium enabling to cover the complexity of sourdough biodiversity; if the MRS with maltose addition would seem more appropriate to estimate the lactobacilli population (Vera et al. 2009), in our study allowed the highest lab growth only for two samples, as reported above.
Isolation at the highest dilution from different culture media was used to achieve an approximate estimation of the dominant sourdough lactobacilli species.
Three colonies were isolated from each medium, excluding those with a number of colonies <10 CFU/g. Isolation was performed at the highest dilution to achieve an approximate estimation of the dominant sourdough lactobacilli species.
Out of 87 isolates, 44 Gram-positive, catalase-negative and rod-shaped microorganisms were presumptively identified as lactobacilli (data not shown) and were subjected to DGGE identification and RAPD-PCR characterisation.
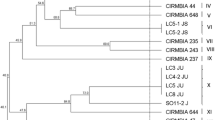
DGGE analysis allowed to obtain the dendrogram shown in Fig. 1. The strains were grouped, according to the migration profile, in eleven clusters and, for each cluster, one to four strains were subjected to sequencing for identification purpose. The results of the sequencing analysis, reported in Table 2, allowed the identification of 14 out of 15 selected strains (marked with an asterisk in Fig. 1).
Combining these results with those obtained from DGGE cluster analysis, it was possible to identify 43 out of 44 strains. In fact, the sole strain D1 resulted an uncultured bacterium clone by sequencing analysis (see Table 2) and clustered alone in the DGGE dendrogram (Fig. 1). In detail, 22 strains were identified as Lb. plantarum, 17 as Lb. brevis, 2 as Lb. casei and 2 as Lb. paracasei ssp. paracasei.
On the basis of the DGGE results, strains belonging to the same species were characterised by RAPD-PCR. Profiles obtained with primers M13 and D8635 highlighted not only a strong biodiversity among strains from the same species but also among strains—from the same species—isolated from the same sourdough (Fig. 2). RAPD-PCR technique revealed, as reported also by other Authors (Andrighetto et al. 2004; Catzeddu et al. 2006), to be a good tool for typing lactobacilli and for obtaining information on genetic diversity. In detail, we found a stronger biodiversity between Lb. plantarum strains than that observed between Lb. brevis strains. In fact, considering the profiles of bands and results given by the cluster analysis, at the least 13 different biotypes were individuated for the former and only 7 for the latter. Also, a great number of strains seemed to be genetically related even if they were isolated from sourdoughs produced in different bakeries and this fact was more evident for Lb. brevis strains. Because a great number of variables can differently affect the microbiota of mature sourdoughs, first of all specific technological parameters of production, very likely it can be assumed that strains genetically related, even if isolated from different doughs, could derive from the same flour probably used by a certain number of bakeries which were studied. In fact, as reported by De Vuyst et al. (2009) the type and the quality of the cereal flour used is indeed the source of autochthonous LAB and plays a key role in establishing stable microbial consortia within a short time. In addition it has been shown that previous introduction of flour into the bakery environment helps to build up a house microbiota that may serve as an important inoculum for subsequent sourdough fermentation, as sourdough and bakery environment isolates are genetically indistinguishable.
Lactobacillus brevis, Lb. plantarum, Lb. paralimentarius, Lb. rossiae and Lb. sanfranciscensis are considered the main species dominating sourdough fermentation processes that are characterised by low incubation temperatures and continuous backslopping (De Vuyst et al. 2009). In our study, the main bacterial species isolated from the eleven sourdoughs were Lb. plantarum, detected in almost all samples, and Lb. brevis, found in six samples (Table 3). In detail, on the one hand Lb. plantarum was found as the predominant species in sourdoughs D, M, N, Q, on the other hand it coexisted with Lb. brevis in samples B, R, W and with Lb. brevis and Lb. casei in sourdough L.
These results are in agreement with other studies on the bacterial population in Sicilian (Randazzo et al. 2005), Sardinian (Catzeddu et al. 2006), Apulian (Ricciardi et al. 2005) and Campanian (Coppola et al. 1996) traditional sourdoughs, which also confirmed that Lb. plantarum has a high prevalence. Iacumin et al. (2009) examined the microbial population of four sourdoughs and found that Lb. plantarum and Lb. brevis are the dominant species occurring in traditional Italian sourdoughs and Lb. plantarum was isolated singly or often in association with Lb. brevis. A similar composition in lactobacilli species was found in the sourdoughs used for the production of Altamura bread, in which 88% of isolates were identified as Lb. plantarum, Lb. casei and Lb. paracasei ssp. paracasei, 12% as Lb. brevis and Leuc. mesenteroides (Ricciardi et al. 2005).
Lb. sanfranciscensis, described as the dominant bacterial species in many traditional Italian baked products (Gobbetti et al. 1994; Vogel et al. 1994; Foschino et al. 2001) was not found in our samples, even if we used SDB and MRS-M media, highly selective substrates to isolate this species, and even though temperature of incubation, pH of the dough and type of technology applied were positive factors of influence on its prevalence.
Conclusion
In conclusion, the present work confirms previous findings, showing that Lb. brevis and Lb. plantarum, occurring singly or in association with other lactic acid bacteria, are the main lactobacilli found in the type I sourdoughs used in Southern Italy.
If DGGE analysis and 16S rRNA gene sequencing result a suitable multiple approach to identify lactobacilli from sourdough, RAPD-PCR analysis represents an important tool in order to individuate genotypic differences between strains from the same species. In fact, RAPD-PCR highlighted a strong biodiversity among strains belonging to the same species and isolated from the same dough. This evidence should be considered as a feature which positively influences the characteristics of natural sourdoughs. This fact suggests that, in order to create a starter culture for traditional bread production, several strains from the same species should be used, with a prevalence of Lb. plantarum and Lb. brevis and a minor presence of other LAB species frequently occurring in traditional sourdoughs. This work represents the start point to select different biotypes which will be assayed for their technological aptitude.
References
Akopyanz N, Bukanov NO, Westblom TU, Kresovich S, Berg DE (1992) DNA diversity among clinical isolates of Helicobacter pylori detected by PCR based RAPD fingerprinting. Nucleic Acids Res 20:5137–5142
Altschul SF, Madden TL, Shaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Andrighetto C, Marcazzan G, Lombardi A (2004) Use of RAPD-PCR and TTGE for the evaluation of biodiversity of whey cultures for Grana Padano cheese. Lett Appl Microbiol 38:400–405
Catzeddu P, Mura E, Parente E, Sanna M, Farris GA (2006) Molecular characterization of lactic acid bacteria from sourdough breads produced in Sardinia (Italy) and multivariate statistical analyses of results. Syst Appl Microbiol 29:138–144
Cocolin L, Manzano M, Cantoni C, Comi G (2001) Denaturing gradient gel electrophoresis analysis of the 16S rRNA gene V1 region to monitor dynamic changes in the bacterial population during fermentation of Italian sausages. Appl Environ Microbiol 67:5113–5121
Coppola S, Pepe O, Masi P, Sepe M (1996) Characterization of leavened doughs for pizza in Naples. Adv Food Sci 18:160–162
Corsetti A, Settanni L (2007) Lactobacilli in sourdough fermentation. Fd Res Int 40:539–558
Corsetti A, Lavermicocca P, Morea M, Baruzzi F, Tosti N, Gobbetti M (2001) Phenotypic and molecular identification and clustering of lactic acid bacteria and yeasts from wheat (species Triticum durum and Triticum aestivum) sourdoughs of southern Italy. Int J Food Microbiol 64:95–104
Corsetti A, Settanni L, Valmorri S, Mastrangelo M, Suzzi G (2007) Identification of subdominant sourdough lactic acid bacteria and their evolution during laboratory-scale fermentation. Food Microbiol 24:592–600
De Vuyst L, Vancanneyt M (2007) Biodiversity and identification of sourdough lactic acid bacteria. Food Microbiol 24:120–127
De Vuyst L, Schrijvers V, Paramithiotis S, Hoste B, Vancanneyt M, Swings J, Kalantzopoulos G, Tsakalidou E, Messens W (2002) The biodiversity of lactic acid bacteria in Greek traditional wheat sourdoughs is reflected in both composition and metabolite formation. Appl Environ Microbiol 68:6059–6069
De Vuyst L, Vrancken G, Ravyts F, Rimaux T, Weckx S (2009) Biodiversity, ecological determinants, and metabolic exploitation of sourdough microbiota. Food Microbiol 26:666–675
Ercolini D (2004) PCR-DGGE fingerprinting: novel strategies for detection of microbes in food. J Microbiol Met 56:297–314
Foschino R, Arrigoni C, Picozzi C, Mora D, Galli A (2001) Phenotypic and genotypic aspects of Lactobacillus sanfranciscensis strains isolated from sourdoughs in Italy. Food Microbiol 18:277–285
Galli A, Franzetti L, Fortina MG (1988) Isolation and identification of sourdough microflora. Microbiol Aliment Nutr 6:345–351
Gobbetti M, Corsetti A, Rossi J, La Rosa F, De Vincenzi S (1994) Identification and clustering of lactic acid bacteria and yeast from wheat sourdough of central Italy. Italian J Food Sci 6:85–94
Huey B, Hall J (1989) Hypervariable DNA fingerprinting in Escherichia coli. Minisatellite probe from bacteriophage M13. J Bacteriol 171:2528–2532
Iacumin L, Cecchini F, Manzano M, Osualdini M, Boscolo D, Orlic S, Comi G (2009) Description of the microflora of sourdoughs by culture-dependent and culture-independent methods. Food Microbiol 26:128–135
Iorizzo M, Coppola R, Sorrentino E, Grazia L (1995) Caratterizzazione microbiologica di paste acide molisane. Ind Aliment 34:1290–1294
Klijn N, Weerkamp AH, de Vos WM (1991) Identification of mesophilic lactic acid bacteria by using polymerase chain reaction-amplified variable regions of 16S rRNA and specific DNA probes. Appl Environ Microbiol 57:3390–3393
Kline L, Sugihara TF (1971) Microorganisms of the San Francisco sour dough bread process. Appl Microbiol 21:459–465
Meroth CB, Walter J, Hertel C, Brandt MJ, Hammes WP (2003) Monitoring the bacterial population dynamics in sourdough fermentation processes by using PCR-denaturing gradient gel electrophoresis. Appl Environ Microbiol 69:475–482
Muyzer G, Smalla K (1998) Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie van Leeuwenhoek 73:127–141
Querol A, Barrio E, Ramon D (1992) A comparative study of different methods of yeast strains characterization. Syst Appl Microbiol 15:439–446
Randazzo CL, Heilig H, Restuccia C, Giudici P, Caggia C (2005) Bacterial populations in traditional sourdough evaluated by molecular methods. J Appl Microbiol 99:251–258
Rantsiou K, Urso R, Iacumin L, Cantoni P, Cattaneo G, Comi G, Cocolin L (2005) Culture dependent and–independent methods to investigate the microbial ecology of Italian fermented sausage. Appl Environ Microbiol 71:1977–1986
Reale A, Tremonte P, Succi M, Sorrentino E, Coppola R (2005) Exploration of lactic acid bacteria ecosystem of sourdoughs from the Molise region. Ann Microbiol 55:17–22
Reale A, Konietzny U, Coppola R, Sorrentino E, Greiner R (2007) The importance of lactic acid bacteria for phytate degradation during cereal dough fermentation. J Agric Food Chem 55:2993–2997
Ricciardi A, Parente E, Piraino P, Paraggio M, Romano P (2005) Phenotypic characterization of lactic acid bacteria from sourdoughs for Altamura bread produced in Apulia (Southern Italy). Int J Food Microbiol 98:63–72
Robert H, Gabriel V, Fontagné-Faucher C (2009) Biodiversity of lactic acid bacteria in French wheat sourdough as determined by molecular characterization using species-specific PCR. Int J Food Microbiol 135:53–59
Rocha MJ, Malacata XF (1999) On the microbiological profile of traditional Portuguese sourdough. J Food Proteomics 62:1416–1429
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, New York, USA
Sheffield VC, Cox DR, Lerman LS, Myers RM (1989) Attachment of a 40-base pairs G+C rich sequence (GC clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc Natl Acad Sci USA 86:297–303
Succi M, Reale A, Andrighetto C, Lombardi A, Sorrentino E, Coppola R (2003) Presence of yeasts in Southern Italian sourdoughs from Triticum aestivum flour. FEMS Microbiol Lett 225:143–148
Temmerman R, Scheirlinck I, Huys G, Swings J (2003) Culture-independent analysis of probiotic products by denaturing gradient gel electrophoresis. Appl Environ Microbiol 69:220–226
Valmorri S, Settanni L, Suzzi G, Gardini F, Vernocchi P, Corsetti A (2006) Application of a novel polyphasic approach to study the lactobacilli composition of sourdoughs from the Abruzzo region (central Italy). Lett Appl Microbiol 43:343–349
Vauterin L, Vauterin P (1992) Computer-aided objective comparison of electrophoretic patterns for grouping and identification of microorganisms. Eur Microbiol 1:37–41
Vera A, Rigobello V, Demarigny Y (2009) Comparative study of culture media used for sourdough lactobacilli. Food Microbiol 26:728–733
Vogel RF, Böcker G, Stolz P, Ehrmann M, Fanta D, Ludwig W, Pot B, Kersters K, Schleifer KH, Hammes WP (1994) Identification of lactobacilli from sourdough and description of Lactobacillus pontis sp. nov. Int J Sys Bacteriol 44:223–229
Vogel RF, Ehrmann MA, Gänzle MG (2002) Development and potential of starter lactobacilli resulting from exploration of the sourdough ecosystem. Antonie Leeuwenhoek 81:631–638
Wintzingerode F, Gobel UB, Stackebrandt E (1997) Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev 21:213–229
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Reale, A., Di Renzo, T., Succi, M. et al. Identification of lactobacilli isolated in traditional ripe wheat sourdoughs by using molecular methods. World J Microbiol Biotechnol 27, 237–244 (2011). https://doi.org/10.1007/s11274-010-0448-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-010-0448-x