Abstract
The thermal and alkaline pH stability of Streptomyces lividans xylanase B was improved greatly by random mutagenesis using DNA shuffling. Positive clones with improved thermal stability in an alkaline buffer were screened on a solid agar plate containing RBB-xylan (blue). Three rounds of directed evolution resulted in the best mutant enzyme 3SlxB6 with a significantly improved stability. The recombinant enzyme exhibited significant thermostability at 70°C for 360 min, while the wild-type lost 50% of its activity after only 3 min. In addition, mutant enzyme 3SlxB6 shows increased stability to treatment with pH 9.0 alkaline buffer. The K m value of 3SlxB6 was estimated to be similar to that of wild-type enzyme; however k cat was slightly decreased, leading to a slightly reduced value of k cat/K m, compared with wild-type enzyme. DNA sequence analysis revealed that eight amino acid residues were changed in 3SlxB6 and substitutions included V3A, T6S, S23A, Q24P, M31L, S33P, G65A, and N93S. The stabilizing effects of each amino acid residue were investigated by incorporating mutations individually into wild-type enzyme. Our results suggest that DNA shuffling is an effective approach for simultaneous improvement of thermal and alkaline pH stability of Streptomyces lividans xylanase B even without structural information.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Enzymes are remarkable catalysts, which can exclusively work on their substrate(s) in mild conditions, discharging little waste. The use of enzymes is considered a great alternative for industrial chemical processes, as environmental problems and the demands for new chemicals rise. However, low stability, narrow substrate specificity and high production costs often limit broad applications of enzymes. Hence improvement of enzyme properties for application to industrial processes or other fields has been a major goal of protein engineering. Despite significant advances in related areas, knowledge-based design of enzymes/proteins with desired properties has not been accomplished in many cases because the current understanding of structure–function relationships is limited. Over the years, The directed evolution method has been proven to be a much more powerful protein engineering tool than conventional rational or random chemical mutagenic approaches (Stemmer 1994; Gonzalez-Blasco et al. 2000; Song and Rhee 2000) and has frequently been used to develop enzymes for such purposes as developing thermostability, substrate specificity (Iffland et al. 2001; Suenaga et al. 2001; Meyer et al. 2002) and organic solvent tolerance (Moore and Arnold 1996; You and Arnold 1996). This approach could overcome unexpected side effects on the structure and function of the target protein that may result from rational design, and can thus be applied to finding novel mutants.
Xylan is a major component of hemicellulose in plant cell walls. It is covalently and noncovalently attached to cellulose, lignin, pectin, and other polysaccharides to maintain cell wall integrity (Hori and Elbein 1985). β-Xylanase (EC 3.2.1.8) hydrolyzes β-1,4-glycosidic linkages within the xylan backbone to yield short-chain xylo-oligosaccharides of varying length. On the basis of primary structure homology, the majority of xylanases have been classified into glycoside hydrolase families 10 and 11 (GH10 and GH11, respectively) (Henrissat and Bairoch 1993). Their useful industrial applications include the conversion of lignocellulosic material to fuels and chemicals (Coughlan and Hazlewood 1993) and the processing of hemicellulose to paper (1992; Eriksson 1990). During the process of pulp bleaching, for example, xylanases have been used instead of chlorine to increase the extractability of lignin for the production of high-quality paper. The use of xylanase to either replace or reduce the amount of chlorine used in pulp bleaching would have a strong positive effect on the environmental impact of the process.
The xylanases used in biobleaching applications are usually family 11 xylanases. This is because a low-molecular-mass enzyme is desirable for fiber penetration, and a lack of cellulase activity is required to maintain pulp yield and strength. The widespread use of xylanase for pulp bleaching, however, has been limited by the high temperature and alkaline pH of pulp-bleaching processes, since most available xylanases are not active under these conditions. There has been a considerable amount of research devoted to identifying new xylanases and improving the properties of wild-types of xylanases to meet the requirements of the pulp-and-paper industry.
Streptomyces lividans xylanase B (SlxB) belongs to family 11 xylanases (Shareck et al. 1991; Biely et al. 1993). The gene encoding SlxB was cloned and expressed in Streptomyces lividans (Shareck et al. 1991). SlxB is a 31 kDa protein comprising an N-terminal catalytic domain and a C-terminal substrate binding domain. In this paper, we describe the bi-directional evolution of SlxB catalytic domain (SlxB-cat) of both thermal and alkaline pH stability. A mutant library was generated by DNA shuffling. The best mutant with improved thermal stability in alkaline solution was screened by using RBB-xylan agar plate after the third round of evolution. Biochemical characteristics of the mutant were investigated. The stabilizing effects of each amino acid mutation in the evolved enzyme were evaluated by site-directed mutagenesis.
Materials and methods
Construction of plasmid pSlxB-cat
SlxB-cat gene from the genomic DNA of Streptomyces lividans 66 mycelium was amplified by using PCR with two primers, SlxB-N (5′-GTGGGATCCGACACGGTCGTCACGACCAACCAGGAGGGC-3′) and SlxB-C (5′-GTGCTGCAG TTACCCGCCGACGTTGATGCTGGAGCTGCC-3′ as N- and C-terminal primers which contain an BamHI and PstI site (underlined), respectively, and the termination codon is in italics. Amplification of DNA was performed using a MiniCycler™ (MJ Research, Watertown, MA, USA) employing the following temperature program: 5 min at 98°C, 25 cycles of 1 min at 98°C, 1 min at 65°C, 1 min at 72°C, followed by 10 min at 72°C. After the first 5 min denaturation step the temperature program was paused, TaKaRa LA Taq™ (1.25 units; Takara Shuzo, Shiga, Japan) was added to the reaction mixture (25 μl), and the temperature program was immediately continued. Using this method, GC-rich target genes were efficiently amplified. The PCR products were subcloned into pCR2.1 (Invitrogen, Carlsbad, CA) and the nucleotide sequences were confirmed with a DNA sequencer (Model 373A, Applied Biosystems, CA, USA). SlxB-cat gene was cleaved by BamHI and PstI, and ligated with pQE30 (Qiagen) using the BamHI and PstI sites to obtain pSlxB-cat.
Random mutagenesis and mutant library construction
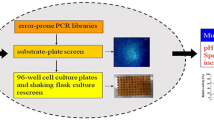
To generate a library of variants of the gene encoding SlxB-cat, we introduced mutations using DNA shuffling. The plasmid pSlxB-cat was digested with DNase I (0.88 unit) at 25°C for 15 min and fragments of 50–100 bp, were extracted from 2% agarose gel, The resultant DNase I digests were mixed and 10 × PCR buffer (Takara Shuzo), MgCl2 (25 mM) and dNTP mixture (2.5 mM each) were added. The subsequent PCR program used was the same as described above, except that annealing was performed at 45°C. The reassembled DNA fragments were amplified by PCR with two primers, SlxB-N and SlxB-C. All PCR procedures were performed using Taq polymerase under the conditions offered by the provider. The amplified DNA fragments were digested with BamHI and PstI, purified by agarose-gel electrophoresis, then ligated into pQE30 between the BamHI and PstI sites. The recombinant plasmids were transformed into Escherichia coli (E. coli) JM109 (Takara Shuzo, Shiga, Japan) and plated on to Luria-Bertani (LB) agar containing 50 μg/ml ampicillin, 20 μg/ml kanamycin and 0.5% 4-O-methyl-d-glucurono-d-xylan-Remazol Brilliant Blue R (RBB-xylan; Sigma). The colonies which were capable of acting on RBB-xylan were picked for screening.
Screening of a mutant library
SlxB variants with improved thermostability in an alkaline solution were primarily screened by using a pH 9.0 alkaline RBB-xylan agar plate. Agar plate contained 0.5% RBB-xylan and 1.5% agar in 50 mM Tris–HCl buffer, pH 9.0. Variants from a mutant library were tooth-picked to an alkaline RBB-xylan agar plate with control colonies. In the first round the RBB-xylan plate was incubated at 60°C for 1 h, then left overnight at room temperature. ‘Successful’ colonies were identified by their ability to form a distinct halo in the RBB-xylan-containing agar. Wild-type SlxB completely lost activity after the above-mentioned steps and no distinct halo occurred on the agar plates. In the second and third round, a mutant library was incubated under conditions where the best mutant selected in the first round lost activity completely: 60°C for 2 h and 3 h, respectively.
Site-directed mutageneses
Site-directed mutageneses were performed, using the ExSite™ PCR-based site-directed mutagenesis kit (Stratagene Cloning Systems, Inc., La Jolla, Calif.). Complementary primers for PCR containing the desired DNA changes (according to the target amino acid position) were constructed and included in reaction mixtures in which 40 ml distilled H2O, 5 ml 10× pfu-turbo polymerase buffer, 1 ml forward primer (125 ng/ml), 1 ml reverse primer (125 ng/ml), 1 ml dNTPs (10 mM), 1 ml plasmid (10–50 ng), and 1 ml pfu-turbo polymerase (2.5 U/ml) were combined in a total reaction volume of 50 ml. Thermocycler settings used was the same as described above, except DpnI (1 ml) was added to the reaction. The resulting plasmids directly from the final reaction mixture were transformed into competent cells of E. coli JM109, and the resultant colonies were DNA sequenced to identify the clones containing the desired substitutions.
Enzyme production and purification
LB-broth (800 ml) supplemented with ampicillin (50 mg/ml) was incubated at 37°C with 80 ml of freshly prepared seed culture of E. coli JM109 harboring recombinant pQE60. Protein production was induced by adding 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to the culture media when the OD of the broth reached to 0.4–0.5 at 660 nm. Cultivation was then continued overnight. Cells were harvested by centrifugation (6,000g, 10 min at 4°C). About 2.5 g of cells (w/w) were subjected to ultrasonication by Sonifier Model 250D (Branson Sonic Power, CT, USA) to extract enzymes, The cell-free lysates were then centrifuged (10,000g, 10 min at 4°C) and each lysate was charged with 1 ml of Ni-NTA resin, which bound to the target proteins. This resin was then packed into 1 ml column and eluted with a linear gradient of 250 mM imidazole in 50 mM Na-phosphate buffer, pH 8.0 at a flow rate of 0.8 ml/min using an FPLC system (Pharmacia, Sweden). The active fractions were combined and dialyzed against 1 l of 20 mM CHES buffer, pH 9.0 for overnight at 4°C. The dialyzed enzyme solutions were reloaded onto a Q-Sepharose column (8 ml) previously equilibrated with either 20 mM CHES buffer, pH 9.0 and the enzymes were eluted with a linear gradient of 0.5 M at a flow rate of 0.5 ml/min. The homogeneity of the purified enzyme fractions was monitored by SDS poly acrylamide gel electrophoresis, and the relevant fractions were pooled and dialyzed against de-ionized water.
Enzyme assay
Crude and purified enzymes were assayed quantitatively using 0.2% of RBB-xylan (Sigma, Germany) as a substrate. Concentrated enzyme solution was diluted with assay buffer, 50 mM McIlvaine, pH 7.0 (a mixture of 0.1 M Citric acid and 0.2 M Na2HPO4), to give a final concentration of 0.05 μg protein/5 μl of solution. About 5 μl of diluted enzyme was applied to a pre-warmed mixture of 20 μl of 50 mM McIlvaine buffer pH 7.0 and 25 μl of 0.4% aqueous solution of RBB-xylan and incubated at 50°C for 15 min. The reaction was stopped by addition of 100 μl of 100% EtOH and kept at room temperature at least for 15 min. The precipitated residual substrate was collected by centrifugation at 12,000g for 2 min and the OD of the supernatant was measured at 595 nm against respective substrate blanks.
Thermal and pH stability
Thermostability was tested by heating purified enzyme samples (10 ± 2 μg of protein/ml) and 50 mM McIlvaine buffer, pH 7.0, at 70°C for various periods of time. The residual activity of the treated enzymes was then measured according to the standard assay method. For determining pH stability, purified enzyme (10 ± 2 μg of protein/ml) were exposed to pH 9.0 level in 50 mM Tris–HCl buffer at 40°C for various periods of time. The residual activities of the treated enzymes were determined.
Analysis of kinetic parameters
Steady-state kinetic parameters were determined as described by Lawson et al. (1996). Soluble birchwood xylan, prepared as described previously (Dupont et al. 1998), was used as substrate. K m and k cat values were determined using substrate concentrations ranging from 0.4 to 10 mg/ml xylan in 30% of 0.2 M McIlvaine buffer, pH 7.0, containing 0.1% BSA. After incubation at 40°C for 30 min, the production of reducing sugar was measured by using p-hydroxybenzoic acid hydrazide (Lever 1972). Protein concentration was measured using DC Protein Assay Reagents (Bio-Rad) with BSA as the standard.
Results
Construction and screening of a mutant library
The recombinant enzyme SlxB-cat consist of 191 amino acid residues and exhibit the same activity as the native enzyme when soluble xylan is used as the substrate. SlxB is known to be composed of a catalytic domain, a binding domain and a linker region (Shareck et al. 1991). It has been reported previously that the removal of the substrate-binding domain from the native enzyme has no effect on the hydrolysis of soluble xylan (Kaneko et al. 2000). SlxB-cat was functionally expressed in E. coli and showed similar properties to those reported for the native enzyme (Kluepfel et al. 1990). Using the random-gene-shuffling technique, the DNA fragments encoding the catalytic domain of SlxB was used in the construction of a mutant library.
From the mutant library generated by the first round of evolution, about 10,000 clones were subjected to the enzyme inactivation steps as described in Materials and methods. For screening of mutant enzyme with higher thermal stability in an alkaline solution than SlxB xylanase, recombinant E. coli colonies produced by the random-gene-shuffling method were placed on to an alkaline RBB-xylan plates (pH 9.0) and treated as described in Material and methods. Theoretically, colonies capable of acting on RBB-xylan after this treatment possessed thermal and alkaline pH stability, because these incubation conditions inactivated SlxB xylanase. In cases where the recombinant enzyme is not stable at 60°C or pH 9.0, only a small halo, due to residual activity, is produced. A small halo may also indicate low specific activity, even if the enzyme is relatively stable at 60°C and pH 9.0. Using this screening method, the variants with improved thermal stability in an alkaline pH range compared to the parental enzyme were easily screened.
Thermal and alkaline pH stability of selected mutants was confirmed by assaying the residual activity after treatment at 70°C and pH 9.0 alkaline buffer for 30 min, respectively, after partial purification. Of the positive variants, 1SlxB18 was found to have the highest thermal stability in pH 9.0 alkaline buffer. In the second round of evolution, the eight best clones from the first round evolution were recombined and used for generation of a mutant library. Colonies were again subjected to the treatment steps under conditions at which the best mutant of the first round completely lost its activity. Screening of 8,600 clones after the enzyme inactivation steps on RBB-xylan plate (pH 9.0, 60°C for 2 h) resulted in a mutant (2SlxB19) with more improved stability than 1SlxB18. After the inactivation steps used for the third round screening, 2SlxB19 show only a small halo on the on RBB-xylan plates. In the same manner, the third round library was generated by using the best 10 clones from the second round library as parental genes. Screening of 6,600 variants on RBB-xylan plate (pH 9.0, 60°C for 3 h) resulted in the best mutant 3SlxB6 possessing the highest thermal and alkaline pH stability compared to that of other positive variants. By a similar procedure, the fourth round mutant library was generated and screened, but no variant with improved thermal stability in pH 9.0 alkaline solution compared to that of 3SlxB6 was found. In this work, 3SlxB6 was finally selected for further study.
To validate the screening system used in this work, the thermal and alkaline pH stabilities of the primary selected mutants from the first round were investigated. As shown in Fig. 1, a large fraction (more than about 80%) of primarily selected variants displayed simultaneously improved thermal and alkaline pH stability compared to that of the wild-type counterpart. From this observation, it is likely that this method on RBB-xylan agar plate is effective for the screening of positive variants with simultaneously improved thermal and pH stability for xylanase.
Thermal and alkaline stability of the primarily screened positive clones from the first round evolution. Thermal and alkaline stability were determined from residual activities after heat (70°C) and alkaline buffer (pH 9.0) treatment for 30 min. Crude extract (1.0 mg/ml protein solution) was used for tests as described in Materials and methods
Thermal and alkaline stability of the evolved 3SlxB6 mutant
Wild-type SlxB and the mutant 3SlxB6 xylanases were purified to homogeneity detected by SDS-PAGE (data not shown) using a nickel-chelating FPLC followed by Q-Sepharose column. The residual activities of both enzymes after heating at 70°C are shown in Fig. 2a. 3SlxB6 was stable at 70°C, even after incubation for 360 min, in contrast with SlxB, which lost 50% of its activity after only 3 min. As for alkaline pH stability, residual enzyme activity was traced in the presence of 50 mM Tris–HCl buffer, pH 9.0 at 40°C for various periods of time, the mutant 3SlxB6 xylanase showed ~95% of the initial activity remained while wild-type enzyme almost completely lost its activity under the same conditions (Fig. 2b). The increment in both was distinctive for the evolved variants 3SlxB6, which confirms that the thermal and alkaline pH stability of xylanase was simultaneously improved by directed evolution.
Thermal and alkaline pH stability curves of purified wild-type and mutant SlxB xylanases. The wild-type and mutant enzymes were heated at 70°C in 50 mM McIlvaine buffer, pH 7.0 (a) or 40°C in 50 mM Tris–HCl buffer, pH 9.0 (b). The residual activity was measured at 40°C with 5 mg/ml soluble xylan in 50 mM McIlvaine buffer, pH 7.0. All data plotted are averages for three independent experiments
Catalytic properties of the evolved 3SlxB6 mutant
Kinetic constants for the wild-type SlxB and mutant 3SlxB6 xylanases are shown in Table 1. The K m value of 3SlxB6 was almost the same as that of wild type, and the k cat was slightly lower than that of wild type (Table 1). The k cat/K m of the 3SlxB6 maintained about 90% of the wild-type enzyme, indicating that the mutant 3SlxB6 was nearly as functional as the wild-type enzyme in performing the hydrolysis of xylan.
Reducing-sugar productivity from soluble xylan
The xylan-hydrolyzing activities of the wild-type and mutant enzymes were determined at various temperatures in pH 7.0 neutral buffer or pH 9.0 alkaline buffer, respectively. The mutant 3SlxB6 xylanase was most active at 75°C in pH 7.0 neutral solution (Fig. 3a) and at 70°C in pH 9.0 alkaline solution (Fig. 3b), 20°C higher than the optimal temperature of wild-type SlxB (55°C in pH 7.0, 50°C in pH 9.0). Remarkably, 3SlxB6 produced seven times more reducing sugar at 70°C than SlxB-cat at 50°C in pH 9.0 alkaline solution, and also three times more reducing sugar at 75°C than SlxB-cat at 55°C in pH 7.0 neutral buffer. This result indicates that the evolved enzyme 3SlxB6 can meet the requirements of biobleaching by its stability and activity under alkaline conditions at high temperatures.
Amino acid substitutions in the mutant 3SlxB6 xylanase
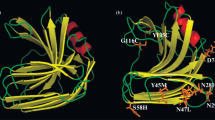
Mutated amino acid residues in 3SlxB6 were identified by sequencing the corresponding gene. As a result, 3SlxB6 was found to possess eight mutations, V3A, T6S, S23A, Q24P, M31L, S33P, G65A, and N93S. To evaluate the contribution of each amino acid change to an increase in thermal and alkaline pH stability, site-directed mutagenesis was conducted for incorporation of each mutation into wild-type enzyme, and the resulting mutants were subjected to stability test. As can be seen in Table 2, S23A, M31L and G65A led to an increase in thermal and alkaline pH stability. In particular, M31L resulted in a significant improvement in both thermal and alkaline pH stability of 3SlxB6. V3A also increased both stabilities to some extent. Especially the mutations G24P and S33P resulted in the highest contribution to thermal stability in the mutations found in 3SlxB6. The other two mutations, T6S and N93S, mainly gave rise to increments in alkaline pH stability rather than thermostability. The thermostability of 3SlxB6 was reinforced as these single thermostabilizing mutations accumulated. However, the cumulative effect on alkaline stability was not observed even though improvement of alkaline pH stability by single mutation was considerable.
Discussion
Directed evolution in vitro is a highly effective strategy in the field of protein engineering. The principle is to remake enzyme gene in vitro under particular evolutionary conditions created artificially in the absence of any knowledge of spatial structure and catalytic mechanism of enzyme by mimicking natural evolution mechanism (random mutation, recombination, and natural selection). Not only can profitable mutations be rapidly accumulated by directed evolution, but also it is a promising approach to combine two or more improved properties evolved separately. In this study we have demonstrated that thermal and alkaline pH stability of xylanase B from Streptomyces lividans can be simultaneously improved by directed evolution. During three subsequent rounds of DNA shuffling, only eight different mutations that led to increased thermal and alkaline pH stability were identified in the 3SlxB6 mutant; these eight mutations accumulated in the final mutant 3SlxB6, which had the highest level of thermal and alkaline pH stability.
Attempts to improve the thermostability of family 11 xylanases have previously been reported (Arase et al. 1993; Wakarchuk et al. 1994), but have been only partly successful. These results showed that the increase in residual activity observed after heating does not always contribute to activity at high temperatures. Although the mutants were thermostable compared with the wild-type, they were less active at low temperatures. It is common for significant conformational changes to occur in enzymes during catalysis (Gerstein et al. 1994). The conformational changes occurring in xylanase from Trichoderma reesei were studied on the basis of crystal structure (Havukainen et al. 1996; Muilu et al. 1998). Structural changes led to the opening and closing of the active site, and this probably plays a role in the function of the enzyme. Therefore amino acid substitution which enhances the thermostability of an enzyme may often change its molecular structure, causing a deterioration in the enzyme activity observed. But our results showed that the k cat values for the purified mutant 3SlxB6 were close to that of wild-type enzyme, indicating that the mutant was as functional as the wild-type enzyme. It is also identified by soluble xylan hydrolysis results, which showed the mutant had an increase in reducing-sugar productivity at high temperatures under pH 9.0 alkaline environment.
In the present study we revealed that significant improvement in thermal and alkaline pH stability was achieved in mutant 3SlxB6 has eight amino acid substitutions, namely Ala3, Ser6, Ala23, Pro24, Leu31, Pro33, Ala65 and Ser93 are predicted to be closely located around the N-terminus. Therefore it is likely that these residues are involved in the structural stability of the N-terminal region, resulting in a significant increase in the half-life of enzymes containing these residues. Site-directed mutagenesis experiments demonstrated that the mutation G24P and S33P resulted in the highest contribution to thermal stability in the mutations found in 3SlxB6. Previous studies revealed that Pro24 is thought to be located between two β-strands, B2 and A2 (Gruber et al. 1998; Torronen and Rouvinen 1997), and seems likely to contribute to structural stability by reducing the entropy of the unfolded state (Schimmel and Flory 1968). Pro33, which is located in the loop between β-strands A2 and A3 (Gruber et al. 1998; Torronen and Rouvinen 1997), is also thought to participate in the stabilization of the enzyme in a similar manner to Pro24. Meanwhile, site-directed mutagenesis experiments also showed that the mutant M31L resulted in a significant improvement in both thermal and alkaline pH stability, and T6S or N93S, mainly gave rise to increments in alkaline pH stability of 3SlxB6. These amino acids change (T6S and N93S) might have reduced the anion–anion repulsion between subunits so that the subunit–subunit interaction was tighter. It should therefore be noted that the significantly enhanced thermal and alkaline pH stability of 3SlxB6 was the result of a cooperative effect of the two different stabilization mechanisms, i.e., the formation of hydrophobic interaction by the M31L substitution and the reduction of anion–anion repulsion by T6S and N93S. Further studies based on site-directed mutagenesis and the three-dimensional structure will elucidate the role of each mutation in the significantly enhanced thermal and alkaline stability.
From the analysis of the mutant library by using alkaline RBB-xylan plate, we found that more than 50% of mutants with an increased thermostability also display improved alkaline pH stability (or vice versa) (data not shown). This observation also can be shown in Fig. 1, it may implies that thermostability is positively correlated with alkaline pH stability in xylanase B from Streptomyces lividans, and these properties could be simultaneously improved. It was found from the mutational analysis that four mutations (V3A, S23A, M31L and G65A) in mutant 3SlxB6 simultaneously increase both thermal and alkaline pH stability of SlxB xylanase, and this supports the above presumption. Previous research had demonstrated that the highly thermophilic family 11 xylanase from Nonomuraea flexuosa had a great number of side chain–side chain polar interactions and several salt bridges (Leskinen et al. 2005). There could be a trend in xylanase structures that acidophilic xylanases have only few and alkalophilic Bacillus circulans xylanase several salt bridges. It is possible that alkaline and thermal adaptation use partly the same mechanisms for improving the stability (Hakulinen et al. 2003).
Conclusion
Our results suggest that DNA shuffling is an effective approach for simultaneous improvement of thermal and alkaline pH stability of SlxB xylanase even without structural information. Three rounds of directed evolution resulted in the best mutant enzyme 3SlxB6 and this evolved enzyme displayed significantly enhanced thermal and alkaline pH stability compared to that of the wild-type counterpart. The evolved enzyme 3SlxB6 is expected to be employed for development of a fully enzymatic process to meet the requirements of the pulp-and-paper industry.
Abbreviations
- SlxB:
-
Streptomyces lividans xylanase B
- GH:
-
Glycoside hydrolase
- LB:
-
Luria-Bertani
- RBB-xylan:
-
4-O-methyl-d-glucurono-d-xylan-Remazol Brilliant Blue R
References
Arase A, Yomo T, Urabe I, Hata Y, Katsube Y, Okada H (1993) Stabilization of xylanase by random mutagenesis. FEBS Lett 316:123–127. doi:10.1016/0014-5793(93)81199-A
Biely P, Kluepfel D, Morosoli R, Shareck F (1993) Mode of action of three endo-beta-1, 4-xylanases of Streptomyces lividans. Biochim Biophys Acta 1162:246–254
Coughlan MP, Hazlewood GP (1993) β-1, 4-D-Xylan-degrading enzyme system: biochemistry, molecular biology and applications. Biotechnol Appl Biochem 17:259–289
Dupont C, Roberge M, Shareck F, Morosoli R, Kluepfel D (1998) Substrate-binding domains of glycanases from Streptomyces lividans: characterization of a new family of xylan-binding domains. Biochem J 330:41–45
Eriksson KEL (1990) Biotechnology in the pulp and paper industry. Wood Sci Technol 24:79–101. doi:10.1007/BF00225309
Gerstein M, Lesk AM, Chothia C (1994) Structural mechanisms for domain movements in proteins. Biochemistry 33:6739–6749. doi:10.1021/bi00188a001
Gonzalez-Blasco G, Sanz-Aparicio J, Gonzalez B, Hermoso JA, Polaina J (2000) Directed evolution of beta-glucosidase A from Paenibacillus polymyxa to thermal resistance. J Biol Chem 275:13708–13712. doi:10.1074/jbc.275.18.13708
Gruber K, Klintschar G, Hayn M, Schlacher A, Steiner W, Kratky C (1998) Thermophilic xylanase from Thermomyces lanuginosus: high-resolution X-ray structure and modeling studies. Biochemistry 37:13475–13485. doi:10.1021/bi980864l
Hakulinen N, Turunen O, Janis J, Leisola M, Rouvinen J (2003) Three-dimensional structures of thermophilic beta-1, 4-xylanases from Chaetomium thermophilum and Nonomuraea flexuosa. Comparison of twelve xylanases in relation to their thermal stability. Eur J Biochem 270:1399–1412. doi:10.1046/j.1432-1033.2003.03496.x
Havukainen R, Torronen A, Laitinen T, Rouvinen J (1996) Covalent binding of three epoxyalkyl xylosides to the active site of endo-1, 4-xylanase II from Trichoderma reesei. Biochemistry 35:9617–9624. doi:10.1021/bi953052n
Henrissat B, Bairoch A (1993) New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J 293:781–788
Hori H, Elbein AD (1985) The biosynthesis of plant cell wall polysaccharides. In: Higuchi T (ed) Biosynthesis and biodegradation of wood components. Academic Press, Inc, Orlando, pp 109–135
Iffland A, Gendreizig S, Tafelmeyer P, Johnsson K (2001) Changing the substrate specificity of cytochrome c peroxidase using directed evolution. Biochem Biophys Res Commun 286:126–132. doi:10.1006/bbrc.2001.5366
Kaneko S, Kuno A, Muramatsu M, Iwamatsu S, Kusakabe I, Hayashi K (2000) Purification and characterization of a family G/11 beta-xylanase from Streptomyces olivaceoviridis E-86. Biosci Biotechnol Biochem 64:447–451. doi:10.1271/bbb.64.447
Kluepfel D, Vats-Mehta S, Aumont F, Shareck F, Morosoli R (1990) Purification and characterization of a new xylanase (xylanase B) produced by Streptomyces lividans 66. Biochem J 267:45–50
Lawson SL, Wakarchuk WW, Withers SG (1996) Effects of both shortening and lengthening the active site nucleophile of Bacillus circulans xylanase on catalytic activity. Biochemistry 35:10110–10118. doi:10.1021/bi960586v
Leskinen S, Mantyla A, Fagerstrom R, Vehmaanpera J, Lantto R, Paloheimo M et al (2005) Thermostable xylanases, Xyn10A and Xyn11A, from the actinomycete Nonomuraea flexuosa: isolation of the genes and characterization of recombinant Xyn11A polypeptides produced in Trichoderma reesei. Appl Microbiol Biotechnol 67:495–505. doi:10.1007/s00253-004-1797-x
Lever M (1972) A new reaction for colorimetric determination of carbohydrates. Anal Biochem 47:273–279. doi:10.1016/0003-2697(72)90301-6
Meyer A, Schmid A, Held M, Westphal AH, Rothlisberger M, Kohler HP et al (2002) Changing the substrate reactivity of 2-hydroxybiphenyl 3-monooxygenase from Pseudomonas azelaica HBP1 by directed evolution. J Biol Chem 277:5575–5582. doi:10.1074/jbc.M110018200
Moore JC, Arnold FH (1996) Directed evolution of a p-nitrobenzyl esterase for aqueous-organic solvents. Nat Biotechnol 14:458–467. doi:10.1038/nbt0496-458
Muilu J, Torronen A, Perakyla M, Rouvinen J (1998) Functional conformational changes of endo-1, 4-xylanase II from Trichoderma reesei: a molecular dynamics study. Proteins 31:434–444. doi:10.1002/(SICI)1097-0134(19980601)31:4<434::AID-PROT9>3.0.CO;2-H
Schimmel PR, Flory PJ (1968) Conformational energies and configurational statistics of copolypeptides containing l-proline. J Mol Biol 34:105–120. doi:10.1016/0022-2836(68)90237-4
Shareck F, Roy C, Yaguchi M, Morosoli R, Kluepfel D (1991) Sequences of three genes specifying xylanases in Streptomyces lividans. Gene 107:75–82. doi:10.1016/0378-1119(91)90299-Q
Song JK, Rhee JS (2000) Simultaneous enhancement of thermostability and catalytic activity of phospholipase A(1) by evolutionary molecular engineering. Appl Environ Microbiol 66:890–894. doi:10.1128/AEM.66.3.890-894.2000
Stemmer WP (1994) Rapid evolution of a protein in vitro by DNA shuffling. Nature 370:389–391. doi:10.1038/370389a0
Suenaga H, Mitsuoka M, Ura Y, Watanabe T, Furukawa K (2001) Directed evolution of biphenyl dioxygenase: emergence of enhanced degradation capacity for benzene, toluene, and alkylbenzenes. J Bacteriol 183:5441–5444. doi:10.1128/JB.183.18.5441-5444.2001
Torronen A, Rouvinen J (1997) Structural and functional properties of low molecular weight endo-1, 4-beta-xylanases. J Biotechnol 57:137–149. doi:10.1016/S0168-1656(97)00095-3
Wakarchuk WW, Sung WL, Campbell RL, Cunningham A, Watson DC, Yaguchi M (1994) Thermostabilization of the Bacillus circulans xylanase by the introduction of disulfide bonds. Protein Eng 7:1379–1386. doi:10.1093/protein/7.11.1379
You L, Arnold FH (1996) Directed evolution of subtilisin E in Bacillus subtilis to enhance total activity in aqueous dimethylformamide. Protein Eng 9:77–83. doi:10.1093/protein/9.1.77
Acknowledgements
This work was sponsored by National Natural Science Foundation of China (30770837) to Qin Wang. We thank Ms. Ying Li for the assistance with the DNA sequence analysis and mutant library construction.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xia, T., Wang, Q. Directed evolution of Streptomyces lividans xylanase B toward enhanced thermal and alkaline pH stability. World J Microbiol Biotechnol 25, 93–100 (2009). https://doi.org/10.1007/s11274-008-9867-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-008-9867-3